Correlation of Galectin Family Expression with Glioblastoma Progression and Survival
Abstract
1. Introduction
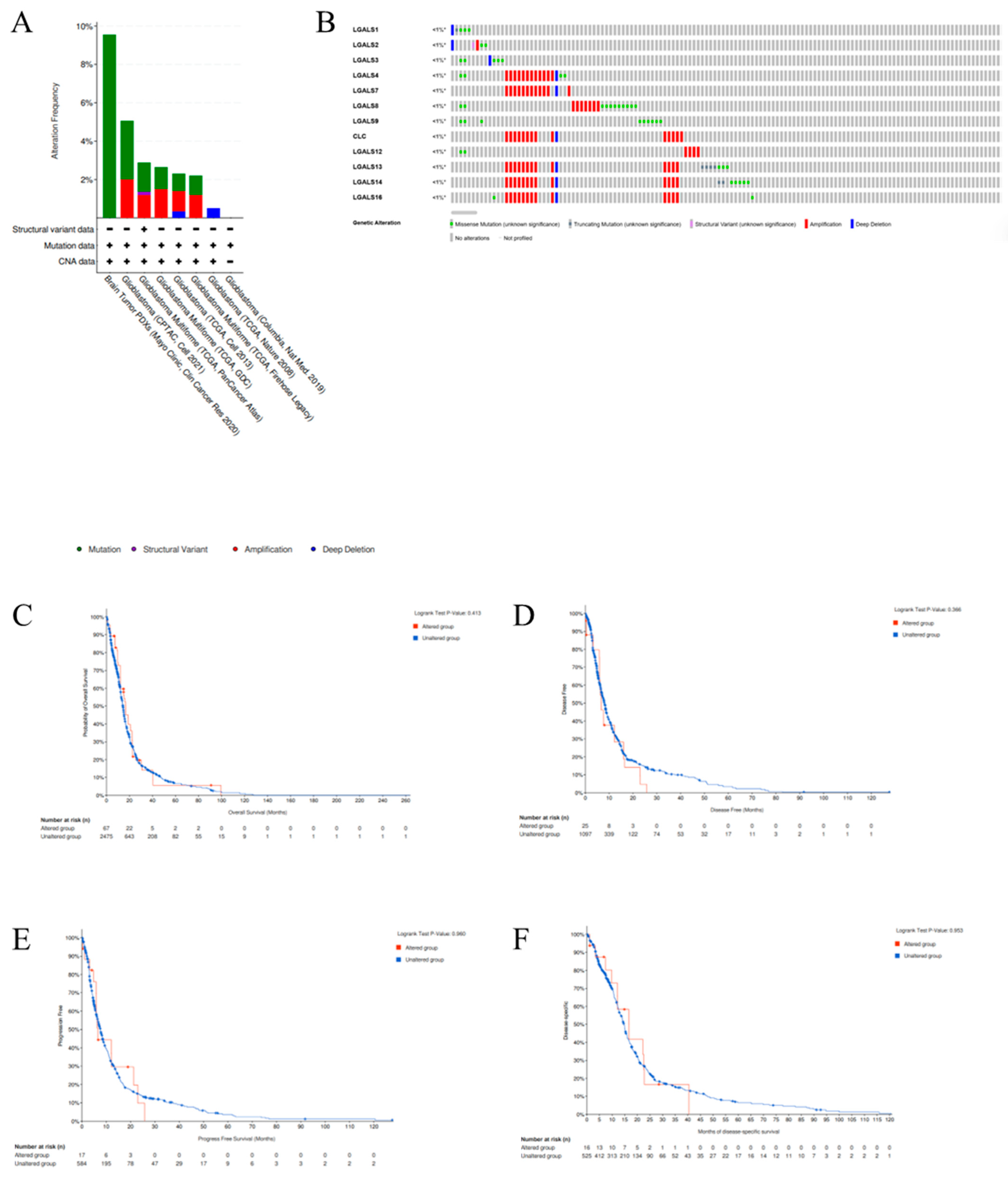
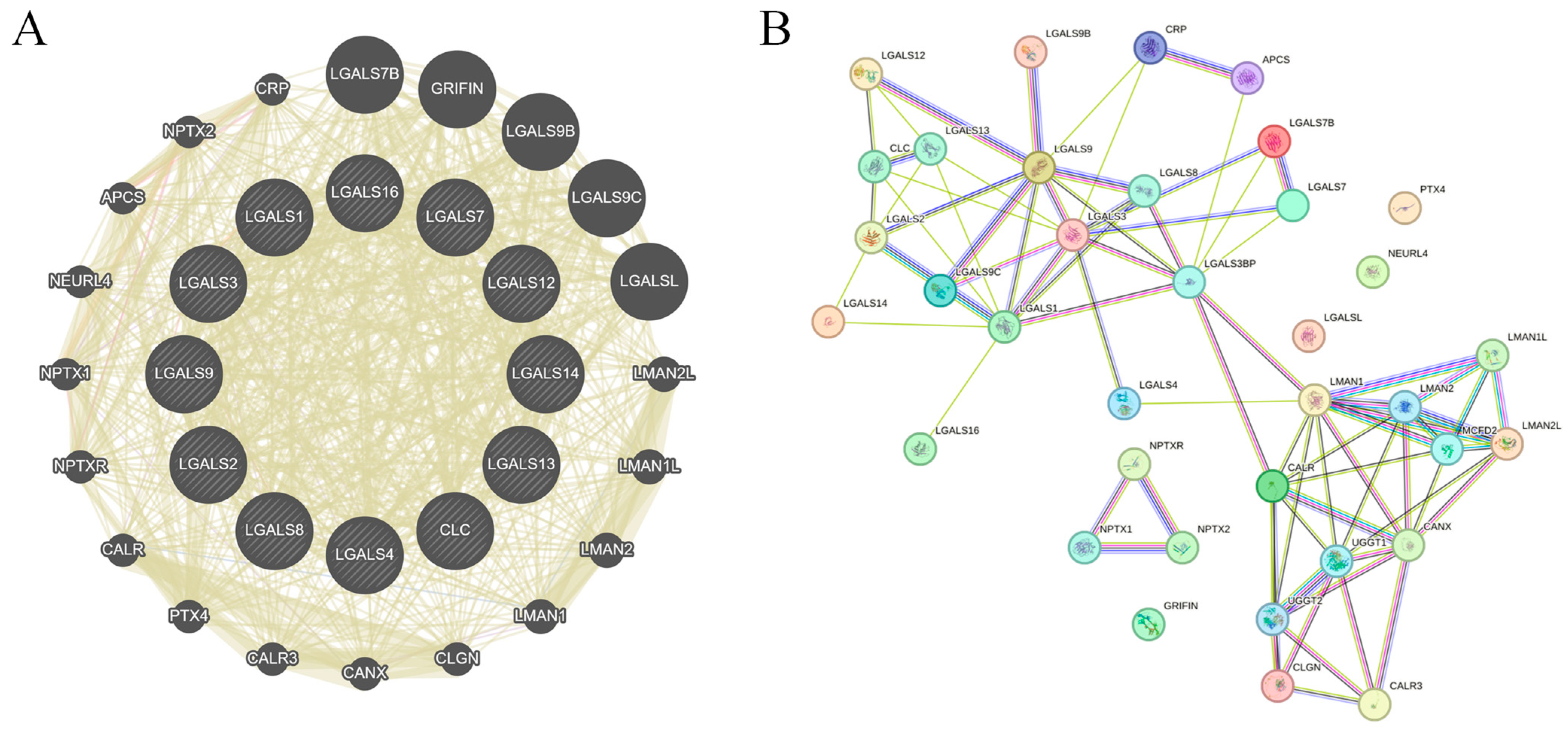
2. Results
3. Discussion
4. Materials and Methods
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ostrom, Q.T.; Bauchet, L.; Davis, F.G.; Deltour, I.; Fisher, J.L.; Langer, C.E.; Pekmezci, M.; Schwartzbaum, J.A.; Turner, M.C.; Walsh, K.M.; et al. The epidemiology of glioma in adults: A “state of the science” review. Neuro-Oncol. 2014, 16, 896–913. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Krex, D.; Klink, B.; Hartmann, C.; von Deimling, A.; Pietsch, T.; Simon, M.; Sabel, M.; Steinbach, J.P.; Heese, O.; Reifenberger, G.; et al. Long-term survival with glioblastoma multiforme. Brain 2007, 130, 2596–2606. [Google Scholar] [CrossRef] [PubMed]
- Dubrow, R.; Darefsky, A.S. Demographic variation in incidence of adult glioma by subtype, United States, 1992–2007. BMC Cancer 2011, 11, 325. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yalamarty, S.S.K.; Filipczak, N.; Li, X.; Subhan, M.A.; Parveen, F.; Ataide, J.A.; Rajmalani, B.A.; Torchilin, V.P. Mechanisms of Resistance and Current Treatment Options for Glioblastoma Multiforme (GBM). Cancers 2023, 15, 2116. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pouyan, A.; Ghorbanlo, M.; Eslami, M.; Jahanshahi, M.; Ziaei, E.; Salami, A.; Mokhtari, K.; Shahpasand, K.; Farahani, N.; Meybodi, T.E.; et al. Glioblastoma multiforme: Insights into pathogenesis, key signaling pathways, and therapeutic strategies. Mol. Cancer 2025, 24, 58. [Google Scholar] [CrossRef] [PubMed]
- Chou, F.C.; Chen, H.Y.; Kuo, C.C.; Sytwu, H.K. Role of Galectins in Tumors and in Clinical Immunotherapy. Int. J. Mol. Sci. 2018, 19, 430. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Thijssen, V.L.; Heusschen, R.; Caers, J.; Griffioen, A.W. Galectin expression in cancer diagnosis and prognosis: A systematic review. Biochim. Biophys. Acta 2015, 1855, 235–247. [Google Scholar] [CrossRef] [PubMed]
