Low Expression of UBE2Z, a Target Protein of miR-500a, Is Associated with Poor Prognosis in Triple-Negative Breast Cancer
Abstract
1. Introduction
2. Results
2.1. Analysis of Differentially Expressed miRNAs and Survival in TNBC
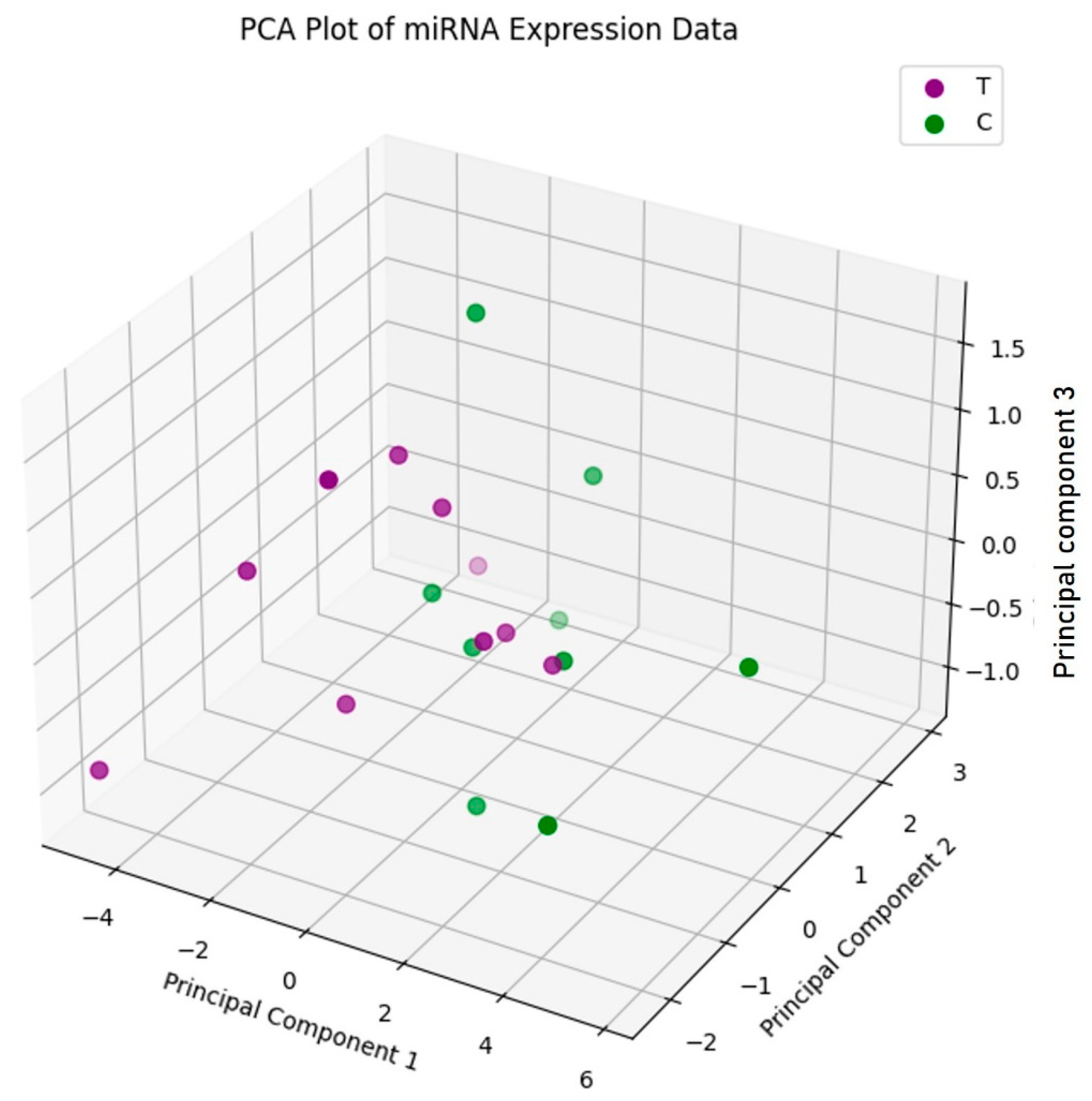
2.2. Principal Component Analysis
2.3. Predicted Target Genes and Functional Analysis
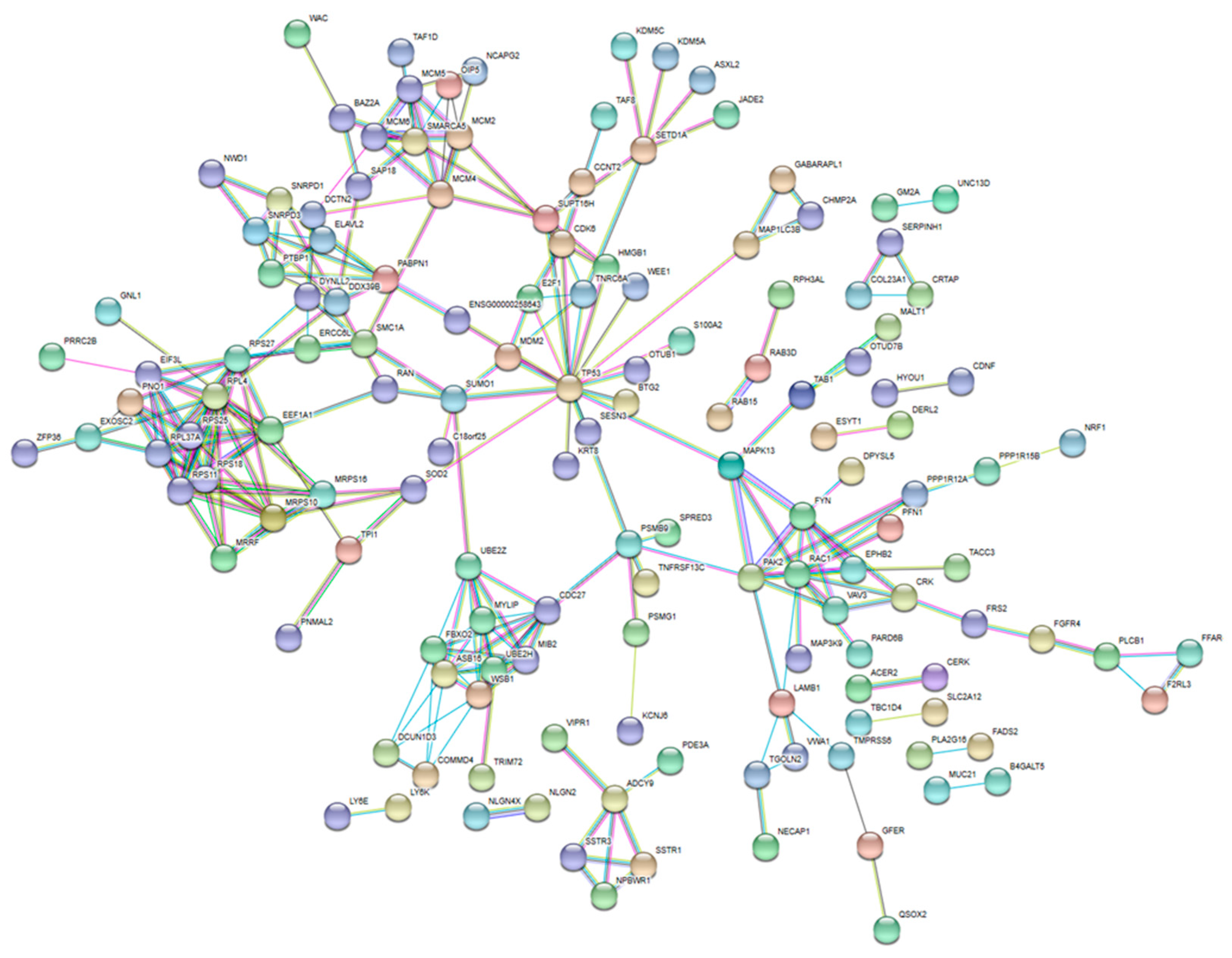
2.4. Analysis of Protein–Protein Interactions and Hub Proteins
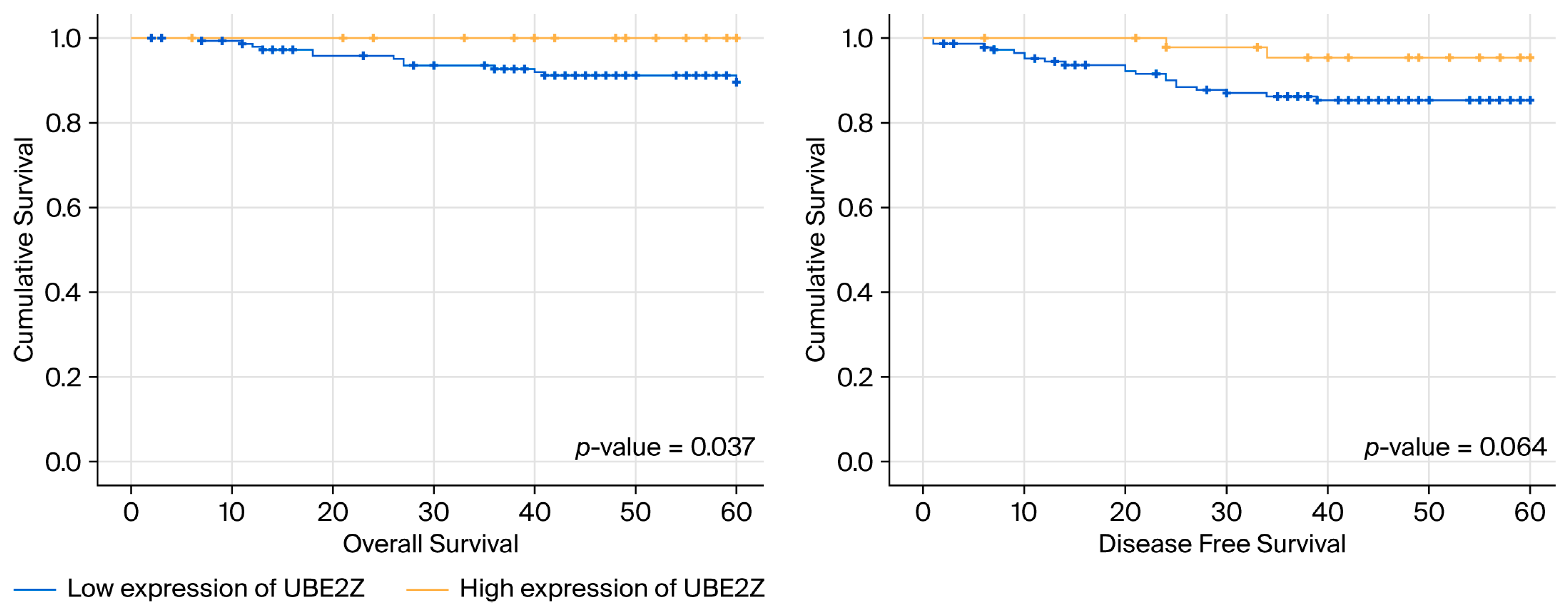
2.5. Associations Among UBE2Z Expression, TNBC Outcomes, and Clinical Features
3. Discussion
4. Materials and Methods
4.1. Sample Selection
4.2. microRNA Array Analysis
4.3. Principal Component Analysis (PCA)
4.4. In Silico Analysis
4.4.1. Prognostic Value of Differentially Expressed miRNAs
4.4.2. Predicted Target Genes and Clinical Implications
4.4.3. Predicted Target Proteins and Implications
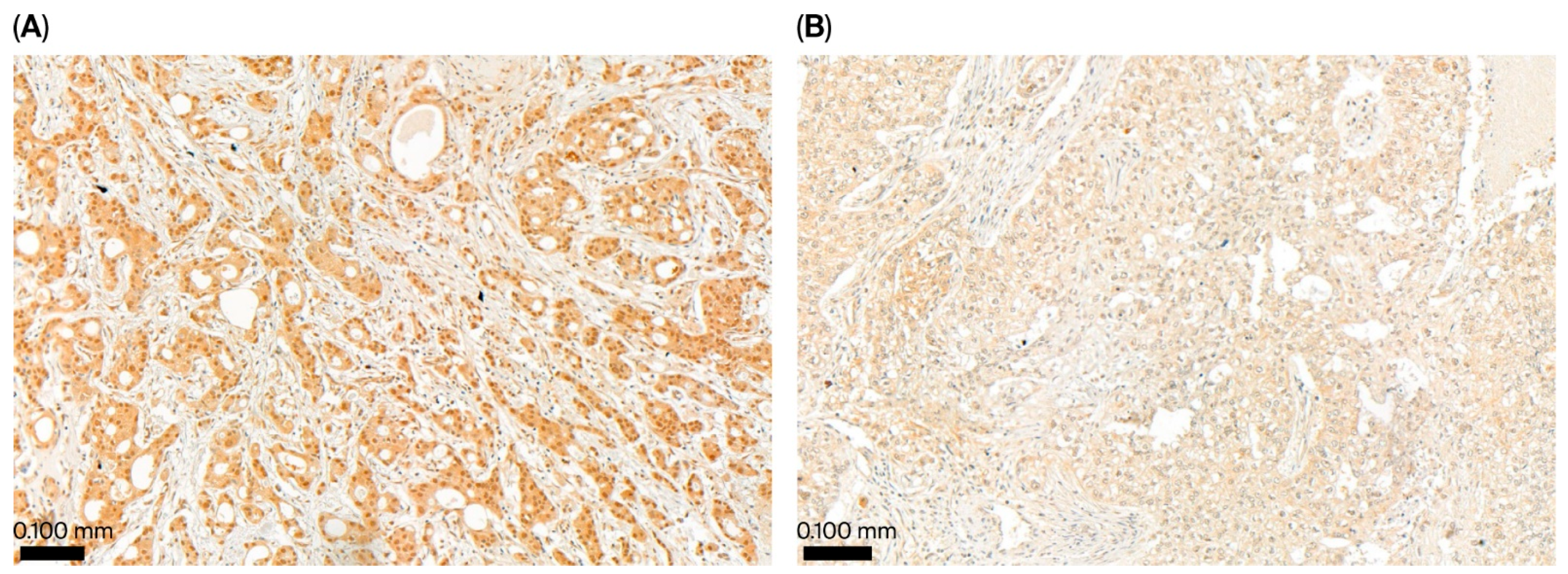
4.5. Immunohistochemistry for UBE2Z
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Goldhirsch, A.; Winer, E.P.; Coates, A.S.; Gelber, R.D.; Piccart-Gebhart, M.; Thürlimann, B.; Senn, H.-J. Personalizing the treatment of women with early breast cancer: Highlights of the St Gallen International expert consensus on the primary therapy of early breast cancer 2013. Ann. Oncol. 2013, 24, 2206–2223. [Google Scholar] [CrossRef]
- Zhu, S.; Wu, Y.; Song, B.; Yi, M.; Yan, Y.; Mei, Q.; Wu, K. Recent advances in targeted strategies for triple-negative breast cancer. J. Hematol. Oncol. 2023, 16, 100. [Google Scholar] [CrossRef]
