Control of Gene Expression by Proteins That Bind Many Alternative Nucleic Acid Structures Through the Same Domain
Abstract
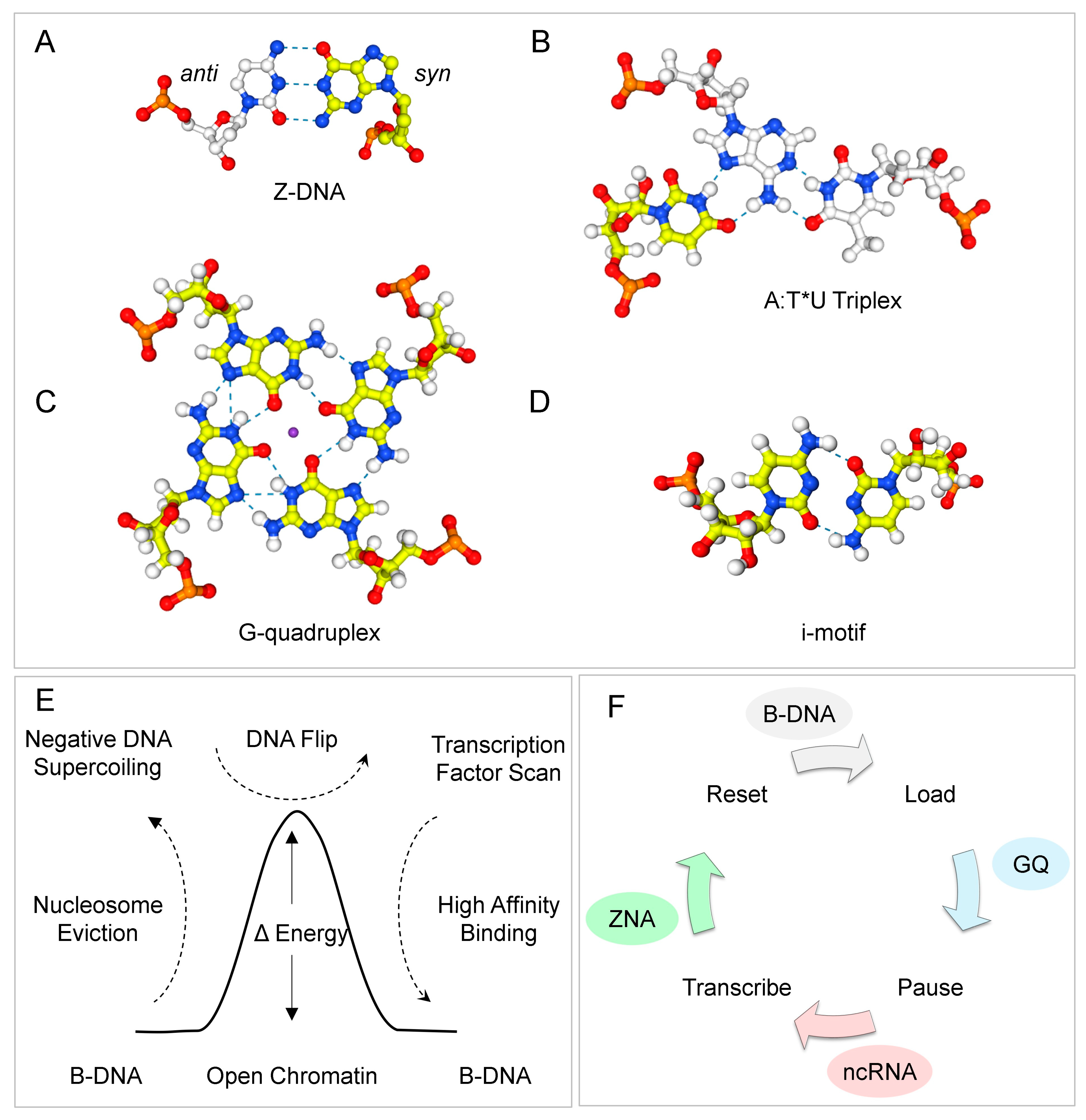
1. Introduction
2. Results
3. Discussion
4. Methods
5. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Felsenfeld, G.; Davies, D.R.; Rich, A. Formation of a Three-Stranded Polynucleotide Molecule. J. Am. Chem. Soc. 1957, 79, 2023–2024. [Google Scholar] [CrossRef]
- Hoogsteen, K. The structure of crystals containing a hydrogen-bonded complex of 1-methylthymine and 9-methyladenine. Acta Crystallogr. 1959, 12, 822–823. [Google Scholar] [CrossRef]
- Sokoloski, J.E.; Godfrey, S.A.; Dombrowski, S.E.; Bevilacqua, P.C. Prevalence of syn nucleobases in the active sites of functional RNAs. RNA 2011, 17, 1775–1787. [Google Scholar] [CrossRef]
- Ellison, M.J.; Kelleher, R.J., 3rd; Wang, A.H.; Habener, J.F.; Rich, A. Sequence-dependent energetics of the B-Z transition in supercoiled DNA containing nonalternating purine-pyrimidine sequences. Proc. Natl. Acad. Sci. USA 1985, 82, 8320–8324. [Google Scholar] [CrossRef]
- Lightfoot, H.L.; Hagen, T.; Tatum, N.J.; Hall, J. The diverse structural landscape of quadruplexes. FEBS Lett. 2019, 593, 2083–2102. [Google Scholar] [CrossRef]
- Frank-Kamenetskii, M.D.; Mirkin, S.M. Triplex DNA structures. Annu. Rev. Biochem. 1995, 64, 65–95. [Google Scholar] [CrossRef]
- Westin, L.; Blomquist, P.; Milligan, J.F.; Wrange, O. Triple helix DNA alters nucleosomal histone-DNA interactions and acts as a nucleosome barrier. Nucleic Acids Res. 1995, 23, 2184–2191. [Google Scholar] [CrossRef]
- Dagneaux, C.; Gousset, H.; Shchyolkina, A.K.; Ouali, M.; Letellier, R.; Liquier, J.; Florentiev, V.L.; Taillandier, E. Parallel and antiparallel A*A-T intramolecular triple helices. Nucleic Acids Res. 1996, 24, 4506–4512. [Google Scholar] [CrossRef] [PubMed]
- Gehring, K.; Leroy, J.L.; Gueron, M. A tetrameric DNA structure with protonated cytosine.cytosine base pairs. Nature 1993, 363, 561–565. [Google Scholar] [CrossRef] [PubMed]
- Herbert, A. Z-DNA and Z-RNA: Methods-Past and Future. Methods Mol. Biol. 2023, 2651, 295–329. [Google Scholar]
- Herbert, A.; Alfken, J.; Kim, Y.G.; Mian, I.S.; Nishikura, K.; Rich, A. A Z-DNA binding domain present in the human editing enzyme, double-stranded RNA adenosine deaminase. Proc. Natl. Acad. Sci. USA 1997, 94, 8421–8426. [Google Scholar] [CrossRef]
- Schade, M.; Turner, C.J.; Lowenhaupt, K.; Rich, A.; Herbert, A. Structure-function analysis of the Z-DNA-binding domain Zα of dsRNA adenosine deaminase type I reveals similarity to the (alpha + beta) family of helix-turn-helix proteins. EMBO J. 1999, 18, 470–479. [Google Scholar] [CrossRef]
- Schwartz, T.; Rould, M.A.; Lowenhaupt, K.; Herbert, A.; Rich, A. Crystal structure of the Zα domain of the human editing enzyme ADAR1 bound to left-handed Z-DNA. Science 1999, 284, 1841–1845. [Google Scholar] [CrossRef] [PubMed]
- Herbert, A. Mendelian disease caused by variants affecting recognition of Z-DNA and Z-RNA by the Zα domain of the double-stranded RNA editing enzyme ADAR. Eur. J. Hum. Genet. 2020, 28, 114–117. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Liu, S.; Sheng, Y.; Zenati, M.; Billiar, T.; Herbert, A.; Wang, Q. ADAR1 Zalpha domain P195A mutation activates the MDA5-dependent RNA-sensing signaling pathway in brain without decreasing overall RNA editing. Cell Rep. 2023, 42, 112733. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Chalk, A.M.; Taylor, S.; Goradia, A.; Heraud-Farlow, J.E.; Walkley, C.R. The phenotype of the most common human ADAR1p150 Zalpha mutation P193A in mice is partially penetrant. EMBO Rep. 2023, 24, e55835. [Google Scholar] [CrossRef]