- Mariño, K.V.; Cagnoni, A.J.; Croci, D.O.; Rabinovich, G.A. Targeting galectin-driven regulatory circuits in cancer and fibrosis. Nat. Rev. Drug Discov. 2023, 22, 295–316. [Google Scholar] [CrossRef]
- Guda, M.R.; Tsung, A.J.; Asuthkar, S.; Velpula, K.K. Galectin-1 activates carbonic anhydrase IX and modulates glioma metabolism. Cell Death Dis. 2022, 13, 574. [Google Scholar] [CrossRef]
- Ikemori, R.Y.; Machado, C.M.; Furuzawa, K.M.; Nonogaki, S.; Osinaga, E.; Umezawa, K.; de Carvalho, M.A.; Verinaud, L.; Chammas, R. Galectin-3 up-regulation in hypoxic and nutrient deprived microenvironments promotes cell survival. PLoS ONE 2014, 9, e111592. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Seguin, L.; Odouard, S.; Corlazzoli, F.; Haddad, S.A.; Moindrot, L.; Calvo Tardón, M.; Yebra, M.; Koval, A.; Marinari, E.; Bes, V.; et al. Macropinocytosis requires Gal-3 in a subset of patient-derived glioblastoma stem cells. Commun. Biol. 2021, 4, 718. [Google Scholar] [CrossRef] [PubMed]
- Toussaint, L.G., 3rd; Nilson, A.E.; Goble, J.M.; Ballman, K.V.; James, C.D.; Lefranc, F.; Kiss, R.; Uhm, J.H. Galectin-1, a gene preferentially expressed at the tumor margin, promotes glioblastoma cell invasion. Mol. Cancer 2012, 11, 32. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yuan, F.; Ming, H.; Wang, Y.; Yang, Y.; Yi, L.; Li, T.; Ma, H.; Tong, L.; Zhang, L.; Liu, P.; et al. Molecular and clinical characterization of Galectin-9 in glioma through 1027 samples. J. Cell. Physiol. 2020, 235, 4326–4334. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Vasta, G.R. Galectins as pattern recognition receptors: Structure, function, and evolution. Adv. Exp. Med. Biol. 2012, 946, 21–36. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Shao, Q.; He, J.; Chen, Z.; Wu, C. Prognostic role of galectins expression in patients with hepatic cancer: A meta-analysis. Medicine 2020, 99, e19622. