Vimentin Dynamics in Viral Infection: Shield or Sabotage?
Abstract
1. Introduction
2. Structure and Function of Vimentin
2.1. Structural Assembly Pathway of Vimentin
2.2. Characteristics of the Vimentin Oligomers
2.3. PTMs Trigger Vimentin Remodeling
3. Cell Surface Vimentin as a Pro-Viral “Shield” Role Facilitating Viral Entry
3.1. Coronavirus
3.2. Flaviviruses
3.3. Hepaciviruses
3.4. Picornaviruses
3.5. Herpesviruses
4. Dual Roles of Vimentin Rearrangement in Viral Replication: Shield or Sabotage
4.1. Large Cytoplasmic DNA Viruses
4.2. Flaviviruses
4.3. Pestivirus
4.4. Picornaviruses
5. Targeting Vimentin for Anti-Viral Therapy
6. Concluding Remarks and Perspectives
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| A549 | Human lung epithelial line |
| ACE2 | Angiotensin-converting enzyme 2 |
| ACN | Antiparallel C–N |
| ASFV | African swine fever virus |
| BHK-21 | Baby hamster kidney cells |
| CaMKII | Calcium/calmodulin-dependent protein kinase II |
| CD81 | Cluster of differentiation 81 |
| Cdc42 | Cell division cycle 42 |
| CDK1 | Cyclin-dependent kinase 1 |
| CDK5 | Cyclin-dependent kinase 5 |
| Cryo-EM | Cryo-electron microscopy |
| CSFV | Classical wine fever virus |
| CSV | Cell surface vimentin |
| DENV | Dengue virus |
| DG-SIGN | Dendritic cell specific ICAM3 grabbing non-integrin receptor |
| DTMUV | Duck Tembusu virus |
| E. coli | Escherichia coli |
| E protein | Envelope protein |
| ER | Endoplasmic reticulum |
| EV71 | Human enterovirus 71 |
| eVIM | Extracellular vimentin |
| FMDV | Foot-and-mouth disease virus |
| HBMECs | Human brain microvascular endothelial cells |
| HCV | Hepatitis C virus |
| HEK-293 | Human embryonic kidney 293 cells |
| HS | Heparan sulfate |
| HTB-11 | Human neuroblastoma cell line |
| hzVSF-v13 | Humanized virus suppressing factor-variant 13 |
| IF | Intermediate filament |
| JEV | Japanese encephalitis virus |
| MCF-10A | Human mammary gland epithelial cells |
| MTOC | Microtubule organizing center |
| MLP | Molecular lipophilicity potential |
| N18 | Mouse neuroblastoma cells |
| PcAbs | Polyclonal antibodies |
| PCV2 | Porcine circovirus type 2 |
| PDB | Protein data bank |
| PK-15 | Porcine kidney epithelial cells |
| PKC | Protein kinase C |
| PLC | Phospholipase C |
| pLDDT | Predicted local distance difference test |
| PLK1 | Polo-like kinase 1 |
| PRV | Pseudorabies virus |
| PSGL-1 | P-selectin glycoprotein ligand-1 |
| PTMs | post-translational modifications |
| RhoA | Ras homolog gene family member A |
| RNAi | RNA interference |
| ROCK | Rho-associated kinase |
| RRBP1 | Ribosome-binding protein 1 |
| S protein | Spike protein |
| SCARB2 | Scavenger receptor B2 |
| TNTs | Tunneling nanotubes |
| ULFs | Unit-length filaments |
| VECs | Vascular endothelial cells |
| VRCs | Viral replication complexes |
| ZIKV | Zika virus |
References
- Herrmann, H.; Aebi, U. Intermediate Filaments: Molecular Structure, Assembly Mechanism, and Integration Into Functionally Distinct Intracellular Scaffolds. Annu. Rev. Biochem. 2004, 73, 749–789. [Google Scholar] [CrossRef]
- Herrmann, H.; Bär, H.; Kreplak, L.; Strelkov, S.V.; Aebi, U. Intermediate filaments: From cell architecture to nanomechanics. Nat. Rev. Mol. Cell Biol. 2007, 8, 562–573. [Google Scholar] [CrossRef]
- Herrmann, H.; Aebi, U. Intermediate Filaments: Structure and Assembly. Cold Spring Harb. Perspect. Biol. 2016, 8, a018242. [Google Scholar] [CrossRef]