- Borri, F.; Granaglia, A. Pathology of triple negative breast cancer. Semin. Cancer Biol. 2021, 72, 136–145. [Google Scholar] [CrossRef]
- Dent, R.; Trudeau, M.; Pritchard, K.I.; Hanna, W.M.; Kahn, H.K.; Sawka, C.A.; Lickley, L.A.; Rawlinson, E.; Sun, P.; Narod, S.A. Triple-negative breast cancer: Clinical features and patterns of recurrence. Clin. Cancer Res. 2007, 13, 4429–4434. [Google Scholar] [CrossRef]
- Denkert, C.; Liedtke, C.; Tutt, A.; von Minckwitz, G. Molecular alterations in triple-negative breast cancer-The road to new treatment strategies. Lancet 2017, 389, 2430–2442. [Google Scholar] [CrossRef] [PubMed]
- Bauer, K.R.; Brown, M.; Cress, R.D.; Parise, C.A.; Caggiano, V. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: A population-based study from the California Cancer Registry. Cancer 2007, 109, 1721–1728. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Castro, A.C.; Lin, N.U.; Polyak, K. Insights into molecular classifications of triple-negative breast cancer: Improving patient selection for treatment. Cancer Discov. 2019, 9, 176–198. [Google Scholar] [CrossRef] [PubMed]
- Burstein, M.D.; Tsimelzon, A.; Poage, G.M.; Covington, K.R.; Contreras, A.; Fuqua, S.A.; Savage, M.I.; Osborne, C.K.; Hilsenbeck, S.G.; Chang, J.C.; et al. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin. Cancer Res. 2015, 21, 1688–1698. [Google Scholar] [CrossRef]
- Carey, L.A.; Winer, E.P.; Viale, G.; Cameron, D.; Gianni, L. Triple-negative breast cancer: Disease entity or title of convenience? Nat. Rev. Clin. Oncol. 2010, 7, 683–692. [Google Scholar] [CrossRef]
