The Conserved GTPase LepA May Contribute to the Final Proper Stabilization of the 3′ Domain of the 30S Subunit During Ribosome Assembly
Abstract
1. Introduction
2. Results
2.1. Characterization of the lepA Null Strain of E. coli
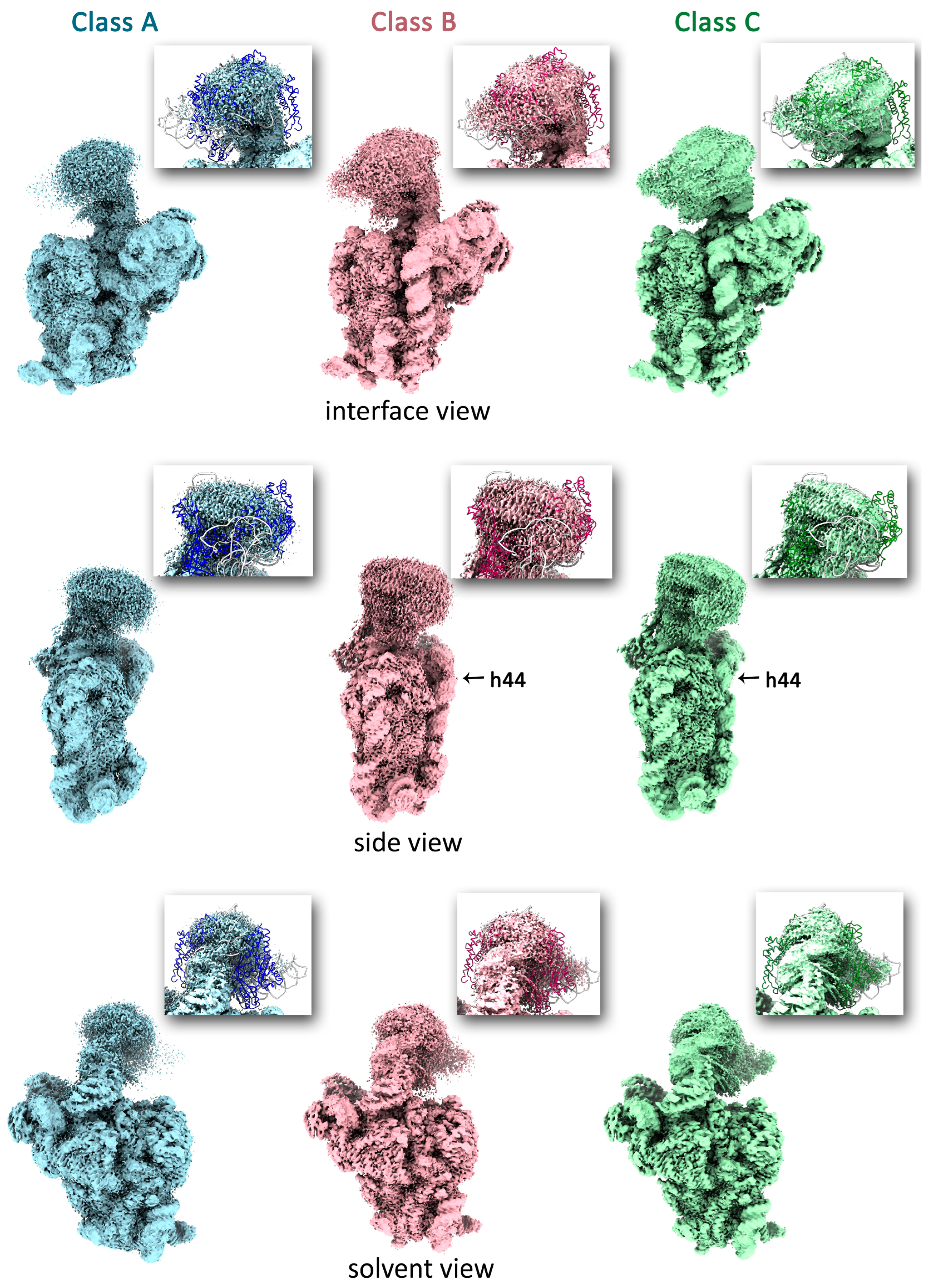
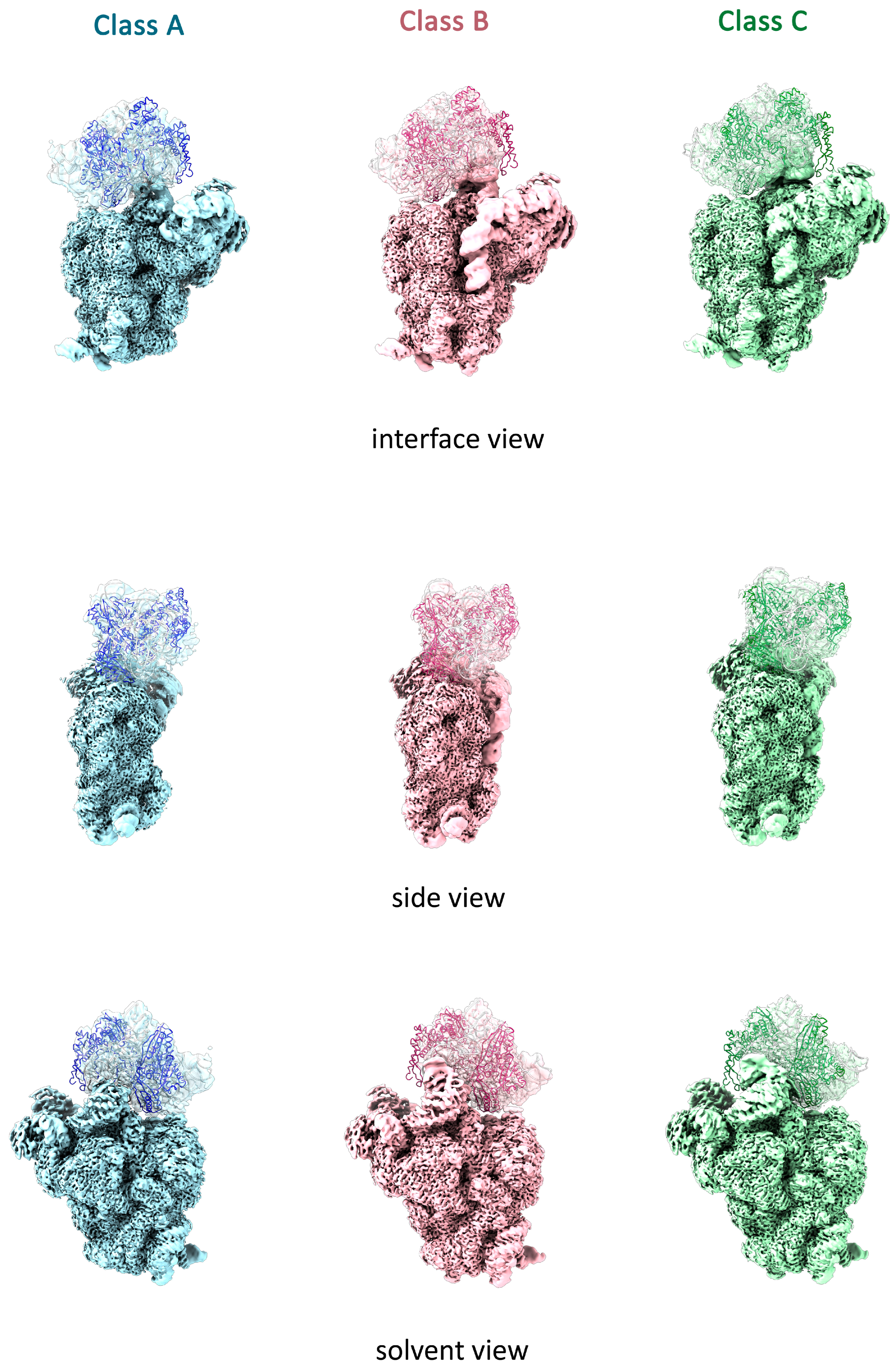
2.2. LepA May Contribute to the Proper Stabilization of the 3′ Domain
3. Discussion
4. Materials and Methods
4.1. E. coli Strains
4.2. Spot Assay
4.3. Ribosome Profile Analysis
4.4. Gel Electrophoresis Assay
4.5. 17S rRNA Measurement with Quantitative PCR
4.6. Isolation of Immature ΔlepA 30S Particles
4.7. Cryo-Sample Preparation
4.8. Cryo-EM Data Collection
4.9. Cryo-EM Data Processing
4.10. Model Building
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pech, M.; Karim, Z.; Yamamoto, H.; Kitakawa, M.; Qin, Y.; Nierhaus, K.H. Elongation factor 4 (EF4/LepA) accelerates protein synthesis at increased Mg2+ concentrations. Proc. Natl. Acad. Sci. USA 2011, 108, 3199–3203. [Google Scholar] [CrossRef]
- Shoji, S.; Janssen, B.D.; Hayes, C.S.; Fredrick, K. Translation factor LepA contributes to tellurite resistance in Escherichia coli but plays no apparent role in the fidelity of protein synthesis. Biochimie 2010, 92, 157–163. [Google Scholar] [CrossRef]
- Bijlsma, J.J.; Lie-A-Ling, M.; Nootenboom, I.C.; Vandenbroucke-Grauls, C.M.; Kusters, J.G. Identification of loci essential for the growth of Helicobacter pylori under acidic conditions. J. Infect. Dis. 2000, 182, 1566–1569. [Google Scholar] [CrossRef]
- Badu-Nkansah, A.; Sello, J.K. Deletion of the elongation factor 4 gene (lepA) in Streptomyces coelicolor enhances the production of the calcium-dependent antibiotic. FEMS Microbiol. Lett. 2010, 311, 147–151. [Google Scholar] [CrossRef]