- Kaiser, W.J.; Upton, J.W.; Mocarski, E.S. Receptor-interacting protein homotypic interaction motif-dependent control of NF-kappa B activation via the DNA-dependent activator of IFN regulatory factors. J. Immunol. 2008, 181, 6427–6434. [Google Scholar] [CrossRef]
- Upton, J.W.; Kaiser, W.J.; Mocarski, E.S. DAI/ZBP1/DLM-1 complexes with RIP3 to mediate virus-induced programmed necrosis that is targeted by murine cytomegalovirus vIRA. Cell Host Microbe 2012, 11, 290–297. [Google Scholar] [CrossRef]
- Lin, J.; Kumari, S.; Kim, C.; Van, T.M.; Wachsmuth, L.; Polykratis, A.; Pasparakis, M. RIPK1 counteracts ZBP1-mediated necroptosis to inhibit inflammation. Nature 2016, 540, 124–128. [Google Scholar] [CrossRef]
- Galluzzi, L.; Vitale, I.; Aaronson, S.A.; Abrams, J.M.; Adam, D.; Agostinis, P.; Alnemri, E.S.; Altucci, L.; Amelio, I.; Andrews, D.W.; et al. Molecular mechanisms of cell death: Recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018, 25, 486–541. [Google Scholar] [CrossRef]
- Devos, M.; Tanghe, G.; Gilbert, B.; Dierick, E.; Verheirstraeten, M.; Nemegeer, J.; de Reuver, R.; Lefebvre, S.; De Munck, J.; Rehwinkel, J.; et al. Sensing of endogenous nucleic acids by ZBP1 induces keratinocyte necroptosis and skin inflammation. J. Exp. Med. 2020, 217, e20191913. [Google Scholar] [CrossRef]
- Zhang, T.; Yin, C.; Boyd, D.F.; Quarato, G.; Ingram, J.P.; Shubina, M.; Ragan, K.B.; Ishizuka, T.; Crawford, J.C.; Tummers, B.; et al. Influenza Virus Z-RNAs Induce ZBP1-Mediated Necroptosis. Cell 2020, 180, 1115–1129. [Google Scholar] [CrossRef]
- Nakahama, T.; Kato, Y.; Shibuya, T.; Inoue, M.; Kim, J.I.; Vongpipatana, T.; Todo, H.; Xing, Y.; Kawahara, Y. Mutations in the adenosine deaminase ADAR1 that prevent endogenous Z-RNA binding induce Aicardi-Goutieres-syndrome-like encephalopathy. Immunity 2021, 54, 1976–1988.e7. [Google Scholar] [CrossRef]
- de Reuver, R.; Verdonck, S.; Dierick, E.; Nemegeer, J.; Hessmann, E.; Ahmad, S.; Jans, M.; Blancke, G.; Van Nieuwerburgh, F.; Botzki, A.; et al. ADAR1 prevents autoinflammation by suppressing spontaneous ZBP1 activation. Nature 2022, 607, 784–789. [Google Scholar] [CrossRef] [PubMed]
- Herbert, A.; Fedorov, A.; Poptsova, M. Mono a Mano: ZBP1’s Love-Hate Relationship with the Kissing Virus. Int. J. Mol. Sci. 2022, 23, 3079. [Google Scholar] [CrossRef]
- Hubbard, N.W.; Ames, J.M.; Maurano, M.; Chu, L.H.; Somfleth, K.Y.; Gokhale, N.S.; Werner, M.; Snyder, J.M.; Lichauco, K.; Savan, R.; et al. ADAR1 mutation causes ZBP1-dependent immunopathology. Nature 2022, 607, 769–775. [Google Scholar] [CrossRef] [PubMed]
- Jiao, H.; Wachsmuth, L.; Wolf, S.; Lohmann, J.; Nagata, M.; Kaya, G.G.; Oikonomou, N.; Kondylis, V.; Rogg, M.; Diebold, M.; et al. ADAR1 averts fatal type I interferon induction by ZBP1. Nature 2022, 607, 776–783. [Google Scholar] [CrossRef]
- Zhang, T.; Yin, C.; Fedorov, A.; Qiao, L.; Bao, H.; Beknazarov, N.; Wang, S.; Gautam, A.; Williams, R.M.; Crawford, J.C.; et al. ADAR1 masks the cancer immunotherapeutic promise of ZBP1-driven necroptosis. Nature 2022, 606, 594–602. [Google Scholar] [CrossRef]
- Yin, C.; Fedorov, A.; Guo, H.; Crawford, J.C.; Rousseau, C.; Zhong, X.; Williams, R.M.; Gautam, A.; Koehler, H.S.; Whisnant, A.W.; et al. Host cell Z-RNAs activate ZBP1 during virus infections. Nature 2025, 648, 707–716. [Google Scholar] [CrossRef]