- Ahn, S.G.; Kim, S.J.; Kim, C.; Jeong, J. Molecular classification of triple-negative breast cancer. J. Breast Cancer 2016, 19, 223–230. [Google Scholar] [CrossRef]
- Liedtke, C.; Bernemann, C.; Kiesel, L.; Rody, A. Genomic profiling in triple-negative breast cancer. Breast Care 2013, 8, 408–413. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Yang, Q.; Yang, H.; Zhang, X. New progress in the role of microRNAs in the diagnosis and prognosis of triple negative breast cancer. Front. Mol. Biosci. 2023, 10, 1162463. [Google Scholar] [CrossRef]
- Ouyang, M.; Li, Y.; Ye, S.; Ma, J.; Lu, L.; Lv, W.; Chang, G.; Li, X.; Li, Q.; Wang, S.; et al. MicroRNA profiling implies new markers of chemoresistance of triple-negative breast cancer. PLoS ONE 2014, 9, e96228. [Google Scholar] [CrossRef] [PubMed]
- Lebert, J.M.; Lester, R.; Powell, E.; Seal, M.; McCarthy, J. Advances in the systemic treatment of triple-negative breast cancer. Curr. Oncol. 2018, 25, S142–S150. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Nedeljković, M.; Damjanović, A. Mechanisms of chemotherapy resistance in triple-negative breast cancer-how we can rise to the challenge. Cells 2019, 8, 957. [Google Scholar] [CrossRef]
- Volovat, S.R.; Volovat, C.; Hordila, I.; Hordila, D.A.; Mirestean, C.C.; Miron, O.T.; Lungulescu, C.; Scripcariu, D.V.; Stolniceanu, C.R.; Konsoulova-Kirova, A.A.; et al. MiRNA and LncRNA as potential biomarkers in triple-negative breast cancer: A review. Front. Oncol. 2020, 10, 526850. [Google Scholar] [CrossRef]
