1. Introduction
Diabetes exerts substantial stress on the brain, particularly within cortical regions involved in metabolic regulation, cognition, and sensory integration. Chronic hyperglycemia, mitochondrial dysfunction, and inflammatory signaling progressively disrupt cortical homeostasis, leading to altered neuronal excitability and transcriptional regulation [
1,
2,
3]. These effects may be further exacerbated when diabetes coexists with obesity, as excess adiposity imposes additional endocrine and metabolic burdens that increase cortical vulnerability [
4]. Accumulating evidence indicates that type 2 diabetes is associated with cortical excitability alterations, neuroinflammatory activation, and synaptic dysregulation; however, the molecular mechanisms underlying these cortical impairments remain insufficiently defined.
Exercise training is well recognized for improving systemic metabolic outcomes in diabetes and obesity, including enhanced glucose regulation, reduced inflammation, and improved cardiovascular function [
5,
6,
7]. Beyond these peripheral effects, exercise training also enhances brain health by promoting mitochondrial capacity, neurotrophic signaling, and synaptic resilience [
8,
9,
10]. Despite these benefits, the molecular pathways through which exercise training modulates cortical function under diabetic–obese conditions remain poorly characterized.
Transcriptomic profiling provides a powerful approach for examining such adaptations. RNA sequencing enables the detection of broad transcriptional shifts across diverse signaling networks, making it well suited for investigating complex metabolic–neuronal interactions within the cortex. Previous studies in high-fat-diet-induced obesity models have reported marked alterations in cortical synaptic, metabolic, and calcium-related gene networks [
11]. In parallel, human and animal studies have demonstrated impaired cortical plasticity and reduced neuronal excitability in diabetic states [
12,
13], suggesting heightened vulnerability of cortical circuits. However, exercise-trained cortical transcriptomic adaptations in diabetic–obese conditions, particularly those involving neuronal signaling pathways, remain largely unexplored.