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mielczarek-Palacz, A.; Kondera-Anasz, Z.; Smycz-Kubańska, M.; Englisz, A.; Janusz, A.; Królewska-Daszczyńska, P.; Wendlocha, D. The role of galectins-1, 3, 7, 8 and 9 as potential diagnostic and therapeutic markers in ovarian cancer (Review). Mol. Med. Rep. 2022, 25, 166. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Martinez-Bosch, N.; Barranco, L.E.; Orozco, C.A.; Moreno, M.; Visa, L.; Iglesias, M.; Oldfield, L.; Neoptolemos, J.P.; Greenhalf, W.; Earl, J.; et al. Increased plasma levels of galectin-1 in pancreatic cancer: Potential use as biomarker. Oncotarget 2018, 9, 32984–32996. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Al-Maghrabi, J.A.; Khabaz, M.N. Clinical significance of galectin-3 expression in urinary bladder carcinoma. J. Int. Med. Res. 2023, 51, 3000605231153323. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Godefa, T.M.; Derks, S.; Thijssen, V.L.J.L. Galectins in Esophageal Cancer: Current Knowledge and Future Perspectives. Cancers 2022, 14, 5790. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Thijssen, V.L.; Barkan, B.; Shoji, H.; Aries, I.M.; Mathieu, V.; Deltour, L.; Hackeng, T.M.; Kiss, R.; Kloog, Y.; Poirier, F.; et al. Tumor cells secrete galectin-1 to enhance endothelial cell activity. Cancer Res. 2010, 70, 6216–6224, Erratum in Cancer Res. 2012, 72, 825. [Google Scholar] [CrossRef] [PubMed]
- Nakahara, S.; Oka, N.; Raz, A. On the role of galectin-3 in cancer apoptosis. Apoptosis 2005, 10, 267–275. [Google Scholar] [CrossRef] [PubMed]
- Nangia-Makker, P.; Nakahara, S.; Hogan, V.; Raz, A. Galectin-3 in apoptosis, a novel therapeutic target. J. Bioenerg. Biomembr. 2007, 39, 79–84. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Barrette, A.M.; Deshpande, K.; Cadiente, M.; Wang, X.; Sun, D. EXTH-73. A novel anti-Galectin-3 monoclonal antibody therapeutic diminishes migration and slows tumor growth to prolong survival in glioblastoma. Neuro-Oncol. 2022, 24, vii226. [Google Scholar] [CrossRef]