- Alieva, I.B.; Shakhov, A.S.; Dayal, A.A.; Churkina, A.S.; Parfenteva, O.I.; Minin, A.A. Unique Role of Vimentin in the Intermediate Filament Proteins Family. Biochemistry 2024, 89, 726–736. [Google Scholar] [CrossRef]
- Herrmann, H.; Foisner, R. Intermediate filaments: Novel assembly models and exciting new functions for nuclear lamins. Cell. Mol. Life Sci. 2003, 60, 1607–1612. [Google Scholar] [CrossRef]
- Coelho-Rato, L.S.; Parvanian, S.; Modi, M.K.; Eriksson, J.E. Vimentin at the core of wound healing. Trends Cell Biol. 2024, 34, 239–254. [Google Scholar] [CrossRef] [PubMed]
- Suprewicz, Ł.; Zakrzewska, M.; Okła, S.; Głuszek, K.; Sadzyńska, A.; Deptuła, P.; Fiedoruk, K.; Bucki, R. Extracellular vimentin as a modulator of the immune response and an important player during infectious diseases. Immunol. Cell Biol. 2024, 102, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Pallari, H.-M.; Eriksson, J.E. Intermediate Filaments as Signaling Platforms. Sci. STKE 2006, 2006, pe53. [Google Scholar] [CrossRef] [PubMed]
- Ostrowska-Podhorodecka, Z.; Ding, I.; Norouzi, M.; McCulloch, C.A. Impact of Vimentin on Regulation of Cell Signaling and Matrix Remodeling. Front. Cell Dev. Biol. 2022, 10, 869069. [Google Scholar] [CrossRef]
- Parvanian, S.; Coelho-Rato, L.S.; Eriksson, J.E.; Patteson, A.E. The molecular biophysics of extracellular vimentin and its role in pathogen–host interactions. Curr. Opin. Cell Biol. 2023, 85, 102233. [Google Scholar] [CrossRef]
- Snider, N.T.; Omary, M.B. Post-translational modifications of intermediate filament proteins: Mechanisms and functions. Nat. Rev. Mol. Cell Biol. 2014, 15, 163–177. [Google Scholar] [CrossRef] [PubMed]
- Ivaska, J.; Vuoriluoto, K.; Huovinen, T.; Izawa, I.; Inagaki, M.; Parker, P.J. PKCɛ-mediated phosphorylation of vimentin controls integrin recycling and motility. EMBO J. 2005, 24, 3834–3845. [Google Scholar] [CrossRef]
- Yasui, Y.; Goto, H.; Matsui, S.; Manser, E.; Lim, L.; Nagata, K. Protein kinases required for segregation of vimentin filaments in mitotic process. Oncogene 2001, 20, 2868–2876. [Google Scholar] [CrossRef]
- Yuan, Z.; Janmey, P.A.; McCulloch, C.A. Structure and function of vimentin in the generation and secretion of extracellular vimentin in response to inflammation. Cell Commun. Signal. 2025, 23, 187. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Miao, C.; Gao, X.; Li, Z.; Eriksson, J.E.; Jiu, Y. Vimentin cage—A double-edged sword in host anti-infection defense. Curr. Opin. Cell Biol. 2024, 86, 102317. [Google Scholar] [CrossRef]
- Wileman, T. Aggresomes and Pericentriolar Sites of Virus Assembly: Cellular Defense or Viral Design? Annu. Rev. Microbiol. 2007, 61, 149–167. [Google Scholar] [CrossRef]
- Thalla, D.G.; Lautenschläger, F. Extracellular vimentin: Battle between the devil and the angel. Curr. Opin. Cell Biol. 2023, 85, 102265. [Google Scholar] [CrossRef]
- Zheng, J.; Li, X.; Zhang, G.; Ren, Y.; Ren, L. Research progress of vimentin in viral infections. Antivir. Res. 2025, 236, 106121. [Google Scholar] [CrossRef]
- Usman, S.; Aldehlawi, H.; Nguyen, T.K.N.; Teh, M.T.; Waseem, A. Impact of N-Terminal Tags on De Novo Vimentin Intermediate Filament Assembly. Int. J. Mol. Sci. 2022, 23, 6349. [Google Scholar] [CrossRef]
- Pogoda, K.; Byfield, F.; Deptuła, P.; Cieśluk, M.; Suprewicz, Ł.; Skłodowski, K.; Shivers, J.L.; van Oosten, A.; Cruz, K.; Tarasovetc, E.; et al. Unique Role of Vimentin Networks in Compression Stiffening of Cells and Protection of Nuclei from Compressive Stress. Nano Lett. 2022, 22, 4725–4732. [Google Scholar] [CrossRef] [PubMed]
