Positive-Strand RNA Viruses Induce LTR Retrotransposon Transcription and Extrachromosomal Circular DNA Generation in Plants
Abstract
1. Introduction
2. Results
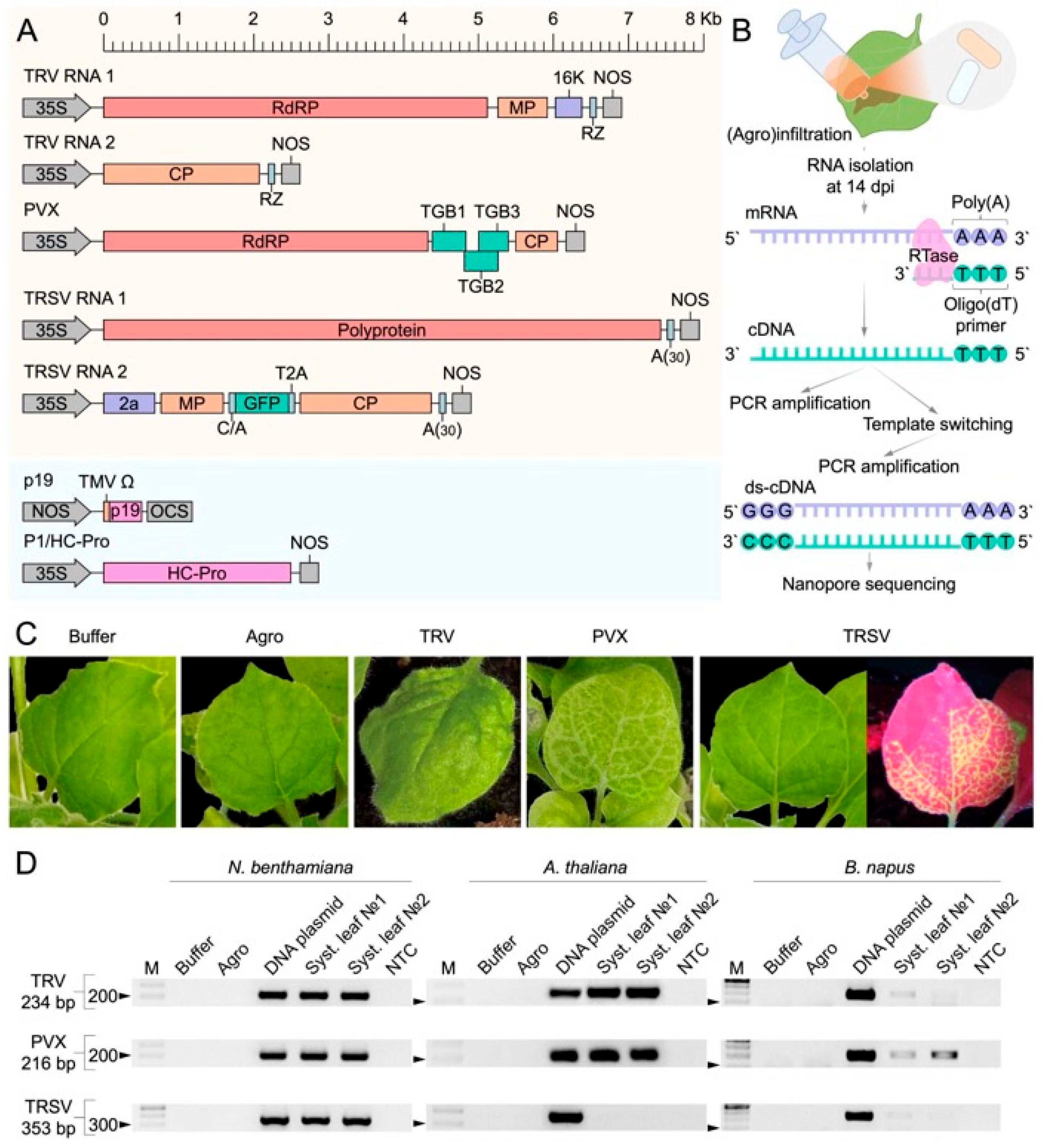
2.1. Experimental Design and Virus Detection in Three Plant Species
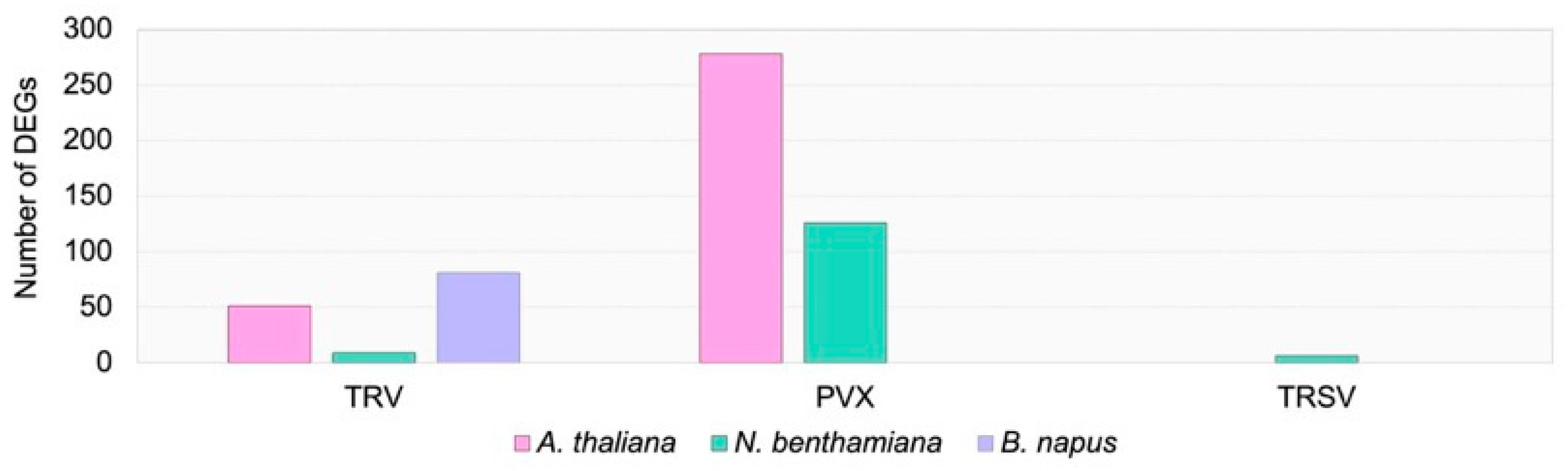
2.2. Transcriptome Response to Viral Infection
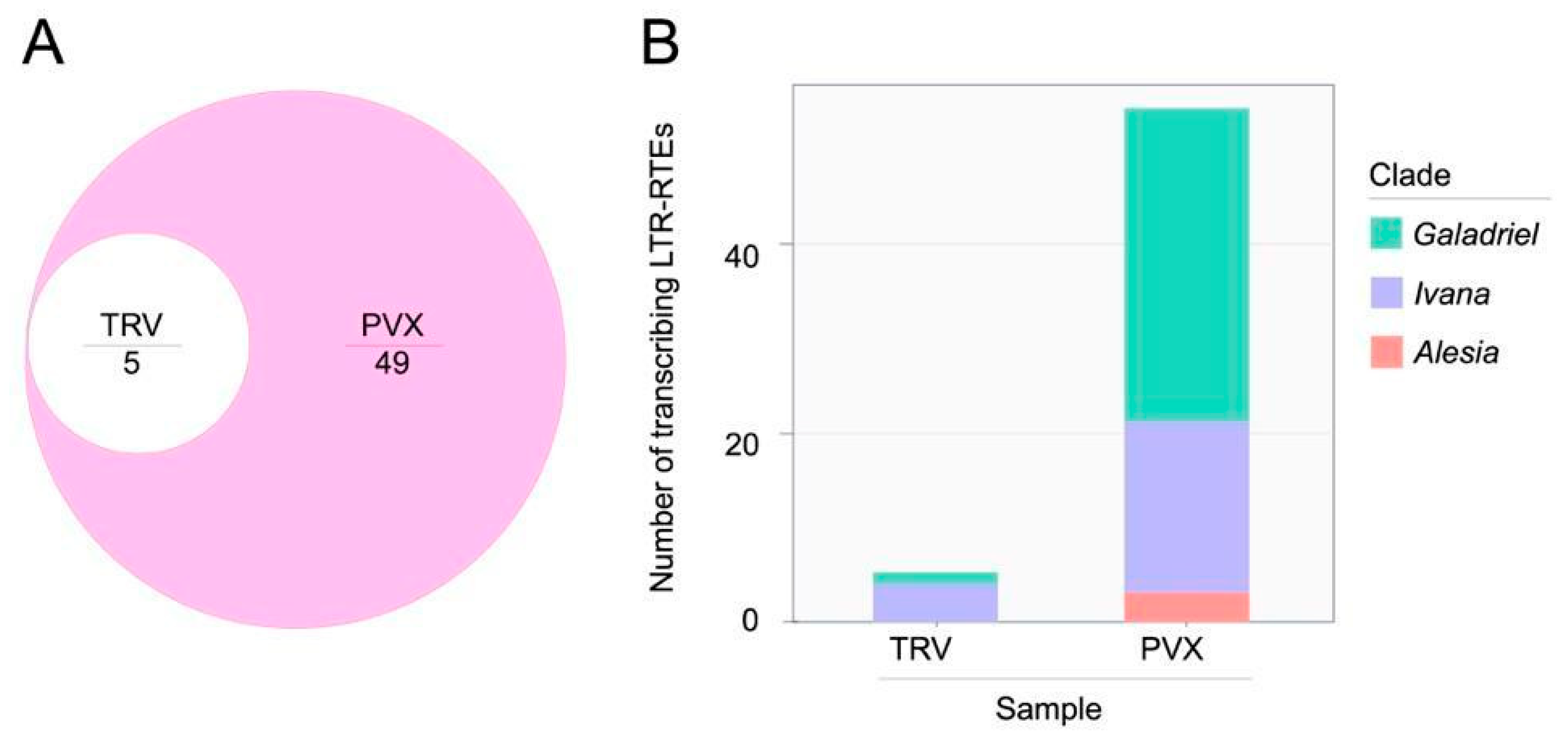
2.3. Numerous LTR Retrotransposons Are Transcribed Under Virus Stress
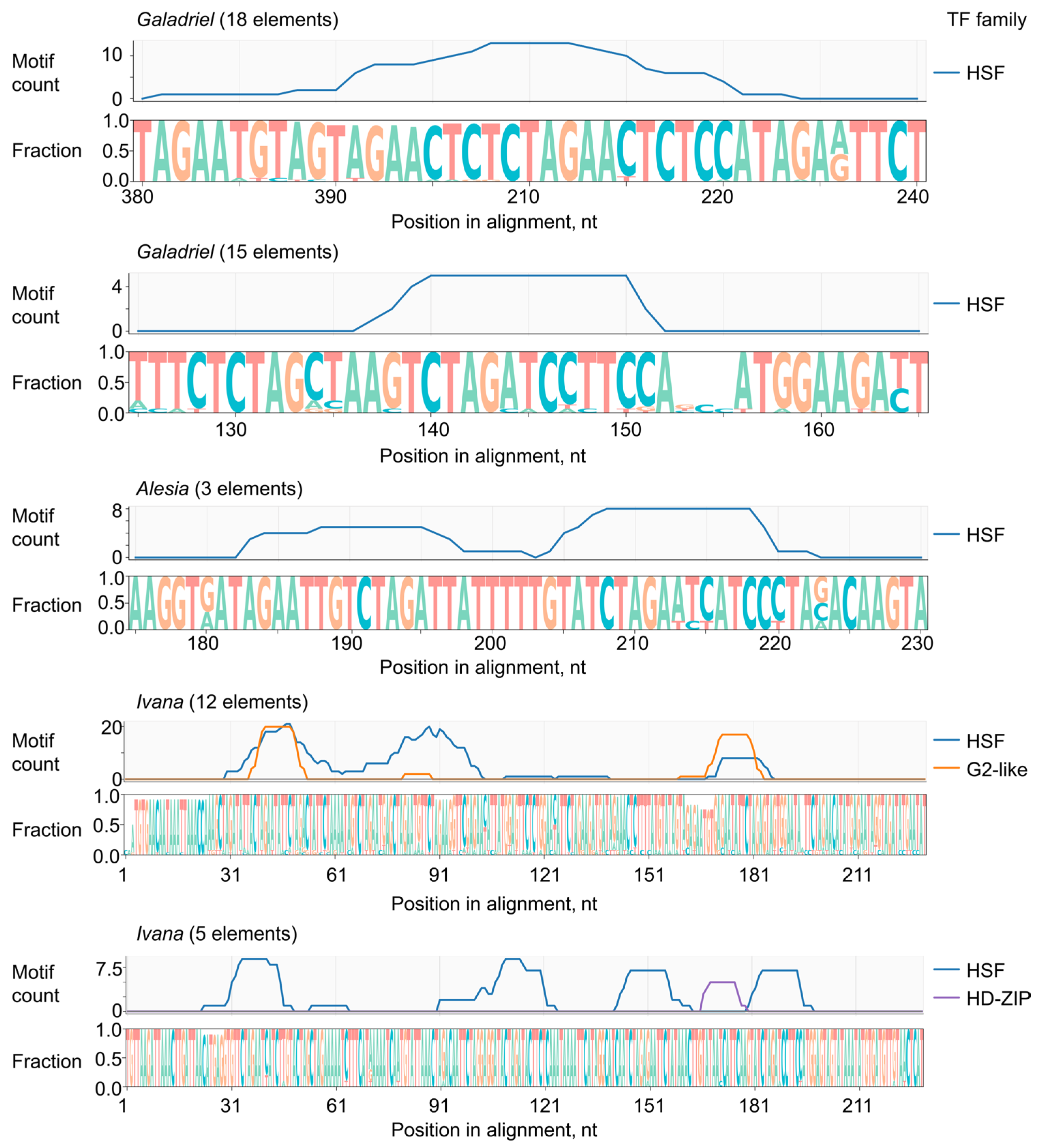
2.4. Putative Regulatory Elements Within LTRs of Virus-Induced LTR-RTEs
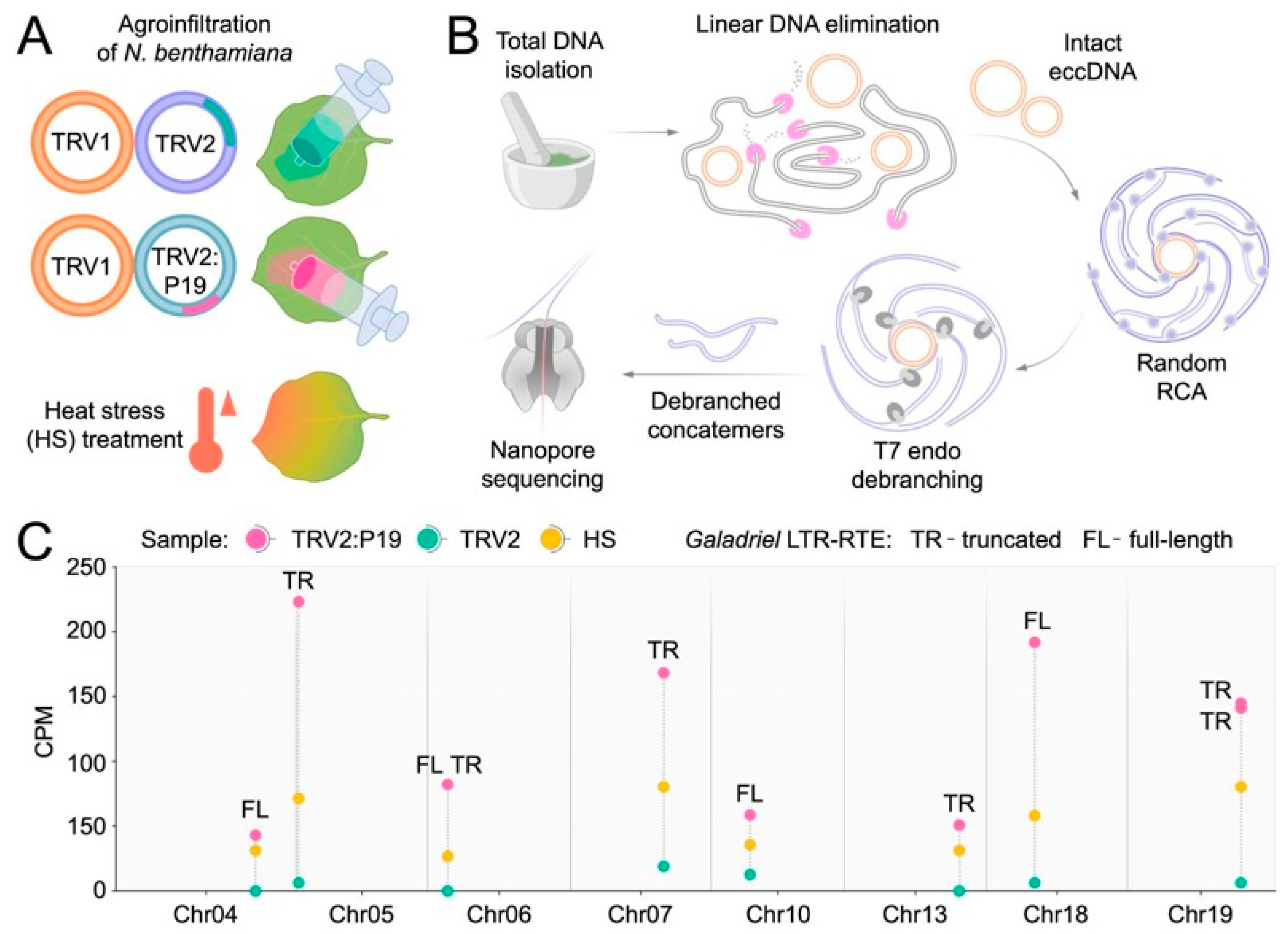
2.5. LTR-RTEs of N. benthamiana Produce Extrachromosomal Circular DNA in Response to a Viral Suppressor of RNA Silencing and Heat Stress
3. Discussion
4. Materials and Methods
4.1. Plant Growth Conditions
4.2. Plants Agroinfiltration
4.3. N. benthamiana Plants Heat Stress Treatment
4.4. RNA Isolation, cDNA Synthesis, and PCR Validation
4.5. Double-Stranded cDNA Synthesis
4.6. DNA Isolation
4.7. eccDNA Isolation and Amplification
4.8. Library Preparation and Nanopore Sequencing
4.9. Genome Assemblies
4.10. LTR-Retrotransposons Annotation
4.11. Gene and LTR-Retrotransposons Expression Analysis
4.12. Putative Transcription Factors Binding Motifs Prediction and Visualization
4.13. Mobilome-Seq Analysis
4.14. Phylogenetic Analysis
4.15. Data Visualization
4.16. Plasmids
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kovalchuk, I.; Kovalchuk, O.; Kalck, V.; Boyko, V.; Filkowski, J.; Heinlein, M.; Hohn, B. Pathogen-induced systemic plant signal triggers DNA rearrangements. Nature 2003, 423, 760–762. [Google Scholar] [CrossRef] [PubMed]
- Lucht, J.M.; Mauch-Mani, B.; Steiner, H.-Y.; Metraux, J.-P.; Ryals, J.; Hohn, B. Pathogen stress increases somatic recombination frequency in Arabidopsis. Nat. Genet. 2002, 30, 311–314. [Google Scholar] [CrossRef] [PubMed]
- Boyko, A.; Kathiria, P.; Zemp, F.J.; Yao, Y.; Pogribny, I.; Kovalchuk, I. Transgenerational changes in the genome stability and methylation in pathogen-infected plants: (Virus-induced plant genome instability). Nucleic Acids Res. 2007, 35, 1714–1725. [Google Scholar] [CrossRef]
- Molinier, J.; Ries, G.; Zipfel, C.; Hohn, B. Transgeneration memory of stress in plants. Nature 2006, 442, 1046–1049. [Google Scholar] [CrossRef]
- Wilkinson, S.W.; Magerøy, M.H.; López Sánchez, A.; Smith, L.M.; Furci, L.; Cotton, T.E.A.; Krokene, P.; Ton, J. Surviving in a hostile world: Plant strategies to resist pests and diseases. Annu. Rev. Phytopathol. 2019, 57, 505–529. [Google Scholar] [CrossRef]
- Ossowski, S.; Schneeberger, K.; Lucas-Lledó, J.I.; Warthmann, N.; Clark, R.M.; Shaw, R.G.; Weigel, D.; Lynch, M. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science 2010, 327, 92–94. [Google Scholar] [CrossRef]
- Hannan Parker, A.; Wilkinson, S.W.; Ton, J. Epigenetics: A catalyst of plant immunity against pathogens. New Phytol. 2022, 233, 66–83. [Google Scholar] [CrossRef]