- Ni, X.; Wu, W.; Sun, X.; Ma, J.; Yu, Z.; He, X.; Cheng, J.; Xu, P.; Liu, H.; Shang, T.; et al. Interrogating glioma-M2 macrophage interactions identifies Gal-9/Tim-3 as a viable target against PTEN-null glioblastoma. Sci. Adv. 2022, 8, eabl5165. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Li, G.; Huang, R.; Fan, W.; Wang, D.; Wu, F.; Zeng, F.; Yu, M.; Zhai, Y.; Chang, Y.; Pan, C.; et al. Galectin-9/TIM-3 as a Key Regulator of Immune Response in Gliomas with Chromosome 1p/19q Codeletion. Front. Immunol. 2021, 12, 800928. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ahmady, F.; Curpen, P.; Perriman, L.; Teixeira, A.F.; Wu, S.; Zhu, H.-J.; Poddar, A.; Aparna Jayachandran Kannourakis, G.; Luwor, R.B. Reduced T and NK Cell Activity in Glioblastoma Patients Correlates with TIM-3 and BAT3 Dysregulation. Cells 2024, 13, 1777. [Google Scholar] [CrossRef]
- Lau, L.S.; Mohammed, N.B.B.; Dimitroff, C.J. Decoding Strategies to Evade Immunoregulators Galectin-1, -3, and -9 and Their Ligands as Novel Therapeutics in Cancer Immunotherapy. Int. J. Mol. Sci. 2022, 23, 15554. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Rabinovich, G.; Alonso, C.; Sotomayor, C.; Durand, S.; Bocco, J.; Riera, C. Molecular mechanisms implicated in galectin-1-induced apoptosis: Activation of the AP-1 transcription factor and downregulation of Bcl-2. Cell Death Differ. 2000, 7, 747–753. [Google Scholar] [CrossRef]
- Brandt, B.; Abou-Eladab, E.F.; Tiedge, M.; Walzel, H. Role of the JNK/c-Jun/AP-1 signaling pathway in galectin-1-induced T-cell death. Cell Death Dis. 2010, 1, e23. [Google Scholar] [CrossRef]
- Seyrek, K.; Richter, M.; Lavrik, I.N. Decoding the sweet regulation of apoptosis: The role of glycosylation and galectins in apoptotic signaling pathways. Cell Death Differ. 2019, 26, 981–993. [Google Scholar] [CrossRef]
- Fukumori, T.; Takenaka, Y.; Yoshii, T.; Kim, H.-R.C.; Hogan, V.; Inohara, H.; Kagawa, S.; Raz, A. CD29 and CD7 mediate galectin-3-induced type II T-cell apoptosis. Cancer Res. 2003, 63, 8302–8311. [Google Scholar]