- Wang, B.W.; Jun-Hao Zhu, J.H.; Javid, B. Clinically relevant mutations in mycobacterial LepA cause rifampicin-specific phenotypic resistance. Sci. Rep. 2020, 10, 8402. [Google Scholar] [CrossRef]
- Fishbein, S.R.S.; Tomasi, F.G.; Wolf, I.D.; Dulberger, C.L.; Wang, A.; Keshishian, H.; Wallace, L.W.; Carr, S.A.; Ioerger, T.R.; Rego, E.H.; et al. The Conserved translation factor LepA is required for optimal synthesis of a porin family in Mycobacterium smegmatis. ASM J. 2021, 203, e00604-20. [Google Scholar] [CrossRef] [PubMed]
- Bauerschmitt, H.; Funes, S.; Herrmann, J.M. The membrane-bound GTPase Guf1 promotes mitochondrial protein synthesis under suboptimal conditions. J. Biol. Chem. 2008, 283, 17139–17146. [Google Scholar] [CrossRef]
- Gao, Y.; Bai, X.; Zhang, D.; Han, C.; Yuan, J.; Liu, W.; Cao, X.; Chen, Z.; Shangguan, F.; Zhu, Z.; et al. Mammalian elongation factor 4 regulates mitochondrial translation essential for spermatogenesis. Nat. Struct. Mol. Biol. 2016, 23, 441–449. [Google Scholar] [CrossRef] [PubMed]
- Ji, D.L.; Lin, H.; Chi, W.; Zhang, L.X. CpLEPA is critical for chloroplast protein synthesis under suboptimal conditions in Arabidopsis thaliana. PLoS ONE 2012, 7, e49746. [Google Scholar] [CrossRef]
- Nierhaus, K.H. Protein synthesis—An elongation factor turn-on. Nature 1996, 379, 491–492. [Google Scholar] [CrossRef]
- Evans, R.N.; Blaha, G.; Bailey, S.; Steitz, T.A. The structure of LepA, the ribosomal back translocase. Proc. Natl. Acad. Sci. USA 2008, 105, 4673–4678. [Google Scholar] [CrossRef]
- Qin, Y.; Polacek, N.; Vesper, O.; Staub, E.; Einfeldt, E.; Wilson, D.N.; Nierhaus, K.H. The highly conserved LepA is a ribosomal elongation factor that back-translocates the ribosome. Cell 2006, 127, 721–733. [Google Scholar] [CrossRef]