- Hassan, A.H.; Mokhtar, M.M.; El Allali, A. Transposable elements: Multifunctional players in the plant genome. Front. Plant Sci. 2023, 14, 1330127. [Google Scholar] [CrossRef]
- Galindo-González, L.; Mhiri, C.; Deyholos, M.K.; Grandbastien, M.-A. LTR-retrotransposons in plants: Engines of evolution. Gene 2017, 626, 14–25. [Google Scholar] [CrossRef]
- Chu, J.; Newman, J.; Cho, J. Molecular mimicry of transposable elements in plants. Plant Cell Physiol. 2025, 66, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Cuerda-Gil, D.; Slotkin, R.K. Non-canonical RNA-directed DNA methylation. Nat. Plants 2016, 2, 16163, Erratum in Nat. Plants 2017, 3, 16211. [Google Scholar] [CrossRef]
- Lewsey, M.G.; Carr, J.P. Plant pathogens: RNA viruses. In Encyclopedia of Microbiology; Elsevier: Amsterdam, The Netherlands, 2009; pp. 443–458. [Google Scholar]
- Voinnet, O. Three decades of mobile RNA silencing within plants: What have we learnt? J. Exp. Bot. 2025, eraf312. [Google Scholar] [CrossRef]
- Li, F.; Li, X.; Zhao, S.; Pan, F.; Li, Z.; Hao, Y.; He, J.; Wang, A.; Kormelink, R.; Zhou, X. Antiviral RNA interference in plants: Increasing complexity and integration with other biological processes. Plant Commun. 2025, 6, 101490. [Google Scholar] [CrossRef]
- Erdmann, R.M.; Picard, C.L. RNA-directed DNA methylation. PLoS Genet. 2020, 16, e1009034. [Google Scholar] [CrossRef]
- Garnelo Gómez, B.; Rosas-Díaz, T.; Shi, C.; Fan, P.; Zhang, D.; Rufián, J.S.; Lozano-Durán, R. The viral silencing suppressor P19 interacts with the receptor-like kinases BAM1 and BAM2 and suppresses the cell-to-cell movement of RNA silencing independently of its ability to bind sRNA. New Phytol. 2021, 229, 1840–1843. [Google Scholar] [CrossRef]
- Beguiristain, T.; Grandbastien, M.A.; Puigdomènech, P.; Casacuberta, J.M. Three Tnt1 subfamilies show different stress-associated patterns of expression in tobacco. Consequences for retrotransposon control and evolution in plants. Plant Physiol. 2001, 127, 212–221. [Google Scholar] [CrossRef]
- Singh, A.; Goswami, S.; Vinutha, T.; Jain, R.K.; Ramesh, S.V.; Praveen, S. Retrotransposons-based genetic regulation underlies the cellular response to two genetically diverse viral infections in tomato. Physiol. Mol. Plant Pathol. 2022, 120, 101839. [Google Scholar] [CrossRef]
- Corrêa, R.L.; Sanz-Carbonell, A.; Kogej, Z.; Müller, S.Y.; Ambrós, S.; López-Gomollón, S.; Gómez, G.; Baulcombe, D.C.; Elena, S.F. Viral fitness determines the magnitude of transcriptomic and epigenomic reprograming of defense responses in plants. Mol. Biol. Evol. 2020, 37, 1866–1881. [Google Scholar] [CrossRef]
- Diezma-Navas, L.; Pérez-González, A.; Artaza, H.; Alonso, L.; Caro, E.; Llave, C.; Ruiz-Ferrer, V. Crosstalk between epigenetic silencing and infection by tobacco rattle virus in Arabidopsis. Mol. Plant Pathol. 2019, 20, 1439–1452. [Google Scholar] [CrossRef]