- Beknazarov, N.; Jin, S.; Poptsova, M. Deep learning approach for predicting functional Z-DNA regions using omics data. Sci. Rep. 2020, 10, 19134. [Google Scholar] [CrossRef] [PubMed]
- Georgakopoulos-Soares, I.; Victorino, J.; Parada, G.E.; Agarwal, V.; Zhao, J.; Wong, H.Y.; Umar, M.I.; Elor, O.; Muhwezi, A.; An, J.Y.; et al. High-throughput characterization of the role of non-B DNA motifs on promoter function. Cell Genom. 2022, 2, 100111. [Google Scholar] [CrossRef]
- Umerenkov, D.; Herbert, A.; Konovalov, D.; Danilova, A.; Beknazarov, N.; Kokh, V.; Fedorov, A.; Poptsova, M. Z-flipon variants reveal the many roles of Z-DNA and Z-RNA in health and disease. Life Sci. Alliance 2023, 6, e202301962. [Google Scholar] [CrossRef]
- Beknazarov, N.; Konovalov, D.; Herbert, A.; Poptsova, M. Z-DNA formation in promoters conserved between human and mouse are associated with increased transcription reinitiation rates. Sci. Rep. 2024, 14, 17786. [Google Scholar] [CrossRef]
- Smeds, L.; Kamali, K.; Kejnovska, I.; Kejnovsky, E.; Chiaromonte, F.; Makova, K.D. Non-canonical DNA in human and other ape telomere-to-telomere genomes. Nucleic Acids Res. 2025, 53, gkaf298. [Google Scholar] [CrossRef] [PubMed]
- Chantzi, N.; Nayak, A.; Baltoumas, F.A.; Aplakidou, E.; Liew, S.W.; Galuh, J.E.; Patsakis, M.; Montgomery, A.; Moeckel, C.; Mouratidis, I.; et al. Quadrupia provides a comprehensive catalog of G-quadruplexes across genomes from the tree of life. Genome Res. 2025, 35, 2578–2600. [Google Scholar] [CrossRef] [PubMed]
- Maizels, N. G4-associated human diseases. EMBO Rep. 2015, 16, 910–922. [Google Scholar] [CrossRef]
- Sauer, M.; Paeschke, K. G-quadruplex unwinding helicases and their function in vivo. Biochem. Soc. Trans. 2017, 45, 1173–1182. [Google Scholar] [CrossRef]
- Spiegel, J.; Adhikari, S.; Balasubramanian, S. The Structure and Function of DNA G-Quadruplexes. Trends Chem. 2020, 2, 123–136. [Google Scholar] [CrossRef] [PubMed]
- Konig, P.; Giraldo, R.; Chapman, L.; Rhodes, D. The crystal structure of the DNA-binding domain of yeast RAP1 in complex with telomeric DNA. Cell 1996, 85, 125–136. [Google Scholar] [CrossRef]
- Traczyk, A.; Liew, C.W.; Gill, D.J.; Rhodes, D. Structural basis of G-quadruplex DNA recognition by the yeast telomeric protein Rap1. Nucleic Acids Res. 2020, 48, 4562–4571. [Google Scholar] [CrossRef] [PubMed]
- Lane, A.N.; Chaires, J.B.; Gray, R.D.; Trent, J.O. Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 2008, 36, 5482–5515. [Google Scholar] [CrossRef] [PubMed]
- Varshney, D.; Spiegel, J.; Zyner, K.; Tannahill, D.; Balasubramanian, S. The regulation and functions of DNA and RNA G-quadruplexes. Nat. Rev. Mol. Cell Biol. 2020, 21, 459–474. [Google Scholar] [CrossRef]
- Sato, K.; Knipscheer, P. G-quadruplex resolution: From molecular mechanisms to physiological relevance. DNA Repair. 2023, 130, 103552. [Google Scholar] [CrossRef]
- Herbert, A. A Compendium of G-Flipon Biological Functions That Have Experimental Validation. Int. J. Mol. Sci. 2024, 25, 10299. [Google Scholar] [CrossRef] [PubMed]
- Herbert, A. The Evolutionary Entanglement of Flipons with Zinc Fingers and Retroelements has Engendered a Large Family of Z-DNA and G-Quadruplex Binding Proteins. Open Biol. 2025, 15, 250171. [Google Scholar] [CrossRef]