- Chen, C.; Lin, C.-J.; Li, S.-Y.; Hu, X.; Shao, Z.-M. Identification of a novel signature with prognostic value in triple-negative breast cancer through clinico-transcriptomic analysis. Ann. Transl. Med. 2022, 10, 1095. [Google Scholar] [CrossRef]
- Ferrari, P.; Scatena, C.; Ghilli, M.; Bargagna, I.; Lorenzini, G.; Nicolini, A. Molecular mechanisms, biomarkers and emerging therapies for chemotherapy resistant TNBC. Int. J. Mol. Sci. 2022, 23, 1665. [Google Scholar] [CrossRef]
- Lü, L.; Mao, X.; Shi, P.; He, B.; Xu, K.; Zhang, S.; Wang, J. MicroRNAs in the prognosis of triple-negative breast cancer: A systematic review and meta-analysis. Medicine 2017, 96, e7085. [Google Scholar] [CrossRef] [PubMed]
- Hong, H.-C.; Chuang, C.-H.; Huang, W.-C.; Weng, S.-L.; Chen, C.-H.; Chang, K.-H.; Liao, K.-W.; Huang, H.-D. A panel of eight microRNAs is a good predictive parameter for triple-negative breast cancer relapse. Theranostics 2020, 10, 8771–8789. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Xing, L.; Wang, M.; Chi, H.; Zhang, L.; Chen, J. Comprehensive analysis of differentially expressed profiles of lncRNAs/mRNAs and miRNAs with associated ceRNA networks in triple-negative breast cancer. Cell Physiol. Biochem. 2018, 50, 473–488. [Google Scholar] [CrossRef] [PubMed]
- Cascione, L.; Gasparini, P.; Lovat, F.; Carasi, S.; Pulvirenti, A.; Ferro, A.; Alder, H.; He, G.; Vecchione, A.; Croce, C.M.; et al. Integrated microRNA and mRNA signatures associated with survival in triple negative breast cancer. PLoS ONE 2013, 8, e55910. [Google Scholar] [CrossRef]
- Haldrup, C.; Kosaka, N.; Ochiya, T.; Borre, M.; Høyer, S.; Orntoft, T.F.; Sorensen, K.D. Profiling of circulating microRNAs for prostate cancer biomarker discovery. Drug Deliv. Transl. Res. 2014, 4, 19–30. [Google Scholar] [CrossRef]