- Panda, K.; Slotkin, R.K. Long-read cDNA sequencing enables a “gene-like” transcript annotation of transposable elements. Plant Cell 2020, 32, 2687–2698. [Google Scholar] [CrossRef]
- Kirov, I.; Merkulov, P.; Polkhovskaya, E.; Konstantinov, Z.; Kazancev, M.; Saenko, K.; Polkhovskiy, A.; Dudnikov, M.; Garibyan, T.; Demurin, Y.; et al. Epigenetic stress and long-read cDNA sequencing of sunflower (Helianthus annuus L.) revealed the origin of the plant retrotranscriptome. Plants 2022, 11, 3579. [Google Scholar] [CrossRef]
- Gullner, G.; Komives, T.; Király, L.; Schröder, P. Glutathione S-transferase enzymes in plant-pathogen interactions. Front. Plant Sci. 2018, 9, 1836. [Google Scholar] [CrossRef]
- Li, K.; Wu, G.; Li, M.; Ma, M.; Du, J.; Sun, M.; Sun, X.; Qing, L. Transcriptome analysis of Nicotiana benthamiana infected by Tobacco curly shoot virus. Virol. J. 2018, 15, 138. [Google Scholar] [CrossRef]
- Wiermer, M.; Feys, B.J.; Parker, J.E. Plant immunity: The EDS1 regulatory node. Curr. Opin. Plant Biol. 2005, 8, 383–389. [Google Scholar] [CrossRef]
- Paudel, D.B.; Montenegro Alonso, A.P.; Chisholm, J.; Xiao, H.; Sanfaçon, H. Transcriptomic changes associated with infection of Nicotiana benthamiana plants with tomato ringspot virus (genus Nepovirus) during the acute symptomatic stage and after symptom recovery. PLoS ONE 2025, 20, e0328517. [Google Scholar] [CrossRef]
- Lin, C.; Zhang, Z.; Zhang, Z.; Long, Y.; Shen, X.; Zhang, J.; Wang, Y. The role of glutathione S-transferase in the regulation of plant growth, and responses to environmental stresses. Phyton 2025, 94, 583–601. [Google Scholar] [CrossRef]
- Bakery, A.; Vraggalas, S.; Shalha, B.; Chauhan, H.; Benhamed, M.; Fragkostefanakis, S. Heat stress transcription factors as the central molecular rheostat to optimize plant survival and recovery from heat stress. New Phytol. 2024, 244, 51–64. [Google Scholar] [CrossRef]
- Lanciano, S.; Carpentier, M.C.; Llauro, C.; Jobet, E.; Robakowska-Hyzorek, D.; Lasserre, E.; Ghesquière, A.; Panaud, O.; Mirouze, M. Sequencing the extrachromosomal circular mobilome reveals retrotransposon activity in plants. PLoS Genet. 2017, 13, e1006630. [Google Scholar] [CrossRef]
- Bacete, L.; Mélida, H.; Miedes, E.; Molina, A. Plant cell wall-mediated immunity: Cell wall changes trigger disease resistance responses. Plant J. 2018, 93, 614–636. [Google Scholar] [CrossRef]
- Ito, H.; Gaubert, H.; Bucher, E.; Mirouze, M.; Vaillant, I.; Paszkowski, J. An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature 2011, 472, 115–119. [Google Scholar] [CrossRef]
- Ito, H.; Yoshida, T.; Tsukahara, S.; Kawabe, A. Evolution of the ONSEN retrotransposon family activated upon heat stress in Brassicaceae. Gene 2013, 518, 256–261. [Google Scholar] [CrossRef]