- Viedma-Poyatos, Á.; Pajares, M.A.; Pérez-Sala, D. Type III intermediate filaments as targets and effectors of electrophiles and oxidants. Redox Biol. 2020, 36, 101582. [Google Scholar] [CrossRef]
- Eldirany, S.A.; Ho, M.; Hinbest, A.J.; Lomakin, I.B.; Bunick, C.G. Human keratin 1/10-1B tetramer structures reveal a knob-pocket mechanism in intermediate filament assembly. EMBO J. 2019, 38, e100741. [Google Scholar] [CrossRef]
- Eibauer, M.; Weber, M.S.; Kronenberg-Tenga, R.; Beales, C.T.; Boujemaa-Paterski, R.; Turgay, Y.; Sivagurunathan, S.; Kraxner, J.; Köster, S.; Goldman, R.D.; et al. Vimentin filaments integrate low-complexity domains in a complex helical structure. Nat. Struct. Mol. Biol. 2024, 31, 939–949. [Google Scholar] [CrossRef]
- Patteson, A.E.; Vahabikashi, A.; Goldman, R.D.; Janmey, P.A. Mechanical and Non-Mechanical Functions of Filamentous and Non-Filamentous Vimentin. BioEssays 2020, 42, 2000078. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Pang, A.H.; Obiero, J.M.; Kulczyk, A.W.; Sviripa, V.M.; Tsodikov, O.V. A crystal structure of coil 1B of vimentin in the filamentous form provides a model of a high-order assembly of a vimentin filament. FEBS J. 2018, 285, 2888–2899. [Google Scholar] [CrossRef]
- Goddard, T.D.; Huang, C.C.; Meng, E.C.; Pettersen, E.F.; Couch, G.S.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 2018, 27, 14–25. [Google Scholar] [CrossRef]
- Testa, B.; Carrupt, P.A.; Gaillard, P.; Billois, F.; Weber, P. Lipophilicity in molecular modeling. Pharm. Res. 1996, 13, 335–343. [Google Scholar] [CrossRef]
- Rosignoli, S.; Paiardini, A. Boosting the Full Potential of PyMOL with Structural Biology Plugins. Biomolecules 2022, 12, 1764. [Google Scholar] [CrossRef]
- Goto, H.; Kosako, H.; Tanabe, K.; Yanagida, M.; Sakurai, M.; Amano, M.; Kaibuchi, K.; Inagaki, M. Phosphorylation of Vimentin by Rho-associated Kinase at a Unique Amino-terminal Site That Is Specifically Phosphorylated during Cytokinesis. J. Biol. Chem. 1998, 273, 11728–11736. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Goto, H.; Yokoyama, T.; Silljé, H.; Hanisch, A.; Uldschmid, A.; Takai, Y.; Oguri, T.; Nigg, E.A.; Inagaki, M. Phosphorylation by Cdk1 induces Plk1-mediated vimentin phosphorylation during mitosis. J. Cell Biol. 2005, 171, 431–436. [Google Scholar] [CrossRef]
- Chan, W.; Kozma, R.; Yasui, Y.; Inagaki, M.; Leung, T.; Manser, E.; Lim, L. Vimentin intermediate filament reorganization by Cdc42: Involvement of PAK and p70 S6 kinase. Eur. J. Cell Biol. 2002, 81, 692–701. [Google Scholar] [CrossRef]
- Eriksson, J.E.; He, T.; Trejo-Skalli, A.V.; Härmälä-Braskén, A.-S.; Hellman, J.; Chou, Y.-H.; Goldman, R.D. Specific in vivo phosphorylation sites determine the assembly dynamics of vimentin intermediate filaments. J. Cell Sci. 2004, 117, 919–932. [Google Scholar] [CrossRef]
- Zhang, Y.; Wen, Z.; Shi, X.; Liu, Y.J.; Eriksson, J.E.; Jiu, Y. The diverse roles and dynamic rearrangement of vimentin during viral infection. J. Cell Sci. 2020, 134, jcs250597. [Google Scholar] [CrossRef]
- Byun, Y.; Chen, F.; Chang, R.; Trivedi, M.; Green, K.J.; Cryns, V.L. Caspase cleavage of vimentin disrupts intermediate filaments and promotes apoptosis. Cell Death Differ. 2001, 8, 443–450. [Google Scholar] [CrossRef]