- Dardalhon, V.; Anderson, A.C.; Karman, J.; Apetoh, L.; Chandwaskar, R.; Lee, D.H.; Cornejo, M.; Nishi, N.; Yamauchi, A.; Quintana, F.J. Tim-3/galectin-9 pathway: Regulation of Th1 immunity through promotion of CD11b+ Ly-6G+ myeloid cells. J. Immunol. 2010, 185, 1383–1392. [Google Scholar] [CrossRef]
- So, J.S.; Kim, H.; Han, K.S. Mechanisms of Invasion in Glioblastoma: Extracellular Matrix, Ca2+ Signaling, and Glutamate. Front. Cell. Neurosci. 2021, 15, 663092. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mohiuddin, E.; Wakimoto, H. Extracellular matrix in glioblastoma: Opportunities for emerging therapeutic approaches. Am. J. Cancer Res. 2021, 11, 3742–3754. [Google Scholar] [PubMed] [PubMed Central]
- Paz, A.; Haklai, R.; Elad-Sfadia, G.; Ballan, E.; Kloog, Y. Galectin-1 binds oncogenic H-Ras to mediate Ras membrane anchorage and cell transformation. Oncogene 2001, 20, 7486–7493. [Google Scholar] [CrossRef] [PubMed]
- Kopitz, J.; André, S.; Von Reitzenstein, C.; Versluis, K.; Kaltner, H.; Pieters, R.J.; Wasano, K.; Kuwabara, I.; Liu, F.-T.; Cantz, M.; et al. Homodimeric galectin-7 (p53-induced gene 1) is a negative growth regulator for human neuroblastoma cells. Oncogene 2003, 22, 6277–6288. [Google Scholar] [CrossRef]
- Rutkowski, M.R.; Stephen, T.L.; Svoronos, N.; Allegrezza, M.J.; Tesone, A.J.; Perales-Puchalt, A.; Brencicova, E.; Escovar-Fadul, X.; Nguyen, J.M.; Cadungog, M.G.; et al. Microbially driven TLR5-dependent signaling governs distal malignant progression through tumor-promoting inflammation. Cancer Cell 2015, 27, 27–40. [Google Scholar] [CrossRef]
- Croci, D.O.; Cerliani, J.P.; Dalotto-Moreno, T.; Méndez-Huergo, S.P.; Mascanfroni, I.D.; Dergan-Dylon, S.; Toscano, M.; Caramelo, J.J.; García-Vallejo, J.J.; Ouyang, J.; et al. Glycosylation-dependent lectin-receptor interactions preserve angiogenesis in anti-VEGF refractory tumors. Cell 2014, 156, 744–758. [Google Scholar] [CrossRef]