- Merkulov, P.; Egorova, E.; Kirov, I. Composition and structure of Arabidopsis thaliana extrachromosomal circular DNAs revealed by nanopore sequencing. Plants 2023, 12, 2178. [Google Scholar] [CrossRef]
- Cavrak, V.V.; Lettner, N.; Jamge, S.; Kosarewicz, A.; Bayer, L.M.; Mittelsten Scheid, O. How a retrotransposon exploits the plant’s heat stress response for its activation. PLoS Genet. 2014, 10, e1004115. [Google Scholar] [CrossRef]
- Alazem, M.; Lin, N.-S. Roles of plant hormones in the regulation of host-virus interactions. Mol. Plant Pathol. 2015, 16, 529–540. [Google Scholar] [CrossRef]
- Duan, K.; Ding, X.; Zhang, Q.; Zhu, H.; Pan, A.; Huang, J. AtCopeg1, the unique gene originated from AtCopia95 retrotransposon family, is sensitive to external hormones and abiotic stresses. Plant Cell Rep. 2008, 27, 1065–1073. [Google Scholar] [CrossRef]
- Zeller, G.; Henz, S.R.; Widmer, C.K.; Sachsenberg, T.; Rätsch, G.; Weigel, D.; Laubinger, S. Stress-induced changes in the Arabidopsis thaliana transcriptome analyzed using whole-genome tiling arrays. Plant J. 2009, 58, 1068–1082. [Google Scholar] [CrossRef]
- Takeda, S.; Sugimoto, K.; Otsuki, H.; Hirochika, H. Transcriptional activation of the tobacco retrotransposon Tto1 by wounding and methyl jasmonate. Plant Mol. Biol. 1998, 36, 365–376. [Google Scholar] [CrossRef]
- Takeda, S.; Sugimoto, K.; Otsuki, H.; Hirochika, H. A 13-bp cis-regulatory element in the LTR promoter of the tobacco retrotransposon Tto1 is involved in responsiveness to tissue culture, wounding, methyl jasmonate and fungal elicitors. Plant J. 1999, 18, 383–393. [Google Scholar] [CrossRef]
- Suoniemi, A.; Anamthawat-Jónsson, K.; Arna, T.; Schulman, A.H. Retrotransposon BARE-1 is a major, dispersed component of the barley (Hordeum vulgare L.) genome. Plant Mol. Biol. 1996, 30, 1321–1329. [Google Scholar] [CrossRef]
- Kimura, Y.; Tosa, Y.; Shimada, S.; Sogo, R.; Kusaba, M.; Sunaga, T.; Betsuyaku, S.; Eto, Y.; Nakayashiki, H.; Mayama, S. OARE-1, a Ty1-copia retrotransposon in oat activated by abiotic and biotic stresses. Plant Cell Physiol. 2001, 42, 1345–1354. [Google Scholar] [CrossRef]
- Rico-Cabanas, L.; Martínez-Izquierdo, J.A. CIRE1, a novel transcriptionally active Ty1-copia retrotransposon from Citrus sinensis. Mol. Genet. Genom. 2007, 277, 365–377. [Google Scholar] [CrossRef]
- Benoit, M.; Drost, H.-G.; Catoni, M.; Gouil, Q.; Lopez-Gomollon, S.; Baulcombe, D.; Paszkowski, J. Environmental and epigenetic regulation of Rider retrotransposons in tomato. PLoS Genet. 2019, 15, e1008370. [Google Scholar] [CrossRef]
- Tapia, G.; Verdugo, I.; Yañez, M.; Ahumada, I.; Theoduloz, C.; Cordero, C.; Poblete, F.; González, E.; Ruiz-Lara, S. Involvement of ethylene in stress-induced expression of the TLC1.1 retrotransposon from Lycopersicon chilense Dun. Plant Physiol. 2005, 138, 2075–2086. [Google Scholar] [CrossRef]