- Meng, Y.; Wang, G.; He, H.; Lau, K.H.; Hurt, A.; Bixler, B.J.; Parham, A.; Jin, S.G.; Xu, X.; Vasquez, K.M.; et al. Z-DNA is remodelled by ZBTB43 in prospermatogonia to safeguard the germline genome and epigenome. Nat. Cell Biol. 2022, 24, 1141–1153. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, R.; Nady, N.; Arora, N.; Hsieh, L.J.; Swigut, T.; Narlikar, G.J.; Wossidlo, M.; Wysocka, J. Zscan4 binds nucleosomal microsatellite DNA and protects mouse two-cell embryos from DNA damage. Sci. Adv. 2020, 6, eaaz9115. [Google Scholar] [CrossRef]
- Iwahara, J.; Levy, Y. Speed-stability paradox in DNA-scanning by zinc-finger proteins. Transcription 2013, 4, 58–61. [Google Scholar]
- Herbert, A. Flipons and the origin of the genetic code. Biol. Lett. 2025, 21, 20240635. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef]
- Kerkhoff, E.; Bister, K.; Klempnauer, K.H. Sequence-specific DNA binding by Myc proteins. Proc. Natl. Acad. Sci. USA 1991, 88, 4323–4327. [Google Scholar] [CrossRef]
- Carroll, P.A.; Freie, B.W.; Mathsyaraja, H.; Eisenman, R.N. The MYC transcription factor network: Balancing metabolism, proliferation and oncogenesis. Front. Med. 2018, 12, 412–425. [Google Scholar] [CrossRef] [PubMed]
- Evans, P.M.; Zhang, W.; Chen, X.; Yang, J.; Bhakat, K.K.; Liu, C. Kruppel-like factor 4 is acetylated by p300 and regulates gene transcription via modulation of histone acetylation. J. Biol. Chem. 2007, 282, 33994–34002. [Google Scholar] [CrossRef] [PubMed]
- Di Giammartino, D.C.; Kloetgen, A.; Polyzos, A.; Liu, Y.; Kim, D.; Murphy, D.; Abuhashem, A.; Cavaliere, P.; Aronson, B.; Shah, V.; et al. KLF4 is involved in the organization and regulation of pluripotency-associated three-dimensional enhancer networks. Nat. Cell Biol. 2019, 21, 1179–1190. [Google Scholar] [CrossRef]
- Soufi, A.; Donahue, G.; Zaret, K.S. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell 2012, 151, 994–1004. [Google Scholar] [CrossRef]
- Petryniak, B.; Staudt, L.M.; Postema, C.E.; McCormack, W.T.; Thompson, C.B. Characterization of chicken octamer-binding proteins demonstrates that POU domain-containing homeobox transcription factors have been highly conserved during vertebrate evolution. Proc. Natl. Acad. Sci. USA 1990, 87, 1099–1103. [Google Scholar] [CrossRef]
- Williams, D.C., Jr.; Cai, M.; Clore, G.M. Molecular basis for synergistic transcriptional activation by Oct1 and Sox2 revealed from the solution structure of the 42-kDa Oct1.Sox2.Hoxb1-DNA ternary transcription factor complex. J. Biol. Chem. 2004, 279, 1449–1457. [Google Scholar] [CrossRef]
- Guan, R.; Lian, T.; Zhou, B.R.; Wheeler, D.; Bai, Y. Structural mechanism of LIN28B nucleosome targeting by OCT4. Mol. Cell 2023, 83, 1970–1982.e6. [Google Scholar] [CrossRef]
- Spiegel, J.; Cuesta, S.M.; Adhikari, S.; Hansel-Hertsch, R.; Tannahill, D.; Balasubramanian, S. G-quadruplexes are transcription factor binding hubs in human chromatin. Genome Biol. 2021, 22, 117. [Google Scholar] [CrossRef]
- Li, S.; Wang, Z.; Wang, X.; Wang, Y.; Pattarayan, D.; Zhang, Y.; Nguyen, P.; Bhuniya, A.; Chen, Y.; Huang, H.; et al. Integrative characterization of MYC RNA-binding function. Cell Genom. 2025, 5, 100878. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, D.J.; Hein, A.E.; Holmes, Z.E.; Wuttke, D.S.; Batey, R.T. The DNA-Binding High-Mobility Group Box Domain of Sox Family Proteins Directly Interacts with RNA In Vitro. Biochemistry 2022, 61, 943–951. [Google Scholar] [CrossRef]
- Tikhanovich, I.; Zhao, J.; Bridges, B.; Kumer, S.; Roberts, B.; Weinman, S.A. Arginine methylation regulates c-Myc-dependent transcription by altering promoter recruitment of the acetyltransferase p300. J. Biol. Chem. 2017, 292, 13333–13344. [Google Scholar] [CrossRef]