- Shoeman, R.L.; Höner, B.; Stoller, T.J.; Mothes, E.; Kesselmeier, C.; Traub, P.; Graves, M.C. Cleavage of the intermediate filament subunit protein vimentin by HIV-1 protease: Utilization of a novel cleavage site and identification of higher order polymers of pepstatin A. Adv. Exp. Med. Biol. 1991, 306, 533–537. [Google Scholar] [CrossRef]
- Snášel, J.; Shoeman, R.; Hořejší, M.; Hrušková-Heidingsfeldová, O.; Sedláček, J.; Ruml, T.; Pichová, I. Cleavage of Vimentin by Different Retroviral Proteases. Arch. Biochem. Biophys. 2000, 377, 241–245. [Google Scholar] [CrossRef]
- Höner, B.; Shoeman, R.L.; Traub, P. Human immunodeficiency virus type 1 protease microinjected into cultured human skin fibroblasts cleaves vimentin and affects cytoskeletal and nuclear architecture. J. Cell Sci. 1991, 100 Pt 4, 799–807. [Google Scholar] [CrossRef]
- Kwak, H.I.; Kang, H.; Dave, J.M.; Mendoza, E.A.; Su, S.C.; Maxwell, S.A.; Bayless, K.J. Calpain-mediated vimentin cleavage occurs upstream of MT1-MMP membrane translocation to facilitate endothelial sprout initiation. Angiogenesis 2012, 15, 287–303. [Google Scholar] [CrossRef]
- Phillips, C.L.; Fu, D.; Herring, L.E.; Armao, D.; Snider, N.T. Calpain-mediated proteolysis of vimentin filaments is augmented in giant axonal neuropathy fibroblasts exposed to hypotonic stress. Front. Cell Dev. Biol. 2022, 10, 1008542. [Google Scholar] [CrossRef]
- Xiong, L.; Ye, X.; Chen, Z.; Fu, H.; Li, S.; Xu, P.; Yu, J.; Wen, L.; Gao, R.; Fu, Y.; et al. Advanced Maternal Age-associated SIRT1 Deficiency Compromises Trophoblast Epithelial-Mesenchymal Transition through an Increase in Vimentin Acetylation. Aging Cell 2021, 20, e13491. [Google Scholar] [CrossRef]
- Chang, W.-H.; Chen, Y.-J.; Hsiao, Y.-J.; Chiang, C.-C.; Wang, C.-Y.; Chang, Y.-L.; Hong, Q.-S.; Lin, C.-Y.; Lin, S.-U.; Chang, G.-C.; et al. Reduced symmetric dimethylation stabilizes vimentin and promotes metastasis in MTAP-deficient lung cancer. EMBO Rep. 2022, 23, e54265. [Google Scholar] [CrossRef]
- Mónico, A.; Guzmán-Caldentey, J.; Pajares, M.A.; Martín-Santamaría, S.; Pérez-Sala, D. Molecular Insight into the Regulation of Vimentin by Cysteine Modifications and Zinc Binding. Antioxidants 2021, 10, 1039. [Google Scholar] [CrossRef]
- Tarbet, H.J.; Dolat, L.; Smith, T.J.; Condon, B.M.; O’Brien, E.T., III; Valdivia, R.H.; Boyce, M. Site-specific glycosylation regulates the form and function of the intermediate filament cytoskeleton. eLife 2018, 7, e31807. [Google Scholar] [CrossRef]
- Snider, N.T.; Ku, N.-O.; Omary, M.B. The sweet side of vimentin. eLife 2018, 7, e35336. [Google Scholar] [CrossRef] [PubMed]
- Hwang, B.; Ise, H. Multimeric conformation of type III intermediate filaments but not the filamentous conformation exhibits high affinity to lipid bilayers. Genes Cells 2020, 25, 413–426. [Google Scholar] [CrossRef]
- Deptuła, P.; Fiedoruk, K.; Wasilewska, M.; Suprewicz, Ł.; Cieśluk, M.; Żeliszewska, P.; Oćwieja, M.; Adamczyk, Z.; Pogoda, K.; Bucki, R. Physicochemical Nature of SARS-CoV-2 Spike Protein Binding to Human Vimentin. ACS Appl. Mater. Interfaces 2023, 15, 34172–34180. [Google Scholar] [CrossRef]
- Lalioti, V.; González-Sanz, S.; Lois-Bermejo, I.; González-Jiménez, P.; Viedma-Poyatos, Á.; Merino, A.; Pajares, M.A.; Pérez-Sala, D. Cell surface detection of vimentin, ACE2 and SARS-CoV-2 Spike proteins reveals selective colocalization at primary cilia. Sci. Rep. 2022, 12, 7063. [Google Scholar] [CrossRef] [PubMed]
- Suprewicz, Ł.; Swoger, M.; Gupta, S.; Piktel, E.; Byfield, F.J.; Iwamoto, D.V.; Germann, D.; Reszeć, J.; Marcińczyk, N.; Carroll, R.J.; et al. Extracellular Vimentin as a Target Against SARS-CoV-2 Host Cell Invasion. Small 2022, 18, 2105640. [Google Scholar] [CrossRef] [PubMed]