- Anandalakshmi, R.; Pruss, G.J.; Ge, X.; Marathe, R.; Mallory, A.C.; Smith, T.H.; Vance, V.B. A viral suppressor of gene silencing in plants. Proc. Natl. Acad. Sci. USA 1998, 95, 13079–13084. [Google Scholar] [CrossRef]
- Wang, L.; Ding, Y.; He, L.; Zhang, G.; Zhu, J.-K.; Lozano-Duran, R. A virus-encoded protein suppresses methylation of the viral genome through its interaction with AGO4 in the Cajal body. eLife 2020, 9, e55542. [Google Scholar] [CrossRef]
- Cheng, X.; Wang, A. The Potyvirus silencing suppressor protein VPg mediates degradation of SGS3 via ubiquitination and autophagy pathways. J. Virol. 2017, 91, 10-1128. [Google Scholar] [CrossRef]
- Kim, E.Y.; Wang, L.; Lei, Z.; Li, H.; Fan, W.; Cho, J. Ribosome stalling and SGS3 phase separation prime the epigenetic silencing of transposons. Nat. Plants 2021, 7, 303–309. [Google Scholar] [CrossRef]
- Kawakatsu, T. RNA-directed DNA methylation links viral disease and plant architecture in rice. Mol. Plant 2020, 13, 814–816. [Google Scholar] [CrossRef]
- Aguilar, E.; Del Toro, F.J.; Brosseau, C.; Moffett, P.; Canto, T.; Tenllado, F. Cell death triggered by the P25 protein in Potato virus X-associated synergisms results from endoplasmic reticulum stress in Nicotiana benthamiana. Mol. Plant Pathol. 2019, 20, 194–210. [Google Scholar] [CrossRef]
- Yang, X.; Luo, X.; Zhang, Y.; Zhang, Z.; OuYang, X.; Shi, X.; Lv, X.; Li, F.; Zhang, S.; Liu, Y.; et al. Tomato chlorosis virus CPm protein is a pathogenicity determinant and suppresses host local RNA silencing induced by single-stranded RNA. Front. Microbiol. 2023, 14, 1151747. [Google Scholar] [CrossRef]
- Matsunaga, W.; Ohama, N.; Tanabe, N.; Masuta, Y.; Masuda, S.; Mitani, N.; Yamaguchi-Shinozaki, K.; Ma, J.F.; Kato, A.; Ito, H. A small RNA mediated regulation of a stress-activated retrotransposon and the tissue specific transposition during the reproductive period in Arabidopsis. Front. Plant Sci. 2015, 6, 48. [Google Scholar] [CrossRef]
- Chiu, M.-H.; Chen, I.H.; Baulcombe, D.C.; Tsai, C.-H. The silencing suppressor P25 of Potato virus X interacts with Argonaute1 and mediates its degradation through the proteasome pathway. Mol. Plant Pathol. 2010, 11, 641–649. [Google Scholar] [CrossRef]
- Várallyay, É.; Oláh, E.; Havelda, Z. Independent parallel functions of p19 plant viral suppressor of RNA silencing required for effective suppressor activity. Nucleic Acids Res. 2014, 42, 599–608. [Google Scholar] [CrossRef]
- Oberlin, S.; Rajeswaran, R.; Trasser, M.; Barragán-Borrero, V.; Schon, M.A.; Plotnikova, A.; Loncsek, L.; Nodine, M.D.; Marí-Ordóñez, A.; Voinnet, O. Innate, translation-dependent silencing of an invasive transposon in Arabidopsis. EMBO Rep. 2022, 23, e53400. [Google Scholar] [CrossRef]
- Nuthikattu, S.; McCue, A.D.; Panda, K.; Fultz, D.; DeFraia, C.; Thomas, E.N.; Slotkin, R.K. The initiation of epigenetic silencing of active transposable elements is triggered by RDR6 and 21-22 nucleotide small interfering RNAs. Plant Physiol. 2013, 162, 116–131. [Google Scholar] [CrossRef]