- Zhang, L.; Shao, G.; Shao, J.; Zhao, J. PRMT5-activated c-Myc promote bladder cancer proliferation and invasion through up-regulating NF-kappaB pathway. Tissue Cell 2022, 76, 101788, Erratum in Tissue Cell 2023, 85, 102221. [Google Scholar] [CrossRef]
- Kumar, P.; Joy, J.; Pandey, A.; Gupta, D. PRmePRed: A protein arginine methylation prediction tool. PLoS ONE 2017, 12, e0183318. [Google Scholar] [CrossRef] [PubMed]
- Behe, M.; Felsenfeld, G. Effects of methylation on a synthetic polynucleotide: The B--Z transition in poly(dG-m5dC).poly(dG-m5dC). Proc. Natl. Acad. Sci. USA 1981, 78, 1619–1623. [Google Scholar] [CrossRef]
- Fujii, S.; Wang, A.H.; van der Marel, G.; van Boom, J.H.; Rich, A. Molecular structure of (m5 dC-dG)3: The role of the methyl group on 5-methyl cytosine in stabilizing Z-DNA. Nucleic Acids Res. 1982, 10, 7879–7892. [Google Scholar] [CrossRef]
- Tan, D.S.; Cheung, S.L.; Gao, Y.; Weinbuch, M.; Hu, H.; Shi, L.; Ti, S.C.; Hutchins, A.P.; Cojocaru, V.; Jauch, R. The homeodomain of Oct4 is a dimeric binder of methylated CpG elements. Nucleic Acids Res. 2023, 51, 1120–1138. [Google Scholar] [CrossRef]
- Wardle, F.C. Master control: Transcriptional regulation of mammalian Myod. J. Muscle Res. Cell Motil. 2019, 40, 211–226. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Yao, Z.; Sarkar, D.; Lawrence, M.; Sanchez, G.J.; Parker, M.H.; MacQuarrie, K.L.; Davison, J.; Morgan, M.T.; Ruzzo, W.L.; et al. Genome-wide MyoD binding in skeletal muscle cells: A potential for broad cellular reprogramming. Dev. Cell 2010, 18, 662–674. [Google Scholar] [CrossRef] [PubMed]
- Kodandapani, R.; Pio, F.; Ni, C.Z.; Piccialli, G.; Klemsz, M.; McKercher, S.; Maki, R.A.; Ely, K.R. A new pattern for helix-turn-helix recognition revealed by the PU.1 ETS-domain-DNA complex. Nature 1996, 380, 456–460. [Google Scholar] [CrossRef]
- Donaldson, L.W.; Petersen, J.M.; Graves, B.J.; McIntosh, L.P. Solution structure of the ETS domain from murine Ets-1: A winged helix-turn-helix DNA binding motif. EMBO J. 1996, 15, 125–134. [Google Scholar] [CrossRef]
- Gutierrez-Hartmann, A.; Duval, D.L.; Bradford, A.P. ETS transcription factors in endocrine systems. Trends Endocrinol. Metab. 2007, 18, 150–158. [Google Scholar] [CrossRef]
- Riege, K.; Kretzmer, H.; Sahm, A.; McDade, S.S.; Hoffmann, S.; Fischer, M. Dissecting the DNA binding landscape and gene regulatory network of p63 and p53. eLife 2020, 9, e63266. [Google Scholar] [CrossRef] [PubMed]
- Hamard, P.J.; Lukin, D.J.; Manfredi, J.J. p53 basic C terminus regulates p53 functions through DNA binding modulation of subset of target genes. J. Biol. Chem. 2012, 287, 22397–22407. [Google Scholar] [CrossRef]
- Vierstra, J.; Lazar, J.; Sandstrom, R.; Halow, J.; Lee, K.; Bates, D.; Diegel, M.; Dunn, D.; Neri, F.; Haugen, E.; et al. Global reference mapping of human transcription factor footprints. Nature 2020, 583, 729–736. [Google Scholar] [CrossRef] [PubMed]
- Rushmore, T.H.; Morton, M.R.; Pickett, C.B. The antioxidant responsive element. Activation by oxidative stress and identification of the DNA consensus sequence required for functional activity. J. Biol. Chem. 1991, 266, 11632–11639. [Google Scholar] [CrossRef] [PubMed]
- Itoh, K.; Chiba, T.; Takahashi, S.; Ishii, T.; Igarashi, K.; Katoh, Y.; Oyake, T.; Hayashi, N.; Satoh, K.; Hatayama, I.; et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 1997, 236, 313–322. [Google Scholar] [CrossRef]