In the present study, we integrated physiological assessments, including body weight, glucose levels and systolic blood pressure, with an in silico analysis of cortical RNA-seq data from diabetic–obese rats subjected to a 12-week swimming training program. Rather than aiming to define definitive molecular mechanisms, this study sought to identify exercise training-modulated neuronal pathways associated with cortical adaptation in diabetic–obese states. Through pathway enrichment and cross-network analyses, we highlight several candidate processes, including glutamatergic synapse regulation, retrograde endocannabinoid signaling, and oxytocin-associated pathways, that may contribute to neuro-metabolic modulation to exercise training. This hypothesis-generating analysis addresses an important knowledge gap regarding how cortical neuronal signaling networks are modulated by exercise training under metabolic dysfunctions.
2. Results
2.1. Overview of Experimental Workflow and Transcriptomic Dataset
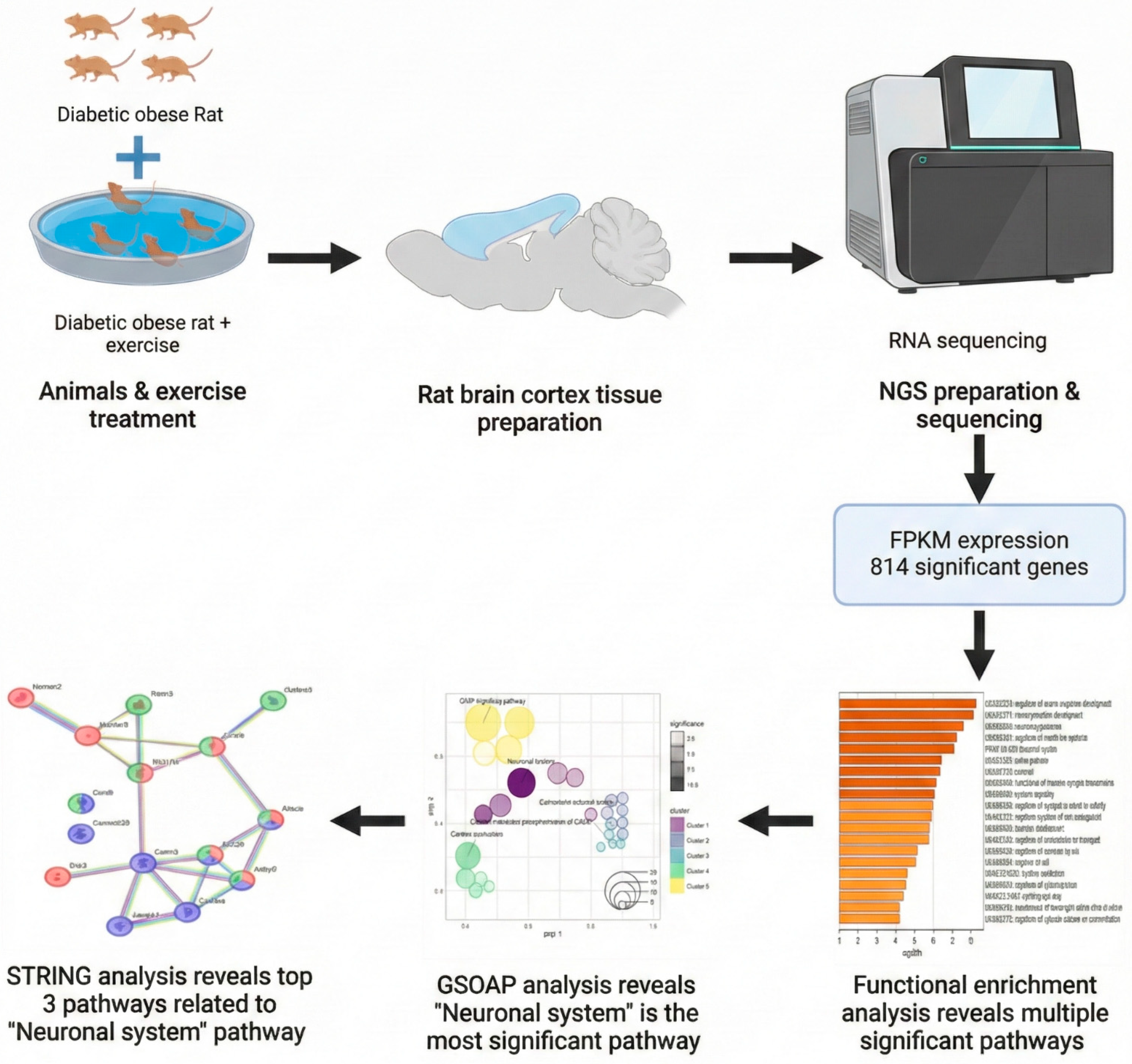
A schematic overview of the experimental design and analytical workflow, including animal grouping, exercise intervention, cortical tissue processing, RNA sequencing, and downstream bioinformatic analyses, is presented in
Figure 1.
RNA sequencing analysis identified a total of 814 genes that were significantly altered by exercise training in obese diabetic rats (p < 0.05). These differentially expressed genes were used for subsequent functional enrichment and network-based analyses.
2.2. Physiological and Metabolic Adaptations After Exercise Training
Exercise training produced clear physiological and metabolic changes in obese diabetic rats. Body weight was significantly lower in the exercise-trained (EX) group compared with the obese sedentary (OB) group, accompanied by a reduction in fasting blood glucose levels (
Table 1). Among blood pressure parameters, only systolic blood pressure showed a significant decrease following exercise training. These findings confirm that the 12-week swimming intervention elicited measurable metabolic and cardiovascular adaptations in ZFDM rats.
2.3. Identification of Neuronal-Related Gene Networks After Exercise Training
Functional enrichment analysis using Metascape, followed by pathway clustering with GSOAP, identified several pathway clusters after exercise training in the cerebral cortex (
Figure 2). Among these clusters, one neuronal-related group comprised 44 genes associated with synaptic transmission, neuromodulation, and intracellular signaling processes.
Protein–protein interaction (PPI) analysis of these 44 genes using the STRING database revealed a densely connected network, indicating extensive molecular connectivity within this neuronal-related cluster (
Figure 2). To provide full network context, a detailed STRING interaction map was generated (
Supplementary Figure S1).