- Trasser, M.; Bohl-Viallefond, G.; Barragán-Borrero, V.; Diezma-Navas, L.; Loncsek, L.; Nordborg, M.; Marí-Ordóñez, A. PTGS is dispensable for the initiation of epigenetic silencing of an active transposon in Arabidopsis. EMBO Rep. 2024, 25, 5780–5809. [Google Scholar] [CrossRef]
- Leone, M.; Zavallo, D.; Venturuzzi, A.; Asurmendi, S. RdDM pathway components differentially modulate Tobamovirus symptom development. Plant Mol. Biol. 2020, 104, 467–481. [Google Scholar] [CrossRef]
- Liu, Q.; Zhao, C.; Sun, K.; Deng, Y.; Li, Z. Engineered biocontainable RNA virus vectors for non-transgenic genome editing across crop species and genotypes. Mol. Plant 2023, 16, 616–631. [Google Scholar] [CrossRef]
- Hirochika, H.; Sugimoto, K.; Otsuki, Y.; Tsugawa, H.; Kanda, M. Retrotransposons of rice involved in mutations induced by tissue culture. Proc. Natl. Acad. Sci. USA 1996, 93, 7783–7788. [Google Scholar] [CrossRef]
- Hirochika, H. Activation of tobacco retrotransposons during tissue culture. EMBO J. 1993, 12, 2521–2528. [Google Scholar] [CrossRef]
- Okamoto, H.; Hirochika, H. Efficient insertion mutagenesis of Arabidopsis by tissue culture-induced activation of the tobacco retrotransposon Tto1. Plant J. 2000, 23, 291–304. [Google Scholar] [CrossRef]
- Brestovitsky, A.; Iwasaki, M.; Cho, J.; Adulyanukosol, N.; Paszkowski, J.; Catoni, M. Specific suppression of long terminal repeat retrotransposon mobilization in plants. Plant Physiol. 2023, 191, 2245–2255. [Google Scholar] [CrossRef]
- Weiss, T.; Kamalu, M.; Shi, H.; Li, Z.; Amerasekera, J.; Zhong, Z.; Adler, B.A.; Song, M.M.; Vohra, K.; Wirnowski, G.; et al. Viral delivery of an RNA-guided genome editor for transgene-free germline editing in Arabidopsis. Nat. Plants 2025, 11, 967–976. [Google Scholar] [CrossRef]
- Qiao, J.-H.; Zang, Y.; Gao, Q.; Liu, S.; Zhang, X.-W.; Hu, W.; Wang, Y.; Han, C.; Li, D.; Wang, X.-B. Transgene- and tissue culture-free heritable genome editing using RNA virus-based delivery in wheat. Nat. Plants 2025, 11, 1252–1259. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Ishikawa, M.; Toki, S.; Ishibashi, K. Heritable tissue-culture-free gene editing in Nicotiana benthamiana through viral delivery of SpCas9 and sgRNA. Plant Cell Physiol. 2024, 65, 1743–1750. [Google Scholar] [CrossRef]
- Merkulov, P.; Serganova, M.; Petrov, G.; Mityukov, V.; Kirov, I. Long-read sequencing of extrachromosomal circular DNA and genome assembly of a Solanum lycopersicum breeding line revealed active LTR retrotransposons originating from S. Peruvianum L. introgressions. BMC Genom. 2024, 25, 404. [Google Scholar] [CrossRef]
- Cheng, C.-Y.; Krishnakumar, V.; Chan, A.P.; Thibaud-Nissen, F.; Schobel, S.; Town, C.D. Araport11: A complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 2017, 89, 789–804. [Google Scholar] [CrossRef] [PubMed]