- Liu, H.; Pan, D.; Pech, M.; Cooperman, B.S. Interrupted catalysis: The EF4 (LepA) effect on back-translocation. J. Mol. Biol. 2010, 396, 1043–1052. [Google Scholar] [CrossRef]
- Balakrishnan, R.; Oman, K.; Shoji, S.; Bundschuh, R.; Fredrick, K. The conserved GTPase LepA contributes mainly to translation initiation in Escherichia coli. Nucleic Acids Res. 2014, 42, 13370–13383. [Google Scholar] [CrossRef]
- Liu, H.; Chen, C.; Zhang, H.; Kaur, J.; Goldman, Y.E.; Cooperman, B.S. The conserved protein EF4 (LepA) modulates the elongation cycle of protein synthesis. Proc. Natl. Acad. Sci. USA 2011, 108, 16223–16228. [Google Scholar] [CrossRef] [PubMed]
- Gagnon, M.G.; Lin, J.; Steitz, T.A. Elongation factor 4 remodels the A-site tRNA on the ribosome. Proc. Natl. Acad. Sci. USA 2016, 113, 4994–4999. [Google Scholar] [CrossRef]
- Gibbs, M.R.; Moon, K.M.; Chen, M.; Balakrishnan, R.; Foster, L.J.; Fredrick, K. Conserved GTPase LepA (Elongation Factor 4) functions in biogenesis of the 30S subunit of the 70S ribosome. Proc. Natl. Acad. Sci. USA 2017, 114, 980–985. [Google Scholar] [CrossRef]
- Campbell, T.L.; Brown, E.D. Genetic interaction screens with ordered overexpression deletion clone sets implicate the Escherichia coli GTPase YjeQin late ribosome biogenesis. J. Bacteriol. 2008, 190, 2537–2545. [Google Scholar] [CrossRef] [PubMed]
- Goto, S.; Kato, S.; Kimura, T.; Muto, A.; Himeno, H. RsgA releases RbfA from 30S ribosome during a late stage of ribosome biosynthesis. EMBO J. 2011, 30, 104–114. [Google Scholar] [CrossRef] [PubMed]
- Leong, V.; Kent, M.; Jomaa, A.; Ortega, J. Escherichia coli rimM and yjeQ null strains accumulate immature 30S subunits of similar structure and protein complement. RNA 2013, 19, 789–802. [Google Scholar] [CrossRef]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006, 2, 2006.0008. [Google Scholar] [CrossRef] [PubMed]