- Lam, F.W.; Brown, C.A.; Ronca, S.E. Recombinant Rod Domain of Vimentin Reduces SARS-CoV-2 Viral Replication by Blocking Spike Protein-ACE2 Interactions. Int. J. Mol. Sci. 2024, 25, 2477. [Google Scholar] [CrossRef]
- Liang, J.-J.; Yu, C.-Y.; Liao, C.-L.; Lin, Y.-L. Vimentin binding is critical for infection by the virulent strain of Japanese encephalitis virus. Cell. Microbiol. 2011, 13, 1358–1370. [Google Scholar] [CrossRef]
- Arrindell, J.; Atmeh, P.A.; Jayet, L.; Sereme, Y.; Mege, J.-L.; Desnues, B. Vimentin is an important ACE2 co-receptor for SARS-CoV-2 in epithelial cells. iScience 2022, 25, 105463. [Google Scholar] [CrossRef]
- Li, Z.; Wu, J.; Zhou, J.; Yuan, B.; Chen, J.; Wu, W.; Mo, L.; Qu, Z.; Zhou, F.; Dong, Y.; et al. A Vimentin-Targeting Oral Compound with Host-Directed Antiviral and Anti-Inflammatory Actions Addresses Multiple Features of COVID-19 and Related Diseases. mBio 2021, 12, e0254221. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.; Wen, Z.; Mei, J.; Huang, X.; Zhao, S.; Zhong, J.; Jiu, Y. Cytoskeletal Vimentin Directs Cell-Cell Transmission of Hepatitis C Virus. Adv. Sci. 2025, 12, 2408917. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Cao, Y.; Su, W.; Huang, S.; Lu, W.; Zhou, Y.; Gao, J.; Zhao, W.; Zhang, B.; Wu, X. Enterovirus A71 VP1 Variation A289T Decreases the Central Nervous System Infectivity via Attenuation of Interactions between VP1 and Vimentin In Vitro and In Vivo. Viruses 2019, 11, 467. [Google Scholar] [CrossRef]
- Li, X.; Zheng, J.; Lv, X.; Han, Y.; Jiang, B.; Zhang, X.; Zhang, G.; Ren, L. Vimentin as a universal receptor for pseudorabies virus infection in pig and human cells. Int. J. Biol. Macromol. 2024, 283, 137638. [Google Scholar] [CrossRef]
- Yu, Y.T.-C.; Chien, S.-C.; Chen, I.-Y.; Lai, C.-T.; Tsay, Y.-G.; Chang, S.C.; Chang, M.-F. Surface vimentin is critical for the cell entry of SARS-CoV. J. Biomed. Sci. 2016, 23, 14. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Paulin, D.; Lacolley, P.; Coletti, D.; Agbulut, O. Vimentin as a target for the treatment of COVID-19. BMJ Open Respir. Res. 2020, 7, e000623. [Google Scholar] [CrossRef]
- Amraei, R.; Xia, C.; Olejnik, J.; White, M.R.; Napoleon, M.A.; Lotfollahzadeh, S.; Hauser, B.M.; Schmidt, A.G.; Chitalia, V.; Mühlberger, E.; et al. Extracellular vimentin is an attachment factor that facilitates SARS-CoV-2 entry into human endothelial cells. Proc. Natl. Acad. Sci. USA 2022, 119, e2113874119. [Google Scholar] [CrossRef]
- Kim, H.M.; Wang, M.; Zhai, C.; Kim, S.; Park, J.; Hong, S.T. Reduction of extracellular vimentin in blood provides protection against SARS-CoV-2 infection. Virulence 2025, 16, 2568052. [Google Scholar] [CrossRef]
- Das, S.; Ravi, V.; Desai, A. Japanese encephalitis virus interacts with vimentin to facilitate its entry into porcine kidney cell line. Virus Res. 2011, 160, 404–408. [Google Scholar] [CrossRef]
- Simanjuntak, Y.; Liang, J.-J.; Lee, Y.-L.; Lin, Y.-L. Japanese Encephalitis Virus Exploits Dopamine D2 Receptor-phospholipase C to Target Dopaminergic Human Neuronal Cells. Front. Microbiol. 2017, 8, 651. [Google Scholar] [CrossRef]
- Kumar, S.; Verma, A.; Yadav, P.; Dubey, S.K.; Azhar, E.I.; Maitra, S.S.; Dwivedi, V.D. Molecular pathogenesis of Japanese encephalitis and possible therapeutic strategies. Arch. Virol. 2022, 167, 1739–1762. [Google Scholar] [CrossRef]