- Rousseau-Gueutin, M.; Belser, C.; Da Silva, C.; Richard, G.; Istace, B.; Cruaud, C.; Falentin, C.; Boideau, F.; Boutte, J.; Delourme, R.; et al. Long-read assembly of the Brassica napus reference genome Darmor-bzh. Gigascience 2020, 9, giaa137. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Guo, L. Data Used in ‘The Complete Genome Assembly of Nicotiana benthamiana Reveals Genetic and Epigenetic Landscape of Centromeres’. 2024. Available online: https://zenodo.org/records/14010728 (accessed on 11 November 2025). [CrossRef]
- Chen, W.; Yan, M.; Chen, S.; Sun, J.; Wang, J.; Meng, D.; Li, J.; Zhang, L.; Guo, L. The complete genome assembly of Nicotiana benthamiana reveals the genetic and epigenetic landscape of centromeres. Nat. Plants 2024, 10, 1928–1943. [Google Scholar] [CrossRef]
- Fernandez-Pozo, N.; Menda, N.; Edwards, J.D.; Saha, S.; Tecle, I.Y.; Strickler, S.R.; Bombarely, A.; Fisher-York, T.; Pujar, A.; Foerster, H.; et al. The Sol Genomics Network (SGN)--from genotype to phenotype to breeding. Nucleic Acids Res. 2015, 43, D1036–D1041. [Google Scholar] [CrossRef]
- Shumate, A.; Salzberg, S.L. Liftoff: Accurate mapping of gene annotations. Bioinformatics 2021, 37, 1639–1643. [Google Scholar] [CrossRef] [PubMed]
- Novák, P.; Hoštáková, N.; Neumann, P.; Macas, J. DANTE and DANTE_LTR: Lineage-centric annotation pipelines for long terminal repeat retrotransposons in plant genomes. NAR Genom. Bioinform. 2024, 6, lqae113. [Google Scholar] [CrossRef]
- Neumann, P.; Novák, P.; Hoštáková, N.; Macas, J. Systematic survey of plant LTR-retrotransposons elucidates phylogenetic relationships of their polyprotein domains and provides a reference for element classification. Mob. DNA 2019, 10, 1. [Google Scholar] [CrossRef]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data Processing, S. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Jalili, V.; Afgan, E.; Gu, Q.; Clements, D.; Blankenberg, D.; Goecks, J.; Taylor, J.; Nekrutenko, A. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2020 update. Nucleic Acids Res. 2020, 48, W395–W402, Erratum in Nucleic Acids Res. 2020, 48, 8205–8207. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef] [PubMed]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental algorithms for scientific computing in Python. Nat. Methods 2020, 17, 261–272, Correction in Nat. Methods 2020, 17, 352. [Google Scholar] [CrossRef] [PubMed]
- Seabold, S.; Perktold, J. Statsmodels: Econometric and statistical modeling with python. In Proceedings of the Python in Science Conference 2010, Austin, TX, USA, 28 June–3 July 2010; pp. 92–96. [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Cock, P.J.A.; Antao, T.; Chang, J.T.; Chapman, B.A.; Cox, C.J.; Dalke, A.; Friedberg, I.; Hamelryck, T.; Kauff, F.; Wilczynski, B.; et al. Biopython: Freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 2009, 25, 1422–1423. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Tian, F.; Yang, D.-C.; Meng, Y.-Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef]