- Katsuoka, F.; Motohashi, H.; Ishii, T.; Aburatani, H.; Engel, J.D.; Yamamoto, M. Genetic evidence that small maf proteins are essential for the activation of antioxidant response element-dependent genes. Mol. Cell Biol. 2005, 25, 8044–8051. [Google Scholar] [CrossRef]
- Dhakshinamoorthy, S.; Jain, A.K.; Bloom, D.A.; Jaiswal, A.K. Bach1 competes with Nrf2 leading to negative regulation of the antioxidant response element (ARE)-mediated NAD(P)H:quinone oxidoreductase 1 gene expression and induction in response to antioxidants. J. Biol. Chem. 2005, 280, 16891–16900. [Google Scholar] [CrossRef]
- Kahmann, J.D.; Wecking, D.A.; Putter, V.; Lowenhaupt, K.; Kim, Y.G.; Schmieder, P.; Oschkinat, H.; Rich, A.; Schade, M. The solution structure of the N-terminal domain of E3L shows a tyrosine conformation that may explain its reduced affinity to Z-DNA in vitro. Proc. Natl. Acad. Sci. USA 2004, 101, 2712–2717. [Google Scholar] [CrossRef]
- Santella, R.M.; Grunberger, D.; Broyde, S.; Hingerty, B.E. Z-DNA conformation of N-2-acetylaminofluorene modified poly(dG-dC).poly(dG-dC) determined by reactivity with anti cytidine antibodies and minimized potential energy calculations. Nucleic Acids Res. 1981, 9, 5459–5467. [Google Scholar] [CrossRef]
- Moller, A.; Nordheim, A.; Kozlowski, S.A.; Patel, D.J.; Rich, A. Bromination stabilizes poly(dG-dC) in the Z-DNA form under low-salt conditions. Biochemistry 1984, 23, 54–62. [Google Scholar] [CrossRef]
- Wang, J.; Wang, S.; Zhong, C.; Tian, T.; Zhou, X. Novel insights into a major DNA oxidative lesion: Its effects on Z-DNA stabilization. Org. Biomol. Chem. 2015, 13, 8996–8999. [Google Scholar] [CrossRef] [PubMed]
- Radzisheuskaya, A.; Shliaha, P.V.; Grinev, V.; Lorenzini, E.; Kovalchuk, S.; Shlyueva, D.; Gorshkov, V.; Hendrickson, R.C.; Jensen, O.N.; Helin, K. PRMT5 methylome profiling uncovers a direct link to splicing regulation in acute myeloid leukemia. Nat. Struct. Mol. Biol. 2019, 26, 999–1012. [Google Scholar] [CrossRef] [PubMed]
- Cao, M.; Nguyen, T.; Song, J.; Zheng, Y.G. Biomedical effects of protein arginine methyltransferase inhibitors. J. Biol. Chem. 2025, 301, 108201. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhu, B.; Winn, R.; Lu, S.; Wang, H. Functional Dynamics of Arginine Mono- and Di-Methylation. Cells 2025, 14, 796. [Google Scholar] [CrossRef]
- Di Gioia, S.A.; Shaaban, S.; Tuysuz, B.; Elcioglu, N.H.; Chan, W.M.; Robson, C.D.; Ecklund, K.; Gilette, N.M.; Hamzaoglu, A.; Tayfun, G.A.; et al. Recessive MYF5 Mutations Cause External Ophthalmoplegia, Rib, and Vertebral Anomalies. Am. J. Hum. Genet. 2018, 103, 115–124. [Google Scholar] [CrossRef]
- Waddington, C.H. Canalization of Development and the Inheritance of Acquired Characters. Nature 1942, 150, 563–565. [Google Scholar] [CrossRef]
- Rodriguez-Martinez, J.A.; Reinke, A.W.; Bhimsaria, D.; Keating, A.E.; Ansari, A.Z. Combinatorial bZIP dimers display complex DNA-binding specificity landscapes. eLife 2017, 6, e19272. [Google Scholar] [CrossRef]
- Hai, T.; Curran, T. Cross-family dimerization of transcription factors Fos/Jun and ATF/CREB alters DNA binding specificity. Proc. Natl. Acad. Sci. USA 1991, 88, 3720–3724. [Google Scholar] [CrossRef]
- van Dam, H.; Castellazzi, M. Distinct roles of Jun: Fos and Jun: ATF dimers in oncogenesis. Oncogene 2001, 20, 2453–2464. [Google Scholar]
- Bejjani, F.; Tolza, C.; Boulanger, M.; Downes, D.; Romero, R.; Maqbool, M.A.; Zine El Aabidine, A.; Andrau, J.C.; Lebre, S.; Brehelin, L.; et al. Fra-1 regulates its target genes via binding to remote enhancers without exerting major control on chromatin architecture in triple negative breast cancers. Nucleic Acids Res. 2021, 49, 2488–2508. [Google Scholar] [CrossRef]