2.4. Pathway-Level Mapping of Convergent Neuronal Signaling Processes
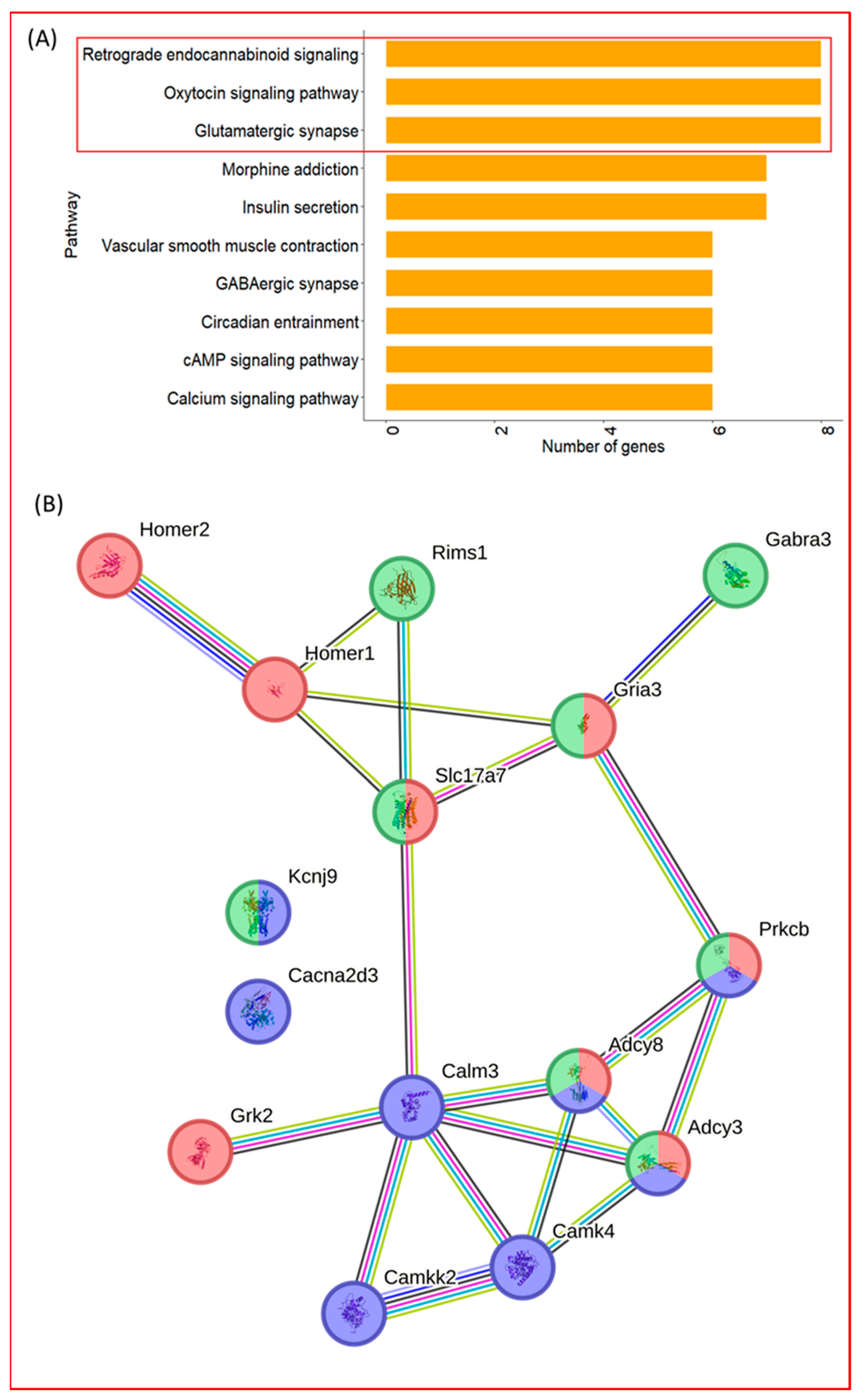
To determine which neuronal signaling pathways were most prominently represented within the neuronal gene set, pathway-specific mapping was performed for the 44 neuronal-related genes. Among the pathways examined, three KEGG signaling pathways contained the highest number of mapped genes: the glutamatergic synapse, oxytocin signaling, and retrograde endocannabinoid signaling (
Figure 3A).
Three genes—
Adcy3,
Adcy8, and
Prkcb—were shared across all three pathways, forming overlapping nodes within the pathway maps. Consistent with this overlap, STRING-based interaction analysis indicated that genes from these pathways formed an interconnected network rather than isolated modules (
Figure 3B).
3. Discussion
Metabolic measurements confirmed that exercise training showed clear physiological improvements in obese diabetic rats. Specifically, exercise training resulted in reduced body weight, lower fasting glucose levels, and decreased systolic blood pressure. These results are consistent with previous findings showing that exercise training improves metabolic status in diabetic–obese conditions [
14,
15]. The presence of these expected metabolic adaptations supports the reliability of the exercise intervention and provides physiological context for interpreting the transcriptomic modulation observed in the cortex.