- Maksimova, E.M.; Korepanov, A.P.; Kravchenko, O.V.; Baymukhametov, T.N.; Myasnikov, A.G.; Vassilenko, K.S.; Afonina, Z.A.; Stolboushkina, E.A. RbfA Is Involved in Two Important Stages of 30S Subunit Assembly: Formation of the Central Pseudoknot and Docking of Helix 44 to the Decoding Center. Int. J. Mol. Sci. 2021, 22, 6140. [Google Scholar] [CrossRef]
- Jomaa, A.; Stewart, G.; Mears, J.A.; Kireeva, I.; Brown, E.D.; Ortega, J. Cryo-electron microscopy structure of the 30S subunit in complex with the YjeQ biogenesis factor. RNA 2011, 17, 2026–2038. [Google Scholar] [CrossRef]
- Guo, Q.; Goto, S.; Chen, Y.; Feng, B.; Xu, Y.; Muto, A.; Himeno, H.; Deng, H.; Lei, J.; Gao, N. Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process. Nucleic Acids Res. 2013, 41, 2609–2620. [Google Scholar] [CrossRef]
- Razi, A.; Davis, J.H.; Hao, Y.; Jahagirdar, D.; Thurlow, B.; Basu, K.; Jain, N.; Gomez-Blanco, J.; Britton, R.A.; Vargas, J.; et al. Role of Era in assembly and homeostasis of the ribosomal small subunit. Nucleic Acids Res. 2019, 47, 8301–8317. [Google Scholar] [CrossRef]
- Yang, Z.; Guo, Q.; Goto, S.; Chen, Y.; Li, N.; Yan, K.; Zhang, Y.; Muto, A.; Deng, H.; Himeno, H.; et al. Structural insights into the assembly of the 30S ribosomal subunit in vivo: Functional role of S5 and location of the 17S rRNA precursor sequence. Protein Cell 2014, 5, 394–407. [Google Scholar] [CrossRef]
- Mulder, A.M.; Yoshioka, C.; Beck, A.H.; Bunner, A.E.; Milligan, R.A.; Potter, C.S.; Bridget Carragher, B.; Williamson, J.R. Visualizing ribosome biogenesis: Parallel assembly pathways for 30S subunit. Science 2010, 330, 673–677. [Google Scholar] [CrossRef]