- Yang, J.; Zou, L.; Yang, Y.; Yuan, J.; Hu, Z.; Liu, H.; Peng, H.; Shang, W.; Zhang, X.; Zhu, J.; et al. Superficial vimentin mediates DENV-2 infection of vascular endothelial cells. Sci. Rep. 2016, 6, 38372. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Wu, Y.; Fan, D.; Gao, N.; Ming, Y.; Wang, P.; An, J. Peptides P4 and P7 derived from E protein inhibit entry of dengue virus serotype 2 via interacting with β3 integrin. Antivir. Res. 2018, 155, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, R.; Gonzalez, A.M.; DeBiase, P.J.; Trejo, H.E.; Goldman, R.D.; Flitney, F.W.; Jones, J.C.R. Recruitment of vimentin to the cell surface by β3 integrin and plectin mediates adhesion strength. J. Cell Sci. 2009, 122, 1390–1400. [Google Scholar] [CrossRef]
- Fan, W.; Qian, P.; Wang, D.; Zhi, X.; Wei, Y.; Chen, H.; Li, X. Integrin αvβ3 promotes infection by Japanese encephalitis virus. Res. Vet. Sci. 2017, 111, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Chastney, M.R.; Kaivola, J.; Leppänen, V.-M.; Ivaska, J. The role and regulation of integrins in cell migration and invasion. Nat. Rev. Mol. Cell Biol. 2025, 26, 147–167. [Google Scholar] [CrossRef]
- Kim, J.; Yang, C.; Kim, E.J.; Jang, J.; Kim, S.J.; Kang, S.M.; Kim, M.G.; Jung, H.; Park, D.; Kim, C. Vimentin filaments regulate integrin–ligand interactions by binding to the cytoplasmic tail of integrin β3. J. Cell Sci. 2016, 129, 2030–2042. [Google Scholar] [CrossRef]
- Outla, Z.; Prechova, M.; Korelova, K.; Gemperle, J.; Gregor, M. Mechanics of cell sheets: Plectin as an integrator of cytoskeletal networks. Open Biol. 2025, 15, 240208. [Google Scholar] [CrossRef]
- Homan, S.M.; Martinez, R.; Benware, A.; LaFlamme, S.E. Regulation of the Association of α6β4 with Vimentin Intermediate Filaments in Endothelial Cells. Exp. Cell Res. 2002, 281, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Koudelka, K.J.; Destito, G.; Plummer, E.M.; Trauger, S.A.; Siuzdak, G.; Manchester, M. Endothelial targeting of cowpea mosaic virus (CPMV) via surface vimentin. PLoS Pathog. 2009, 5, e1000417. [Google Scholar] [CrossRef]
- Du, N.; Cong, H.; Tian, H.; Zhang, H.; Zhang, W.; Song, L.; Tien, P. Cell Surface Vimentin Is an Attachment Receptor for Enterovirus 71. J. Virol. 2014, 88, 5816–5833. [Google Scholar] [CrossRef]
- Kobayashi, K.; Koike, S. Cellular receptors for enterovirus A71. J. Biomed. Sci. 2020, 27, 23. [Google Scholar]
- Heath, C.M.; Windsor, M.; Wileman, T. Aggresomes Resemble Sites Specialized for Virus Assembly. J. Cell Biol. 2001, 153, 449–456. [Google Scholar] [CrossRef]
- Stefanovic, S.; Windsor, M.; Nagata, K.; Inagaki, M.; Wileman, T. Vimentin Rearrangement during African Swine Fever Virus Infection Involves Retrograde Transport along Microtubules and Phosphorylation of Vimentin by Calcium Calmodulin Kinase II. J. Virol. 2005, 79, 11766–11775. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.; Yang, X.; Yang, X.; Cao, Z.; Wei, N.; Lin, X.; Shi, M.; Cao, R. Japanese encephalitis virus NS1 and NS1’ proteins induce vimentin rearrangement via the CDK1-PLK1 axis to promote viral replication. J. Virol. 2024, 98, e0019524. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Liu, X.; Li, Q.; Wang, J.; Ruan, W. Proteomic analyses identify intracellular targets for Japanese encephalitis virus nonstructural protein 1 (NS1). Virus Res. 2021, 302, 198495. [Google Scholar] [CrossRef]
- Teo, C.S.H.; Chu, J.J.H. Cellular Vimentin Regulates Construction of Dengue Virus Replication Complexes through Interaction with NS4A Protein. J. Virol. 2014, 88, 1897–1913. [Google Scholar] [CrossRef]