- Walsh, K.; Gualberto, A. MyoD binds to the guanine tetrad nucleic acid structure. J. Biol. Chem. 1992, 267, 13714–13718. [Google Scholar] [CrossRef]
- Yafe, A.; Shklover, J.; Weisman-Shomer, P.; Bengal, E.; Fry, M. Differential binding of quadruplex structures of muscle-specific genes regulatory sequences by MyoD, MRF4 and myogenin. Nucleic Acids Res. 2008, 36, 3916–3925. [Google Scholar] [CrossRef]
- Pipier, A.; Devaux, A.; Lavergne, T.; Adrait, A.; Coute, Y.; Britton, S.; Calsou, P.; Riou, J.F.; Defrancq, E.; Gomez, D. Constrained G4 structures unveil topology specificity of known and new G4 binding proteins. Sci. Rep. 2021, 11, 13469. [Google Scholar] [CrossRef]
- Meier-Stephenson, V. G4-quadruplex-binding proteins: Review and insights into selectivity. Biophys. Rev. 2022, 14, 635–654. [Google Scholar] [PubMed]
- Morange, M. What history tells us IX. Z-DNA: When nature is not opportunistic. J. Biosci. 2007, 32, 657–661. [Google Scholar] [CrossRef]
- Brazdova, M.; Tichy, V.; Helma, R.; Bazantova, P.; Polaskova, A.; Krejci, A.; Petr, M.; Navratilova, L.; Ticha, O.; Nejedly, K.; et al. p53 Specifically Binds Triplex DNA In Vitro and in Cells. PLoS ONE 2016, 11, e0167439. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Deng, W. Dynamic transcription regulation at the single-molecule level. Dev. Biol. 2022, 482, 67–81. [Google Scholar] [CrossRef] [PubMed]
- Choi, I.; Baek, I. Single-molecule imaging for investigating the transcriptional control. Mol. Cells 2025, 48, 100179. [Google Scholar] [CrossRef]
- Li, S.; Zheng, E.B.; Zhao, L.; Liu, S. Nonreciprocal and Conditional Cooperativity Directs the Pioneer Activity of Pluripotency Transcription Factors. Cell Rep. 2019, 28, 2689–2703.e4. [Google Scholar] [CrossRef]
- Chen, Y.; Tokuda, J.M.; Topping, T.; Sutton, J.L.; Meisburger, S.P.; Pabit, S.A.; Gloss, L.M.; Pollack, L. Revealing transient structures of nucleosomes as DNA unwinds. Nucleic Acids Res. 2014, 42, 8767–8776. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Tokuda, J.M.; Topping, T.; Meisburger, S.P.; Pabit, S.A.; Gloss, L.M.; Pollack, L. Asymmetric unwrapping of nucleosomal DNA propagates asymmetric opening and dissociation of the histone core. Proc. Natl. Acad. Sci. USA 2017, 114, 334–339. [Google Scholar] [CrossRef] [PubMed]
- Herbert, A. Nucleosomes and flipons exchange energy to alter chromatin conformation, the readout of genomic information, and cell fate. Bioessays 2022, 44, e2200166. [Google Scholar] [CrossRef]
- Herbert, A. Flipons and small RNAs accentuate the asymmetries of pervasive transcription by the reset and sequence-specific microcoding of promoter conformation. J. Biol. Chem. 2023, 299, 105140. [Google Scholar] [CrossRef]
- Rodriguez, J.; Ren, G.; Day, C.R.; Zhao, K.; Chow, C.C.; Larson, D.R. Intrinsic Dynamics of a Human Gene Reveal the Basis of Expression Heterogeneity. Cell 2019, 176, 213–226.e18. [Google Scholar] [CrossRef]
- Popp, A.P.; Hettich, J.; Gebhardt, J.C.M. Altering transcription factor binding reveals comprehensive transcriptional kinetics of a basic gene. Nucleic Acids Res. 2021, 49, 6249–6266. [Google Scholar] [CrossRef]
- Darzacq, X.; Shav-Tal, Y.; de Turris, V.; Brody, Y.; Shenoy, S.M.; Phair, R.D.; Singer, R.H. In vivo dynamics of RNA polymerase II transcription. Nat. Struct. Mol. Biol. 2007, 14, 796–806. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, J.; Larson, D.R. Transcription in Living Cells: Molecular Mechanisms of Bursting. Annu. Rev. Biochem. 2020, 89, 189–212. [Google Scholar] [CrossRef]