- De Laurentiis, E.I.; Wieden, H.J. Identification of two structural elements important for ribosome-dependent GTPase activity of elongation factor 4 (EF4/LepA). Sci. Rep. 2015, 5, 8573. [Google Scholar] [CrossRef] [PubMed]
- Shetty, S.; Varshney, U. An evolutionarily conserved element in initiator tRNAs prompts ultimate steps in ribosome maturation. Proc. Natl. Acad. Sci. USA 2016, 113, E6126–E6134. [Google Scholar] [CrossRef] [PubMed]
- Strunk, B.S.; Novak, M.N.; Young, C.L.; Karbstein, K. Joining of 60S subunits and a translation-like cycle in 40S ribosome maturation. Cell 2012, 150, 111–121. [Google Scholar] [CrossRef]
- Blattner, F.R.; Plunkett, G., 3rd; Bloch, C.A.; Perna, N.T.; Burland, V.; Riley, M.; Collado-Vides, J.; Glasner, J.D.; Rode, C.K.; Mayhew, G.F.; et al. The complete genome sequence of Escherichia coli K-12. Science 1997, 277, 1453–1462. [Google Scholar] [CrossRef] [PubMed]
- Mastronarde, D.N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 2005, 152, 36–51. [Google Scholar] [CrossRef] [PubMed]
- Tegunov, D.; Cramer, P. Real-time cryo-electron microscopy data preprocessing with Warp. Nat. Methods 2019, 16, 1146–1152. [Google Scholar] [CrossRef] [PubMed]
- Punjani, A.; Rubinstein, J.L.; Fleet, D.J.; Brubaker, M.A. cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 2017, 14, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Zivanov, J.; Nakane, T.; Scheres, S.H.W. A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. UCrJ 2019, 6, 5–17. [Google Scholar] [CrossRef]
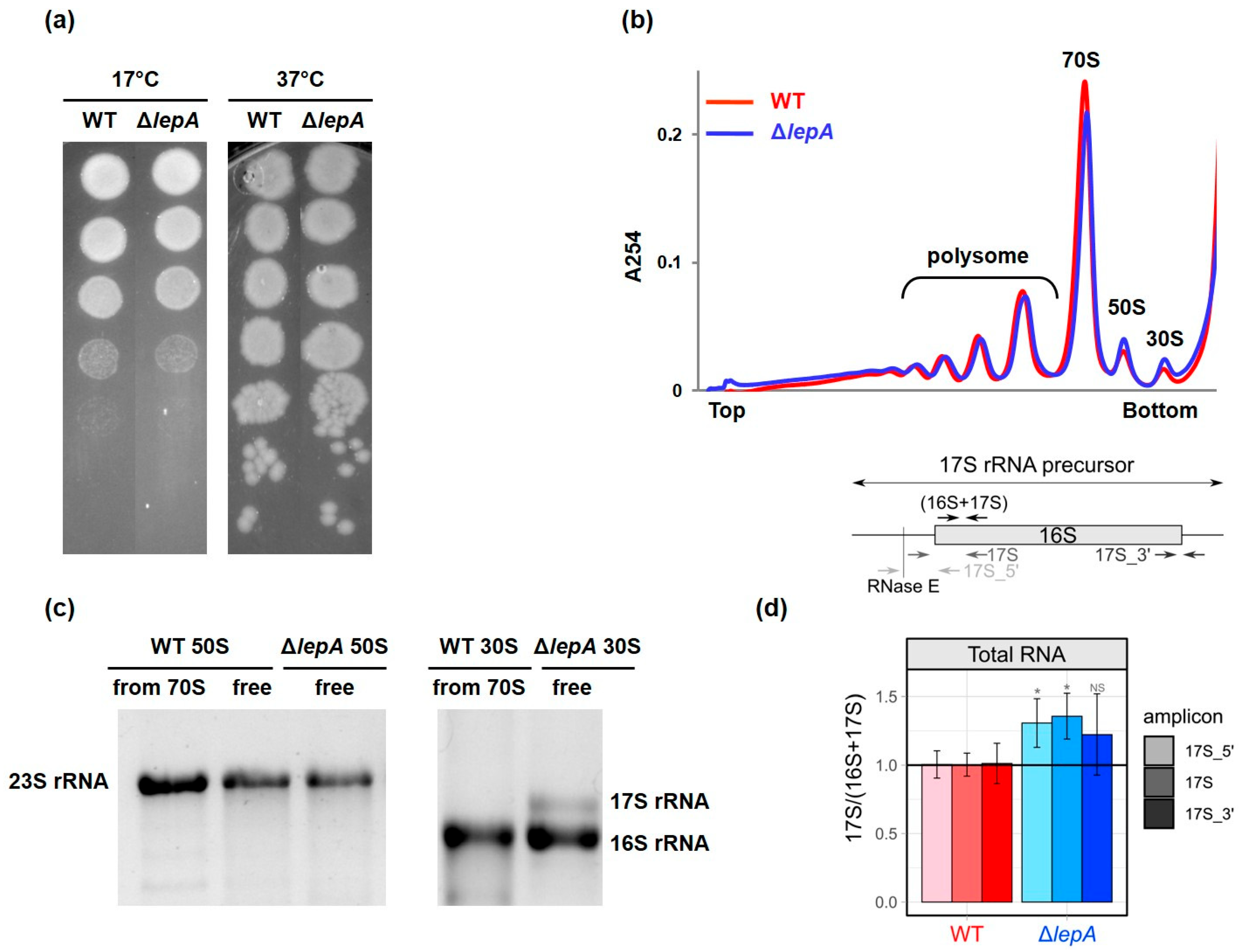
| Amplicon | Lenght, bp | Position (Relative to the 5′ End of Processed 16S rRNA) | Primer Name | Sequence |
|---|---|---|---|---|
| 16S + 17S | 100 | 6–27 | Fw(16S rRNA) | GAAGAGTTTGATCATGGCTCAG |