- Demotte, N.; Stroobant, V.; Courtoy, P.J.; Van Der Smissen, P.; Colau, D.; Luescher, I.F.; Hivroz, C.; Nicaise, J.; Squifflet, J.-L.; Mourad, M.; et al. Restoring the association of the T cell receptor with CD8 reverses anergy in human tumor-infiltrating lymphocytes. Immunity 2008, 28, 414–424. [Google Scholar] [CrossRef]
- Camby, I.; Belot, N.; Lefranc, F.; Sadeghi, N.; de Launoit, Y.; Kaltner, H.; Musette, S.; Darro, F.; Danguy, A.; Salmon, I.; et al. Galectin-1 modulates human glioblastoma cell migration into the brain through modifications to the actin cytoskeleton and the levels of expression of small GTPases. J. Neuropathol. Exp. Neurol. 2002, 61, 585–597. [Google Scholar] [CrossRef] [PubMed]
- Camby, I.; Decaestecker, C.; Lefranc, F.; Kaltner, H.; Gabius, H.J.; Kiss, R. Galectin-1 knocking down in human U87 glioblastoma cells alters their gene expression pattern. Biochem. Biophys. Res. Commun. 2005, 335, 27–35. [Google Scholar] [CrossRef]
- Jung, T.Y.; Jung, S.; Ryu, H.H.; Jeong, Y.I.; Jin, Y.H.; Jin, S.G.; Kim, I.Y.; Kang, S.S.; Kim, H.S. Role of galectine-1 in migration and invasion of human glioblastoma multiforme cell lines. J. Neurosurg. 2008, 109, 273–284. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.V.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404, Erratum in Cancer Discov. 2012, 2, 960. [Google Scholar] [CrossRef]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38, W214–W220. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Li, T.; Fan, J.; Wang, B.; Traugh, N.; Chen, Q.; Liu, J.S.; Li, B.; Liu, X.S. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017, 77, e108–e110. [Google Scholar] [CrossRef] [PubMed]
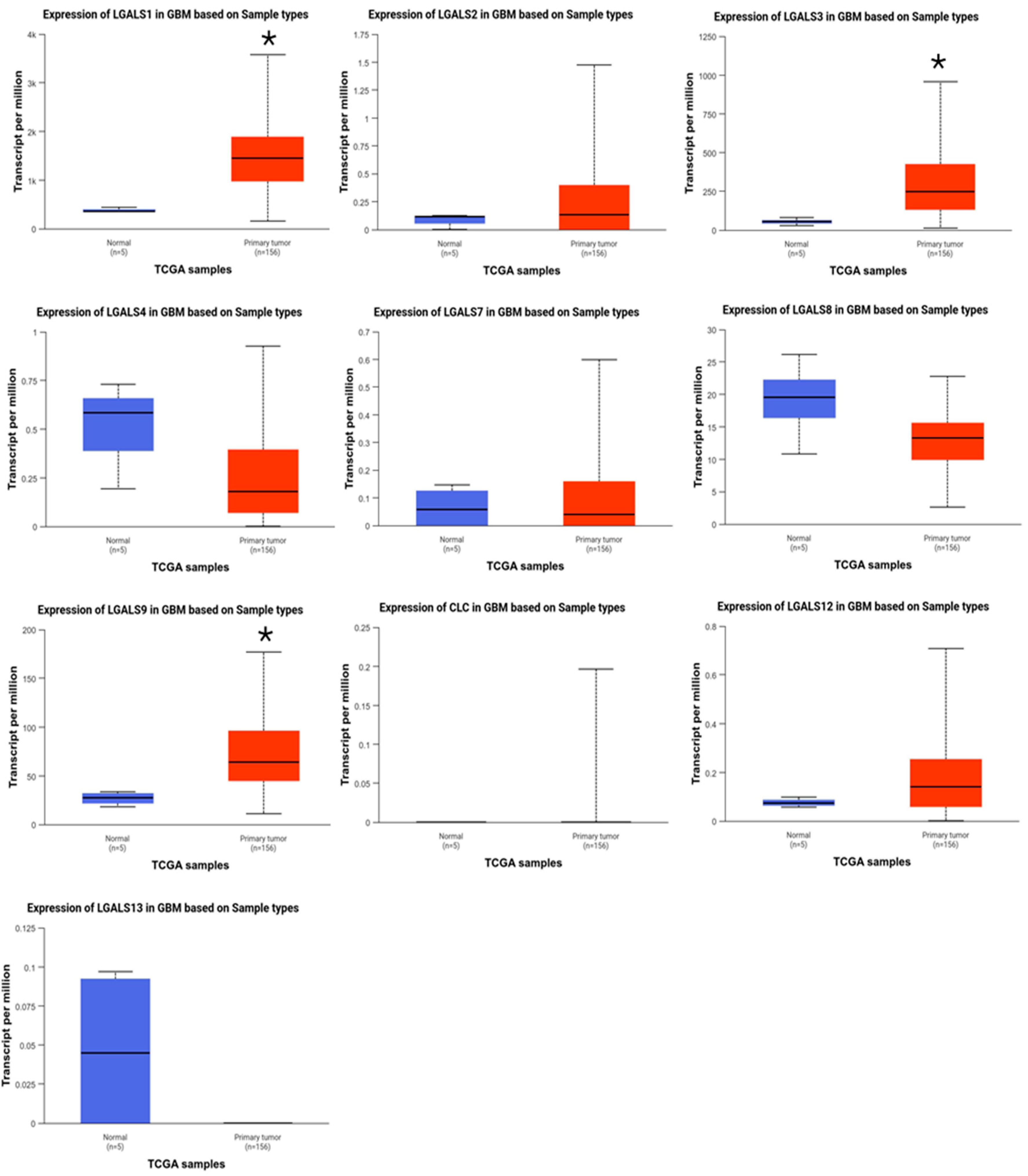
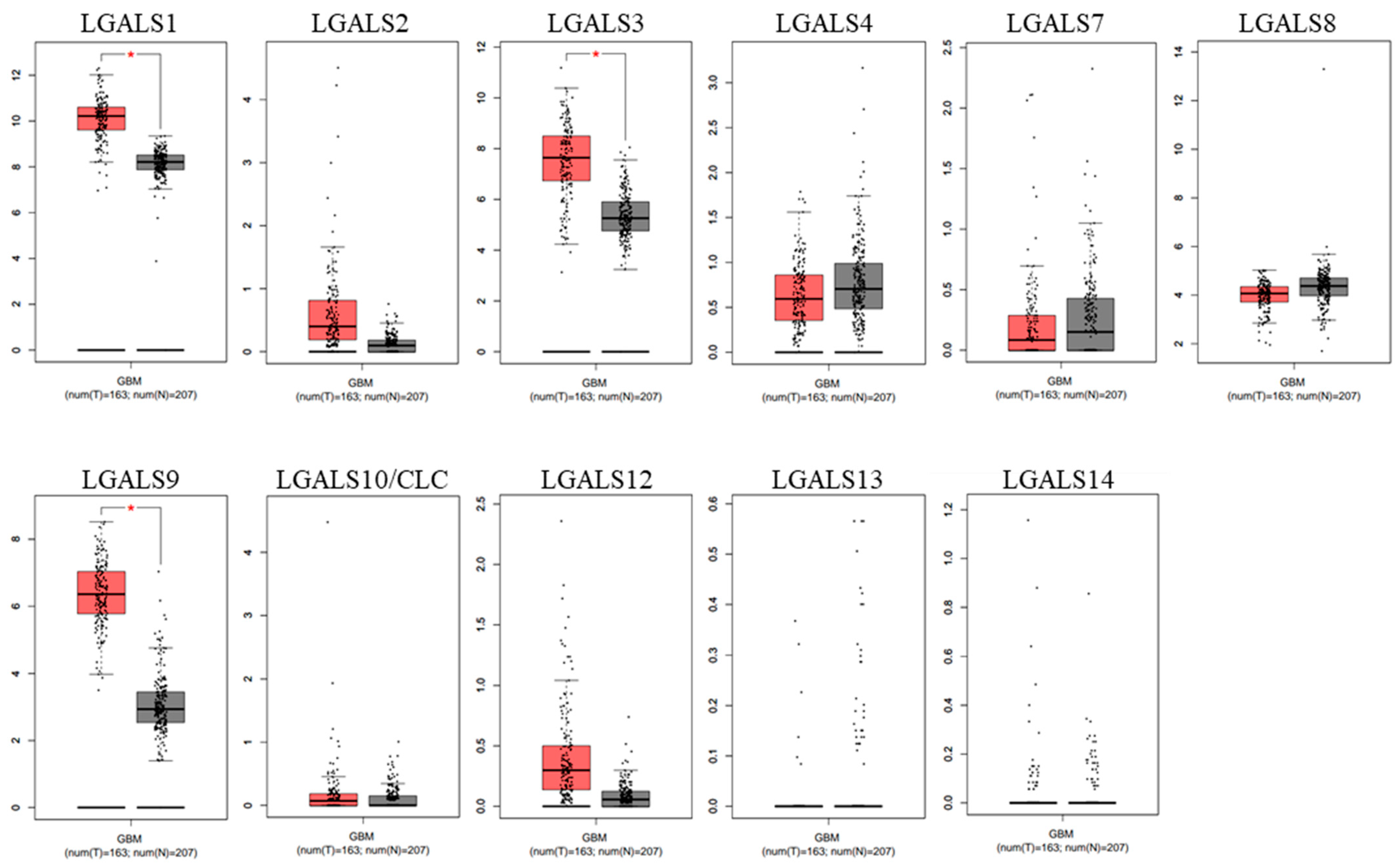
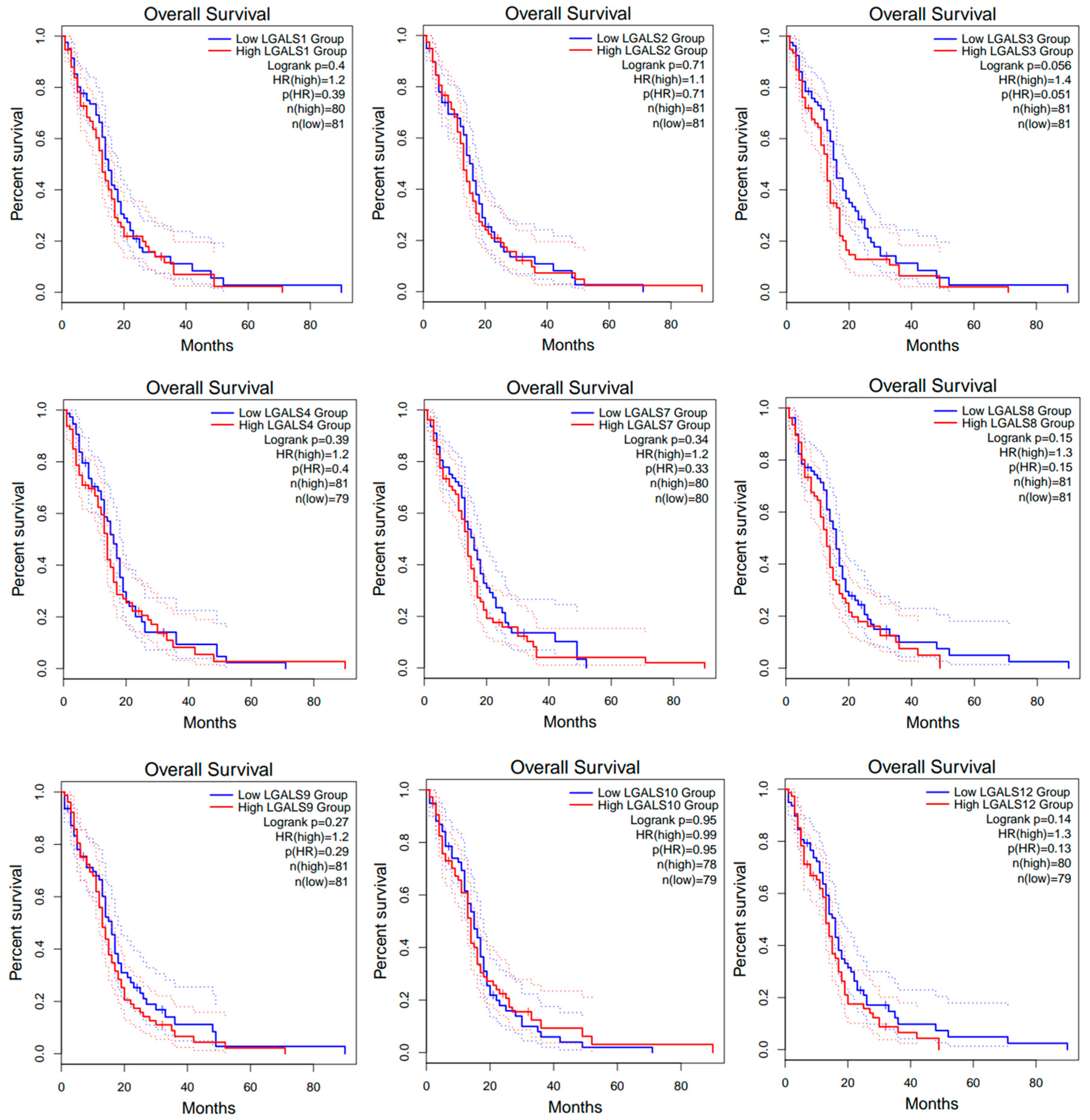
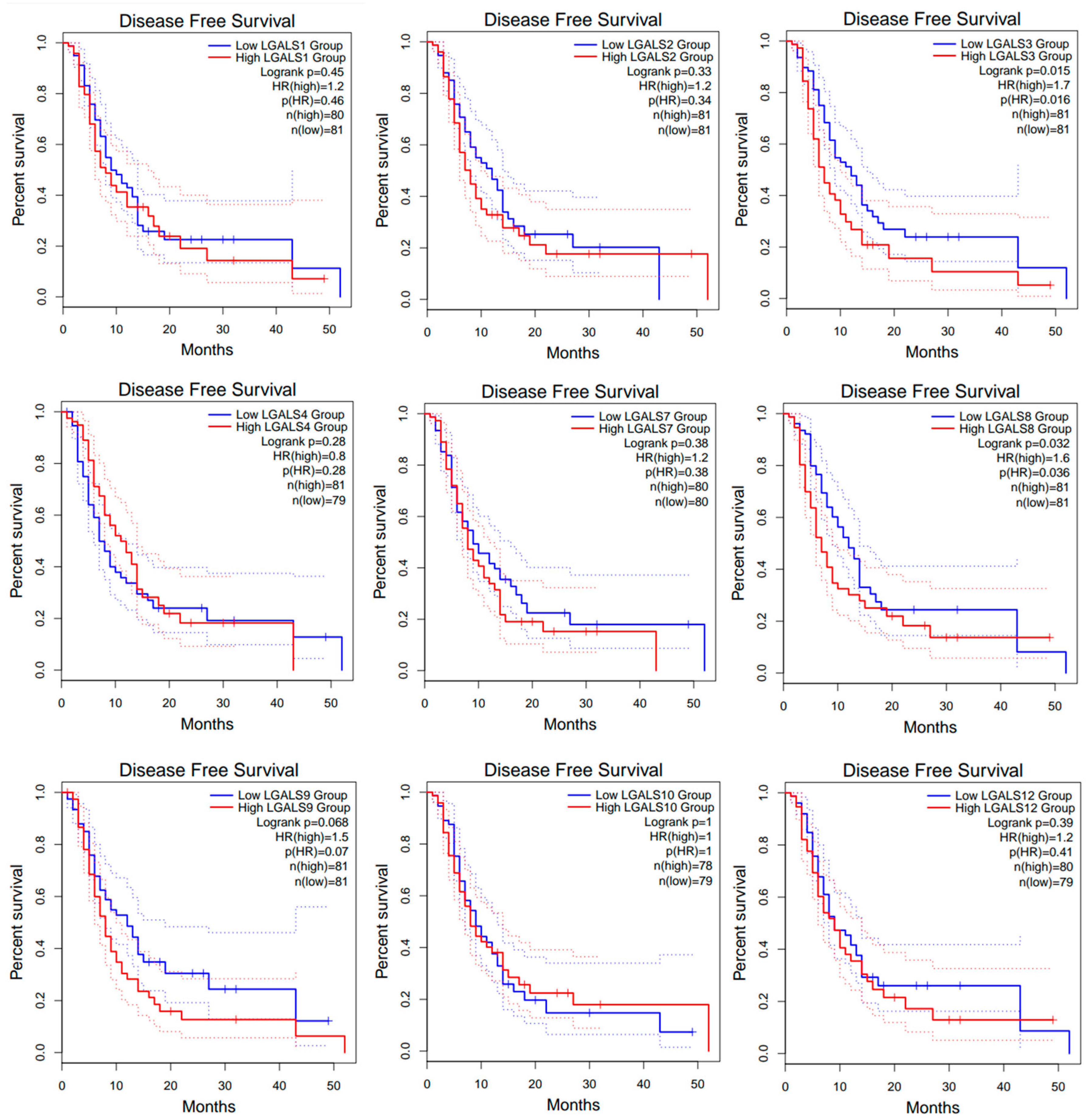
| Category | Term | Gene Count | % | p-Value |
|---|---|---|---|---|