- Hadavi, R.; Mohammadi-Yeganeh, S.; Razaviyan, J.; Koochaki, A.; Kokhaei, P.; Bandegi, A. Expression of bioinformatically candidate miRNAs including, miR-576-5p, miR-501-3p and miR-3143, targeting PI3K pathway in triple-negative breast cancer. Galen. Med. J. 2019, 8, e1646. [Google Scholar] [CrossRef]
- Shi, X.; Wang, B.; Chen, X.; Zheng, Y.; Ding, Y.; Wang, C. Upregulation of ubiquitin-conjugating enzyme E2Z is associated with human hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 2020, 523, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Madden, S.F.; Clarke, C.; Gaule, P.; Aherne, S.T.; O’DOnovan, N.; Clynes, M.; Crown, J.; Gallagher, W.M. BreastMark: An integrated approach to mining publicly available transcriptomic datasets relating to breast cancer outcome. Breast Cancer Res. 2013, 15, R52. [Google Scholar] [CrossRef]
| Expression in Comparison with Non-Relapse Patients | miRNA ID | Fold Change | p-Value |
|---|---|---|---|
| Overexpressed | hsa-miR-500a-3p | 2.395126 | 0.039 |
| hsa-miR-501-3p | 2.101952 | 0.037 | |
| hsa-miR-502-3p | 2.483641 | 0.047 | |
| Underexpressed | hsa-miR-6798-5p | −2.161684 | 0.050 |
| hsa-miR-7150 | −2.336029 | 0.047 |
| Category | Term | Gene No. | p-Value |
|---|---|---|---|
| KEGG pathway | Cell cycle | 9 | 1.2 × 10−3 |
| Rap1 signaling pathway | 10 | 0.0010 | |
| Neurotrophin signaling pathway | 7 | 0.0170 | |
| Glioma | 5 | 0.0250 | |
| Chronic myeloid leukemia | 5 | 0.0350 | |
| Biocarta | Tumor suppressor Arf inhibits ribosomal biogenesis | 4 | 0.0087 |