- Lei, S.; Tian, Y.-P.; Xiao, W.-D.; Li, S.; Rao, X.-C.; Zhang, J.-L.; Yang, J.; Hu, X.-M.; Chen, W. ROCK is Involved in Vimentin Phosphorylation and Rearrangement Induced by Dengue Virus. Cell Biochem. Biophys. 2013, 67, 1333–1342. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, S.; Li, Y.; Feng, F.; Li, M.; Xue, Y.; Cui, J.; Xu, T.; Jin, X.; Jiu, Y. Host cytoskeletal vimentin serves as a structural organizer and an RNA-binding protein regulator to facilitate Zika viral replication. Proc. Natl. Acad. Sci. USA 2022, 119, e2113909119. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Lou, J.; Liu, Y.; Liu, C.; Chen, J.; Yang, M.; Ye, Y.; Go, Y.Y.; Zhou, B. Intracellular Vimentin Regulates the Formation of Classical Swine Fever Virus Replication Complex through Interaction with NS5A Protein. J. Virol. 2023, 97, e01770-22. [Google Scholar] [CrossRef]
- Haolong, C.; Du, N.; Hongchao, T.; Yang, Y.; Wei, Z.; Hua, Z.; Wenliang, Z.; Lei, S.; Po, T. Enterovirus 71 VP1 activates calmodulin-dependent protein kinase II and results in the rearrangement of vimentin in human astrocyte cells. PLoS ONE 2013, 8, e73900. [Google Scholar] [CrossRef]
- Bao, G.; Fan, S.; Hu, C.; Li, C.; Ma, F.; Wang, G.; Fan, H.; Wang, Q. CDK5-mediated rearrangement of vimentin during Duck Tembusu virus infection inhibits viral replication. Vet. Microbiol. 2024, 292, 110071. [Google Scholar] [CrossRef]
- Gladue, D.P.; O’Donnell, V.; Baker-Branstetter, R.; Holinka, L.G.; Pacheco, J.M.; Sainz, I.F.; Lu, Z.; Ambroggio, X.; Rodriguez, L.; Borca, M.V. Foot-and-Mouth Disease Virus Modulates Cellular Vimentin for Virus Survival. J. Virol. 2013, 87, 6794. [Google Scholar] [CrossRef]
- Ma, X.; Ling, Y.; Li, P.; Sun, P.; Cao, Y.; Bai, X.; Li, K.; Fu, Y.; Zhang, J.; Li, D.; et al. Cellular Vimentin Interacts with Foot-and-Mouth Disease Virus Nonstructural Protein 3A and Negatively Modulates Viral Replication. J. Virol. 2020, 94, 16. [Google Scholar] [CrossRef]
- Wang, Z.-J.; Xu, C.-M.; Song, Z.-B.; Wang, M.; Liu, Q.-Y.; Jiang, P.; Li, Y.-F.; Bai, J.; Wang, X.-W. Vimentin modulates infectious porcine circovirus type 2 in PK-15 cells. Virus Res. 2018, 243, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Porter, L.A.; Donoghue, D.J. Cyclin B1 and CDK1: Nuclear localization and upstream regulators. Prog. Cell Cycle Res. 2003, 5, 335–347. [Google Scholar] [PubMed]
- Sahlgren, C.M.; Pallari, H.-M.; He, T.; Chou, Y.-H.; Goldman, R.D.; Eriksson, J.E. A nestin scaffold links Cdk5/p35 signaling to oxidant-induced cell death. EMBO J. 2006, 25, 4808–4819. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Tang, L.; Dai, F.; Meng, G.; Yin, R.; Xu, X.; Yao, W. Raf-1/CK2 and RhoA/ROCK signaling promote TNF-α-mediated endothelial apoptosis via regulating vimentin cytoskeleton. Toxicology 2017, 389, 74–84. [Google Scholar] [CrossRef]
- Wang, J.; Wang, Y.; Liu, J.; Ding, L.; Zhang, Q.; Li, X.; Cao, H.; Tang, J.; Zheng, S.J. A critical role of N-myc and STAT interactor (Nmi) in foot-and-mouth disease virus (FMDV) 2C-induced apoptosis. Virus Res. 2012, 170, 59–65. [Google Scholar] [CrossRef]
- Kvansakul, M. Viral Infection and Apoptosis. Viruses 2017, 9, 356. [Google Scholar] [CrossRef]