| GOTERM_MF_DIRECT | Carbohydrate binding | 23 | 74.2 | 8.00 × 10−38 |
| GOTERM_MF_DIRECT | Galactoside binding | 6 | 19.4 | 3.70 × 10−13 |
| GOTERM_MF_DIRECT | Unfolded protein binding | 6 | 19.4 | 1.60 × 10−6 |
| GOTERM_MF_DIRECT | Lactose binding | 3 | 9.7 | 7.10 × 10−6 |
| GOTERM_MF_DIRECT | D-mannose binding | 4 | 12.9 | 8.70 × 10−6 |
| GOTERM_MF_DIRECT | Complement component C1q complex binding | 3 | 9.7 | 1.30 × 10−4 |
| GOTERM_BP_DIRECT | Protein folding | 7 | 22.6 | 9.10 × 10−8 |
| GOTERM_BP_DIRECT | Negative regulation of CD4-positive, alpha-beta T cell proliferation | 4 | 12.9 | 6.00 × 10−7 |
| GOTERM_BP_DIRECT | Negative regulation of type II interferon production | 4 | 12.9 | 3.10 × 10−5 |
| GOTERM_BP_DIRECT | ERAD pathway | 4 | 12.9 | 2.80 × 10−4 |
| GOTERM_BP_DIRECT | Positive regulation of gene expression | 6 | 19.4 | 4.90 × 10−4 |
| GOTERM_BP_DIRECT | Endoplasmic reticulum to Golgi vesicle-mediated transport | 4 | 12.9 | 1.80 × 10−2 |
| GOTERM_CC_DIRECT | Collagen-containing extracellular matrix | 9 | 29 | 3.40 × 10−8 |
| GOTERM_CC_DIRECT | COPII-coated ER to Golgi transport vesicle | 4 | 12.9 | 2.50 × 10−5 |
| GOTERM_CC_DIRECT | Extracellular space | 11 | 35.5 | 1.70 × 10−4 |
| GOTERM_CC_DIRECT | Endoplasmic reticulum–Golgi intermediate compartment | 4 | 12.9 | 1.70 × 10−4 |
| GOTERM_CC_DIRECT | Endoplasmic reticulum–Golgi intermediate compartment membrane | 4 | 12.9 | 1.20 × 10−4 |
| GOTERM_CC_DIRECT | Endoplasmic reticulum membrane | 8 | 25.8 | 7.60 × 10−3 |
| KEGG_PATHWAY | Protein processing in endoplasmic reticulum | 5 | 16.1 | 1.30 × 10−7 |
| KEGG_PATHWAY | Antigen processing and presentation | 2 | 6.5 | 3.60 × 10−2 |
| KEGG_PATHWAY | Phagosome | 2 | 6.5 | 7.00 × 10−2 |
| KEGG_PATHWAY | Human T cell leukemia virus 1 infection | 2 | 6.5 | 9.70 × 10−2 |
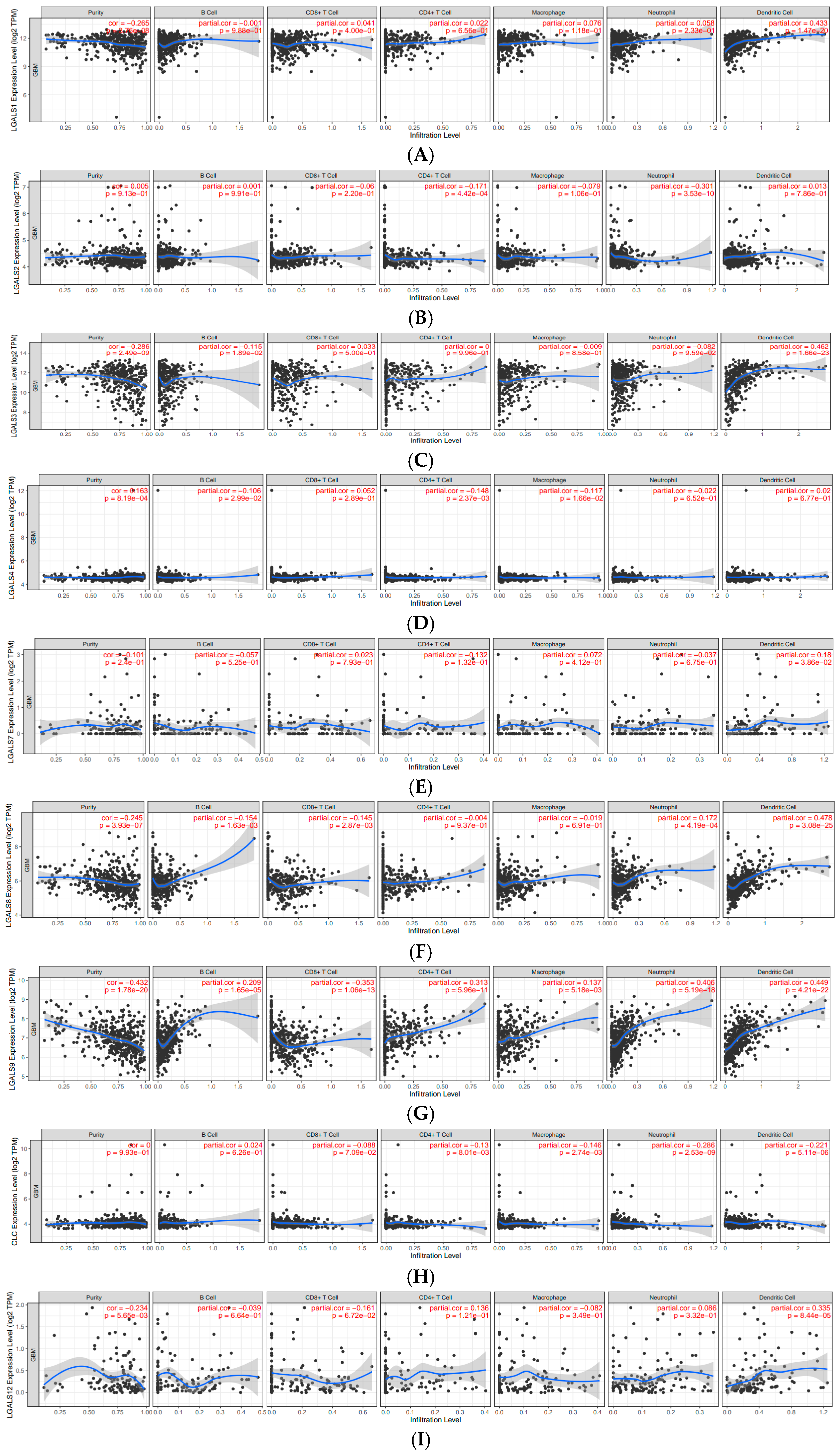
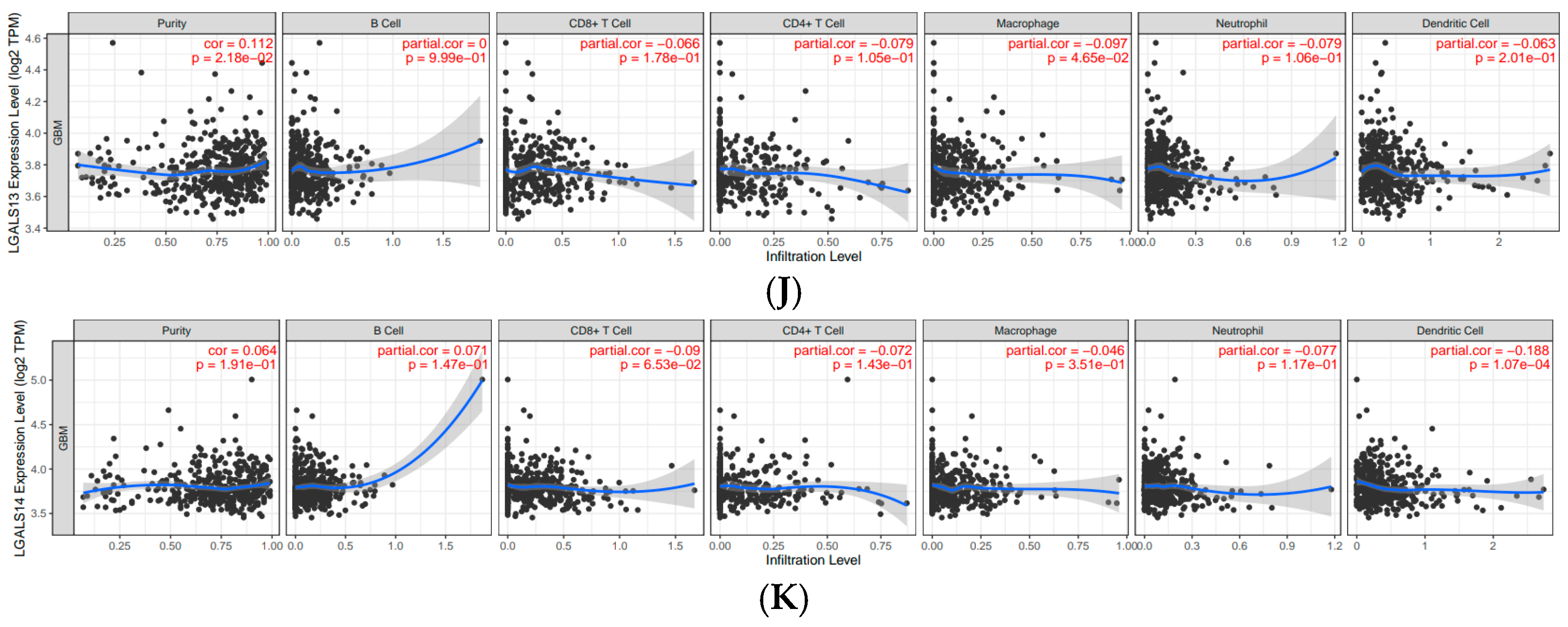
| Galectin Family | B Cell | CD8+ T Cell | CD4+ T Cell | Macrophage | Neutrophil | Dendritic Cell |