| Sumoylation by RanBP2 regulates transcriptional repression | 3 | 0.0330 |
| UBE2Z Expression | Low (n = 148) | High (n = 47) | p-Value |
|---|---|---|---|
| Histologic grade | 0.805 | ||
| Grade 1 | 2 (1.4%) | 0 | |
| Grade 2 | 22 (14.9%) | 7 (14.9%) | |
| Grade 3 | 124 (83.8%) | 40 (85.1%) | |
| T stage | 0.238 | ||
| Early (T1) | 62 (42.2%) | 25 (53.2%) | |
| Advanced (T2–T4) | 85 (57.8%) | 22 (46.8%) | |
| LN metastasis | 0.008 | ||
| Negative (N0) | 98 (67.1%) | 41 (87.2%) | |
| Positive (N1–N3) | 48 (32.9%) | 6 (12.8%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Kim, D.; Choi, S.-Y. Low Expression of UBE2Z, a Target Protein of miR-500a, Is Associated with Poor Prognosis in Triple-Negative Breast Cancer. Int. J. Mol. Sci. 2026, 27, 361. https://doi.org/10.3390/ijms27010361
Kim D, Choi S-Y. Low Expression of UBE2Z, a Target Protein of miR-500a, Is Associated with Poor Prognosis in Triple-Negative Breast Cancer. International Journal of Molecular Sciences. 2026; 27(1):361. https://doi.org/10.3390/ijms27010361
Chicago/Turabian StyleKim, Donghyun, and Song-Yi Choi. 2026. "Low Expression of UBE2Z, a Target Protein of miR-500a, Is Associated with Poor Prognosis in Triple-Negative Breast Cancer" International Journal of Molecular Sciences 27, no. 1: 361. https://doi.org/10.3390/ijms27010361
APA StyleKim, D., & Choi, S.-Y. (2026). Low Expression of UBE2Z, a Target Protein of miR-500a, Is Associated with Poor Prognosis in Triple-Negative Breast Cancer. International Journal of Molecular Sciences, 27(1), 361. https://doi.org/10.3390/ijms27010361