- Buels, R.; Yao, E.; Diesh, C.M.; Hayes, R.D.; Munoz-Torres, M.; Helt, G.; Goodstein, D.M.; Elsik, C.G.; Lewis, S.E.; Stein, L.; et al. JBrowse: A dynamic web platform for genome visualization and analysis. Genome Biol. 2016, 17, 66. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534, Corrigendum in Mol. Biol. Evol. 2020, 37, 2461. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2. Use R! 2016; Springer: Cham, Switzerland, 2016. [Google Scholar] [CrossRef]
- Hunter, J.D. Matplotlib: A 2D Graphics Environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]
- Zhang, D.; Dey, R.; Lee, S. Fast and robust ancestry prediction using principal component analysis. Bioinformatics 2020, 36, 3439–3446. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Fang, L.; Wei, X.-Y.; Liu, L.-Z.; Zhou, L.-X.; Tian, Y.-P.; Geng, C.; Li, X.-D. A tobacco ringspot virus-based vector system for gene and microRNA function studies in cucurbits. Plant Physiol. 2021, 186, 853–864, Erratum in Plant Physiol. 2021, 3, 1835. [Google Scholar] [CrossRef]
- Uranga, M.; Aragonés, V.; García, A.; Mirabel, S.; Gianoglio, S.; Presa, S.; Granell, A.; Pasin, F.; Daròs, J.-A. RNA virus-mediated gene editing for tomato trait breeding. Hortic. Res. 2024, 11, uhad279. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Merkulov, P.; Bolotina, A.; Vlasova, A.; Ivakhnenko, A.; Prokofeva, A.; Perevozchikov, D.; Kamarauli, E.; Soloviev, A.; Kirov, I. Positive-Strand RNA Viruses Induce LTR Retrotransposon Transcription and Extrachromosomal Circular DNA Generation in Plants. Int. J. Mol. Sci. 2026, 27, 286. https://doi.org/10.3390/ijms27010286
Merkulov P, Bolotina A, Vlasova A, Ivakhnenko A, Prokofeva A, Perevozchikov D, Kamarauli E, Soloviev A, Kirov I. Positive-Strand RNA Viruses Induce LTR Retrotransposon Transcription and Extrachromosomal Circular DNA Generation in Plants. International Journal of Molecular Sciences. 2026; 27(1):286. https://doi.org/10.3390/ijms27010286
Chicago/Turabian StyleMerkulov, Pavel, Anna Bolotina, Anastasia Vlasova, Anna Ivakhnenko, Alena Prokofeva, Danil Perevozchikov, Elizaveta Kamarauli, Alexander Soloviev, and Ilya Kirov. 2026. "Positive-Strand RNA Viruses Induce LTR Retrotransposon Transcription and Extrachromosomal Circular DNA Generation in Plants" International Journal of Molecular Sciences 27, no. 1: 286. https://doi.org/10.3390/ijms27010286
APA StyleMerkulov, P., Bolotina, A., Vlasova, A., Ivakhnenko, A., Prokofeva, A., Perevozchikov, D., Kamarauli, E., Soloviev, A., & Kirov, I. (2026). Positive-Strand RNA Viruses Induce LTR Retrotransposon Transcription and Extrachromosomal Circular DNA Generation in Plants. International Journal of Molecular Sciences, 27(1), 286. https://doi.org/10.3390/ijms27010286