- Le, S.N.; Brown, C.R.; Harvey, S.; Boeger, H.; Elmlund, H.; Elmlund, D. The TAFs of TFIID Bind and Rearrange the Topology of the TATA-Less RPS5 Promoter. Int. J. Mol. Sci. 2019, 20, 3290. [Google Scholar] [CrossRef]
- Herbert, A. The ancient Z-DNA and Z-RNA specific Zalpha fold has evolved modern roles in immunity and transcription through the natural selection of flipons. R. Soc. Open Sci. 2024, 11, 240080. [Google Scholar] [CrossRef]
- Mayer, A.; di Iulio, J.; Maleri, S.; Eser, U.; Vierstra, J.; Reynolds, A.; Sandstrom, R.; Stamatoyannopoulos, J.A.; Churchman, L.S. Native elongating transcript sequencing reveals human transcriptional activity at nucleotide resolution. Cell 2015, 161, 541–554. [Google Scholar] [CrossRef] [PubMed]
- Ramaker, R.C.; Hardigan, A.A.; Goh, S.T.; Partridge, E.C.; Wold, B.; Cooper, S.J.; Myers, R.M. Dissecting the regulatory activity and sequence content of loci with exceptional numbers of transcription factor associations. Genome Res. 2020, 30, 939–950. [Google Scholar] [CrossRef]
- Hemphill, W.O.; Fenske, R.; Gooding, A.R.; Cech, T.R. PRC2 direct transfer from G-quadruplex RNA to dsDNA has implications for RNA-binding chromatin modifiers. Proc. Natl. Acad. Sci. USA 2023, 120, e2220528120. [Google Scholar] [CrossRef]
- Frankle, J.; Carbin, M. The Lottery Ticket Hypothesis: Finding Sparse, Trainable Neural Networks. arXiv 2019, arXiv:1803.03635. [Google Scholar] [CrossRef]
- Herbert, A.; Cherednichenko, O.; Lybrand, T.P.; Egli, M.; Poptsova, M. Zalpha and Zbeta Localize ADAR1 to Flipons That Modulate Innate Immunity, Alternative Splicing, and Nonsynonymous RNA Editing. Int. J. Mol. Sci. 2025, 26, 2422. [Google Scholar] [CrossRef] [PubMed]
- Kroft, C.W.; Krall, J.B.; Warchol, M.; Welty, R.; Herbert, A.; Henen, M.A.; Vogeli, B. Zα and Zβ domains of ADAR1 and ZBP1 bind to G-quadruplexes with low micromolar affinity. bioRxiv 2025. [Google Scholar] [CrossRef]
- Schade, M.; Turner, C.J.; Kuhne, R.; Schmieder, P.; Lowenhaupt, K.; Herbert, A.; Rich, A.; Oschkinat, H. The solution structure of the Zα domain of the human RNA editing enzyme ADAR1 reveals a prepositioned binding surface for Z-DNA. Proc. Natl. Acad. Sci. USA 1999, 96, 12465–12470. [Google Scholar] [CrossRef] [PubMed]
- Herbert, A. Flipons enable genomes to learn by intermediating the exchange of energy for information. J. R. Soc. Interface 2025, 22, 20250049. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Herbert, A. Control of Gene Expression by Proteins That Bind Many Alternative Nucleic Acid Structures Through the Same Domain. Int. J. Mol. Sci. 2026, 27, 272. https://doi.org/10.3390/ijms27010272
Herbert A. Control of Gene Expression by Proteins That Bind Many Alternative Nucleic Acid Structures Through the Same Domain. International Journal of Molecular Sciences. 2026; 27(1):272. https://doi.org/10.3390/ijms27010272
Chicago/Turabian StyleHerbert, Alan. 2026. "Control of Gene Expression by Proteins That Bind Many Alternative Nucleic Acid Structures Through the Same Domain" International Journal of Molecular Sciences 27, no. 1: 272. https://doi.org/10.3390/ijms27010272
APA StyleHerbert, A. (2026). Control of Gene Expression by Proteins That Bind Many Alternative Nucleic Acid Structures Through the Same Domain. International Journal of Molecular Sciences, 27(1), 272. https://doi.org/10.3390/ijms27010272