- Korolowicz, K.E.; Suresh, M.; Li, B.; Huang, X.; Yon, C.; Kallakury, B.V.; Lee, K.; Park, S.; Kim, Y.-W.; Menne, S. Combination Treatment with the Vimentin-Targeting Antibody hzVSF and Tenofovir Suppresses Woodchuck Hepatitis Virus Infection in Woodchucks. Cells 2021, 10, 2321. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Chen, Z.; Xie, L.; Xu, R.; Chen, L.; Jia, T.; Shi, W.; Wang, Y.; Song, Y.; Han, Q.; et al. A fusion protein of vimentin with Fc fragment inhibits Japanese encephalitis virus replication. Front. Vet. Sci. 2024, 11, 1368725. [Google Scholar] [CrossRef] [PubMed]
- Sookhoo, J.R.V.; Schiffman, Z.; Ambagala, A.; Kobasa, D.; Pardee, K.; Babiuk, S. Protein Expression Platforms and the Challenges of Viral Antigen Production. Vaccines 2024, 12, 1344. [Google Scholar] [CrossRef] [PubMed]
- Arrindell, J.; Desnues, B. Vimentin: From a cytoskeletal protein to a critical modulator of immune response and a target for infection. Front. Immunol. 2023, 14, 1224352. [Google Scholar] [CrossRef]

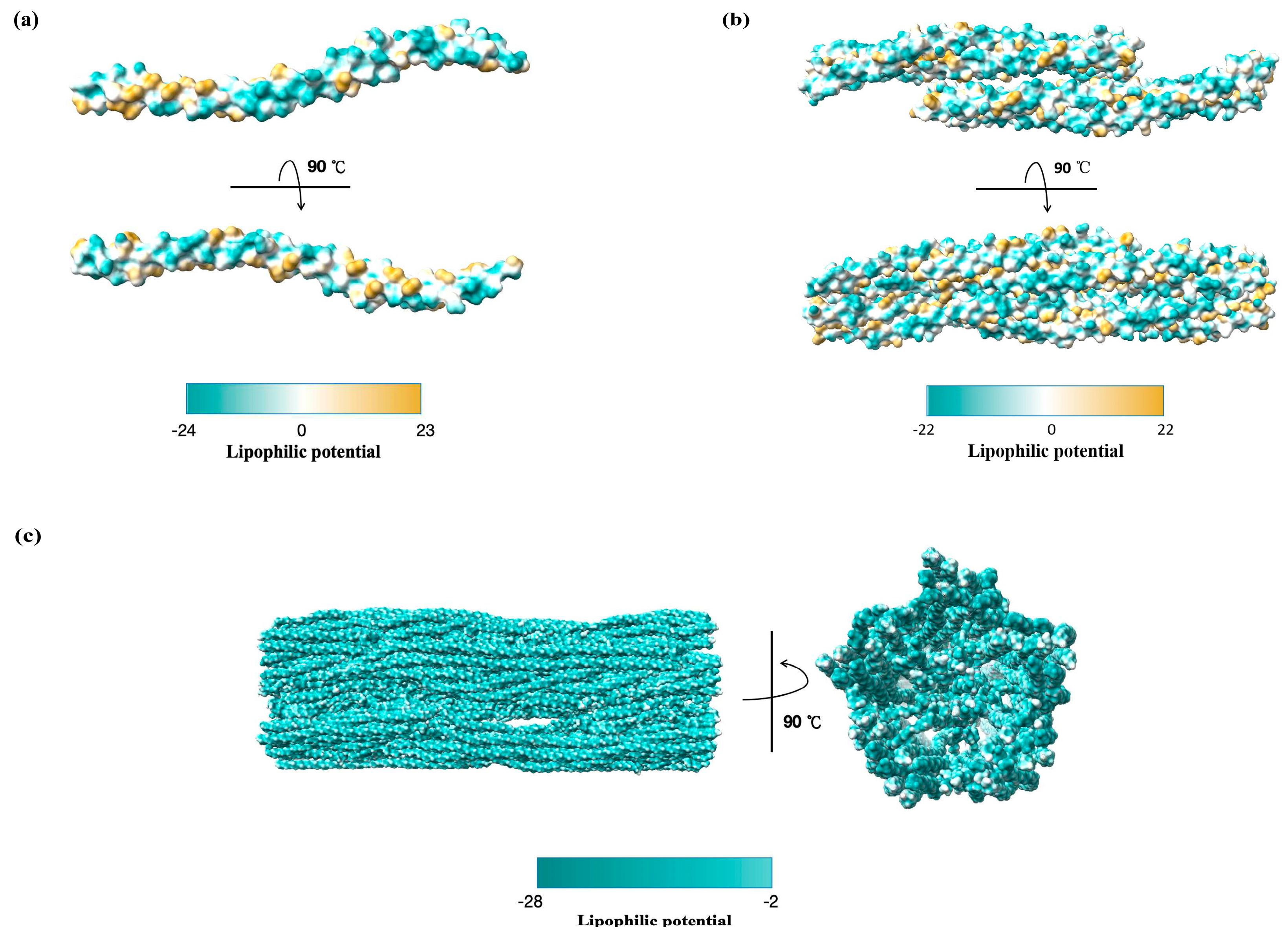
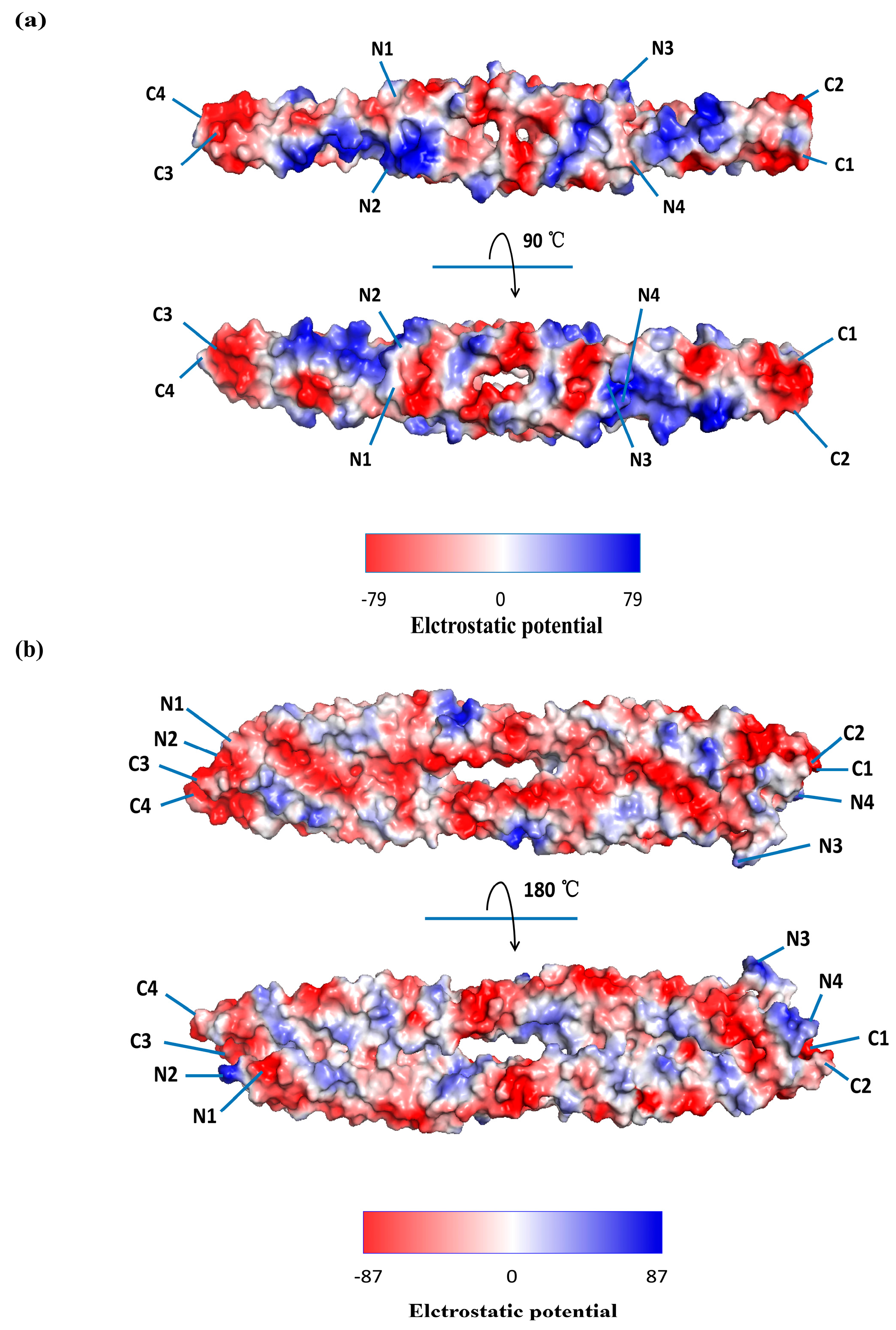
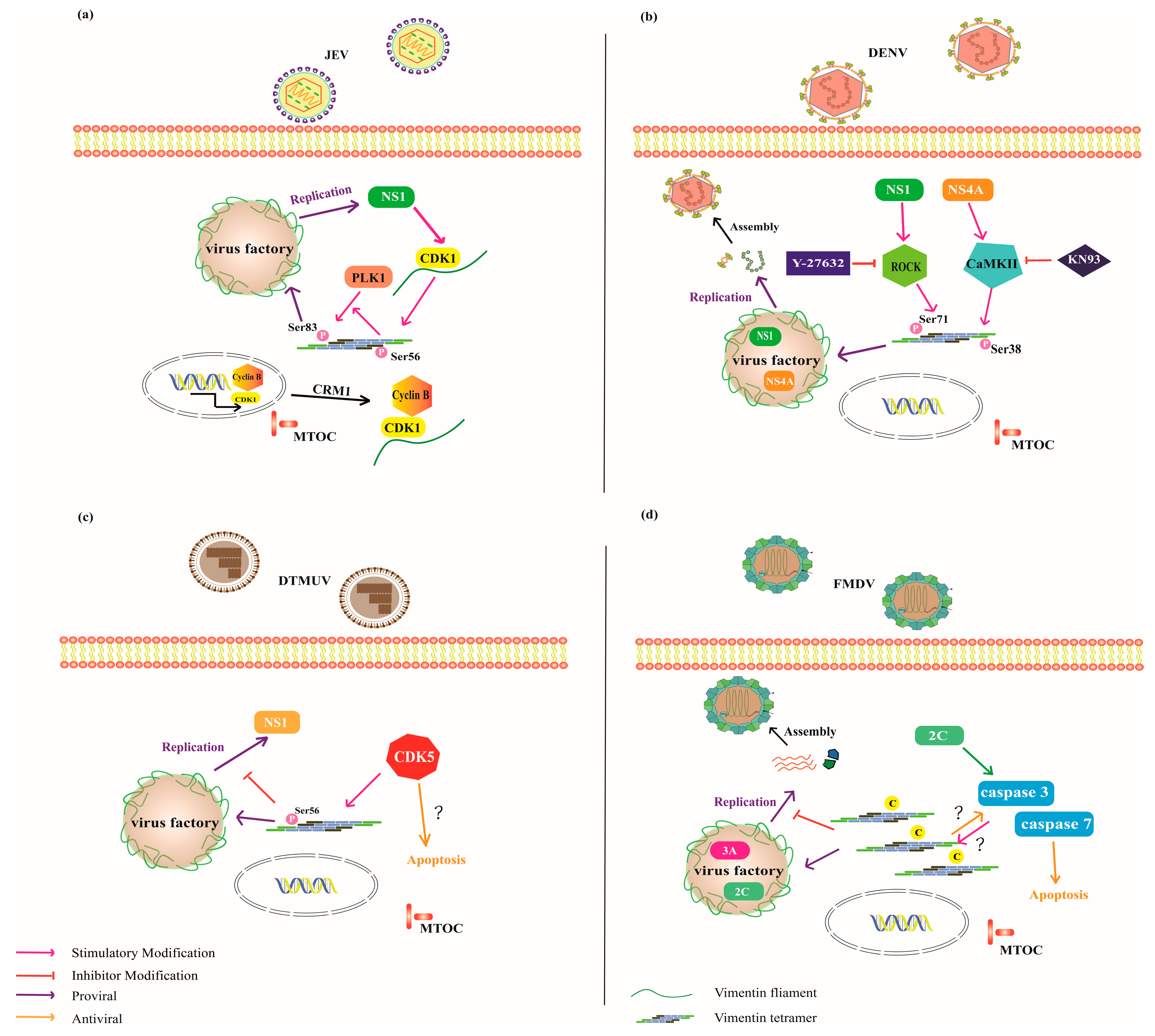
| Virus Species | Genetic Material | Receptor | Co-Location Viral Protein | Binding Domain on CSV | Cell Line/Animal Model | Model of Interaction | Function | Ref. |
|---|---|---|---|---|---|---|---|---|
| SARS-CoV-2 | (+)ssRNA | ACE2 | Spike protein | Tail/Rod | A549/Vero E6 | Hydrogen bonds/Electrostatic contacts | Synergistic ACE2 combination | [47,48,49,50] |
| JEV | (+)ssRNA | vβ3 integrin | E protein | Head/Tail | BHK-21/HTB-11/N18 | Electrostatic contacts | Virus adsorption/endocytosis | [51] |
| DENV | (+)ssRNA | DG-SIGN | E(EDIII) | Rod | VECs | Electrostatic contacts | Enhance cell recognition | [52,53] |
| HCV | (+)ssRNA | CD81 | E1 protein | Head | Huh-7.5.1 | - | Cell-to-cell transmission of virus | [54] |
| EV71 | (+)ssRNA | SCARB2/PSGL-1 | VP1 | Head | HBMECs/Mice | - | Assist in virus localization | [55] |
| PRV | (+)dsDNA | HS | gD/gH | Rod | HEK-293/PK-15 | Hydrogen bonds | Virus adsorption | [56] |
| Strategy | Virus Species | Co-Location Viral Protein | Signaling Pathway Involved | Vimentin Phosphorylation Sites | Binding Domain on Vimentin | Ref. |
|---|---|---|---|---|---|---|
| Shield (pro-viral function) | Vaccinia virus | p39 | - | - | - | [75] |
| Iridoviruses | - | - | - | - | [75] | |
| ASFV | - | CaMKII | Ser82 | - | [76] | |
| JEV | NS1; NS1′ | CDK1-PLK1 | Ser56; Ser83 | - | [77,78] | |
| DENV | NS4A; NS1 | CaMKII; ROCK | Ser38; Ser71 | - | [79,80] | |
| ZIKV | RRBP1 | - | Non-Phosphorylation Ser38; Ser39; Ser56 | - | [81] | |
| CSFV | NS5A | RhoA/ROCK | Ser72 | Rod | [82] | |
| EV71 | VP1; 3C | CaMKII | Ser82 | [83] | ||
| Sabotage (anti-viral) | DTMUV | NS1 | CDK5 | Ser56 | - | [84] |
| FMDV | 2C; 3A | - | - | - | [85,86] | |
| PCV2 | Cap protein | NF-κB; Caspase-3 | - | - | [87] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Ling, Y.; Ling, X.; Liu, Z. Vimentin Dynamics in Viral Infection: Shield or Sabotage? Int. J. Mol. Sci. 2026, 27, 388. https://doi.org/10.3390/ijms27010388
Ling Y, Ling X, Liu Z. Vimentin Dynamics in Viral Infection: Shield or Sabotage? International Journal of Molecular Sciences. 2026; 27(1):388. https://doi.org/10.3390/ijms27010388
Chicago/Turabian StyleLing, Ying, Xuanyi Ling, and Zaixin Liu. 2026. "Vimentin Dynamics in Viral Infection: Shield or Sabotage?" International Journal of Molecular Sciences 27, no. 1: 388. https://doi.org/10.3390/ijms27010388
APA StyleLing, Y., Ling, X., & Liu, Z. (2026). Vimentin Dynamics in Viral Infection: Shield or Sabotage? International Journal of Molecular Sciences, 27(1), 388. https://doi.org/10.3390/ijms27010388