| 87–105 | Rw(16S rRNA) | CCACTCGTCAGCAAAGAAG | ||
| 17S_5′ | 125 | (−77)–(−98) | Fw(17S rRNA_5′) | ACGGATTCTTAACGTCGCAAG |
| 5–27 | Rw(17S rRNA_5′) | CTGAGCCATGATCAAACTCTTCA | ||
| 17S | 142 | (−11)–(−37) | Fw(17S rRNA) | TCATTACGAAGTTTAATTCTTTGAGCG |
| 87–105 | Rw(16S rRNA) | CCACTCGTCAGCAAAGAAG | ||
| 17S_3′ | 110 | 1456–1474 | Fw(17S rRNA_3′) | AGGGCGCTTACCACTTTGT |
| 1538–1562 | Rw(17S rRNA_3′) | CTGCAAAGTACGCTTCTTTAAGGTAAGG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Kravchenko, O.; Maksimova, E.; Baymukhametov, T.; Eliseeva, I.; Stolboushkina, E. The Conserved GTPase LepA May Contribute to the Final Proper Stabilization of the 3′ Domain of the 30S Subunit During Ribosome Assembly. Int. J. Mol. Sci. 2026, 27, 489. https://doi.org/10.3390/ijms27010489
Kravchenko O, Maksimova E, Baymukhametov T, Eliseeva I, Stolboushkina E. The Conserved GTPase LepA May Contribute to the Final Proper Stabilization of the 3′ Domain of the 30S Subunit During Ribosome Assembly. International Journal of Molecular Sciences. 2026; 27(1):489. https://doi.org/10.3390/ijms27010489
Chicago/Turabian StyleKravchenko, Olesya, Elena Maksimova, Timur Baymukhametov, Irina Eliseeva, and Elena Stolboushkina. 2026. "The Conserved GTPase LepA May Contribute to the Final Proper Stabilization of the 3′ Domain of the 30S Subunit During Ribosome Assembly" International Journal of Molecular Sciences 27, no. 1: 489. https://doi.org/10.3390/ijms27010489
APA StyleKravchenko, O., Maksimova, E., Baymukhametov, T., Eliseeva, I., & Stolboushkina, E. (2026). The Conserved GTPase LepA May Contribute to the Final Proper Stabilization of the 3′ Domain of the 30S Subunit During Ribosome Assembly. International Journal of Molecular Sciences, 27(1), 489. https://doi.org/10.3390/ijms27010489