|---|---|---|---|---|---|---|
| LGALS1 | −0.001 | 0.041 | 0.022 | 0.076 | 0.058 | 0.433 |
| LGALS2 | 0.001 | −0.06 | −0.171 | −0.079 | −0.301 | 0.013 |
| LGALS3 | −0.115 | 0.033 | 0 | −0.009 | −0.082 | 0.462 |
| LGALS4 | −0.106 | 0.052 | −0.148 | −0.117 | −0.022 | 0.02 |
| LGALS7 | −0.057 | 0.023 | −0.132 | 0.072 | −0.037 | 0.18 |
| LGALS8 | −0.154 | −0.145 | −0.004 | −0.019 | 0.172 | 0.478 |
| LGALS9 | 0.209 | −0.353 | 0.313 | 0.137 | 0.406 | 0.449 |
| CLC | 0.024 | −0.088 | −0.13 | −0.146 | −0.286 | −0.221 |
| LGALS12 | −0.039 | −0.161 | 0.136 | −0.082 | 0.086 | 0.335 |
| LGALS13 | 0 | −0.066 | −0.079 | −0.097 | −0.079 | −0.063 |
| LGALS14 | 0.071 | −0.09 | −0.072 | −0.046 | −0.077 | −0.188 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Curpen, P.; Ahmady, F.; Carnie, B.M.H.; Anderson, G.E.C.; Kannourakis, G.; Sharma, A.; Achuthan, A.A.; Luwor, R.B. Correlation of Galectin Family Expression with Glioblastoma Progression and Survival. Int. J. Mol. Sci. 2026, 27, 417. https://doi.org/10.3390/ijms27010417
Curpen P, Ahmady F, Carnie BMH, Anderson GEC, Kannourakis G, Sharma A, Achuthan AA, Luwor RB. Correlation of Galectin Family Expression with Glioblastoma Progression and Survival. International Journal of Molecular Sciences. 2026; 27(1):417. https://doi.org/10.3390/ijms27010417
Chicago/Turabian StyleCurpen, Peter, Farah Ahmady, Blaine M. H. Carnie, Grace E. C. Anderson, George Kannourakis, Amit Sharma, Adrian A. Achuthan, and Rodney B. Luwor. 2026. "Correlation of Galectin Family Expression with Glioblastoma Progression and Survival" International Journal of Molecular Sciences 27, no. 1: 417. https://doi.org/10.3390/ijms27010417
APA StyleCurpen, P., Ahmady, F., Carnie, B. M. H., Anderson, G. E. C., Kannourakis, G., Sharma, A., Achuthan, A. A., & Luwor, R. B. (2026). Correlation of Galectin Family Expression with Glioblastoma Progression and Survival. International Journal of Molecular Sciences, 27(1), 417. https://doi.org/10.3390/ijms27010417