High-throughput RNA-seq analysis revealed substantial transcriptional changes associated with exercise training, identifying 814 differentially expressed genes. Among the pathway clusters highlighted by GSOAP, the neuronal system emerged as the most prominent. Given the cortical origin of the samples, exercise training modulation of neuron-related genes is plausible; however, the broad 44-gene cluster required further resolution to identify more interpretable biological patterns. To refine these findings, we examined the protein–protein interaction structure of the neuronal gene set using STRING. Three pathways—glutamatergic synapse, retrograde endocannabinoid signaling, and oxytocin signaling—showed the strongest representation and formed an interconnected network, unified through shared nodes such as
Adcy3,
Adcy8, and
Prkcb (
Figure 3B).
3.1. Transcriptomic Signatures Related to Glutamatergic, Excitability, and Glucose Regulatory Processes After Exercise Training
Across the three neuronal-related pathways,
Adcy3,
Adcy8, and
Prkcb were consistently present, serving as shared nodes that linked multiple signaling processes within the exercise training-modulated network (
Figure 3B). Additional cross-pathway overlaps, including
Slc17a7,
Gria3, and
Kcnj9, further illustrated the interconnected nature of excitatory, retrograde, and neuromodulatory gene groups. These patterns indicate that exercise training may influence network-level organization rather than individual pathways in isolation.
Interpretation of these relationships must remain cautious, as transcriptomic data do not establish functional or mechanistic consequences. Nonetheless, the observed pathway signatures are broadly consistent with neural adaptations reported in metabolic or exercise-related contexts. For example, glutamatergic dysregulation has been described in diabetic cortical conditions, supporting the relevance of excitatory signaling within this network [
16]. Exercise training has also been shown to enhance endocannabinoid tone in metabolic and neural settings, further aligning with the observed enrichment of endocannabinoid-related genes [
17]. In addition, studies in diabetic neuropathic pain models demonstrate that exercise training can attenuate neuroinflammatory and excitability-related disturbances [
18], suggesting that multiple neuronal conditions in diabetes-associated environments may be improved after exercise training.
Altogether, these observations prompted further consideration of how exercise training-associated transcriptional changes may converge across multiple neuronal signaling pathways in the diabetic–obese cortex (
Figure 4). Exercise training potentially reduced glutamate through
Grk2,
Adcy complex,
Slc17a7, and
Kcnj9. Reduced glutamate levels may contribute to decreased neuronal excitability, potentially through downregulation of
Gria,
Homer,
Adcy complex, and
Prkcb gene expression. Exercise training also affected the oxytocin signaling-related pathway which led to lower glucose uptake. Glucose uptake was reduced during exercise due to lower gene expression of
Cacna2d3,
CaMK4,
Prkcb,
Calm3, and
CaMkk2. While the present findings remain hypothesis-generating and require future mechanistic validation, the convergence of glutamatergic, endocannabinoid, and oxytocin signaling pathways identifies several molecular candidates that warrant further investigation. These pathway-level insights may guide subsequent studies aiming to clarify how exercise influences cortical neuronal regulation under diabetic–obese conditions.
3.2. Integration of Physiological and Cortical Transcriptional Adaptations
Exercise training produced clear metabolic improvements, including reductions in body weight and fasting glucose and lower systolic blood pressure. These physiological changes are consistent with previous evidence showing that exercise training enhances metabolic status in diabetic–obese conditions [
4,
14,
15]. Such systemic adaptations may coincide with transcriptional changes observed in the cortex, although direct relationships were not assessed in the present study.
Within the cortical transcriptome, exercise training was associated with shifts involving glutamatergic synaptic signaling, retrograde endocannabinoid pathways, and oxytocin-related neuromodulatory processes. Although these associations do not establish causality, the direction of several transcriptional patterns is observed alongside neural adaptations previously described in diabetic or exercise contexts [
16,
17].
Taken together, these observations suggest that systemic benefits induced by exercise training may coincide with cortical signaling adjustments, providing a conceptual framework for understanding how exercise training could support neuronal adaptation under diabetic–obese conditions. However, these inferences remain hypothesis-generating, and future mechanistic studies will be required to determine whether such transcriptomic shifts have functional consequences.
3.3. Limitations
Several limitations should be acknowledged. First, the transcriptomic analysis was based on four biological replicates per group, which is common in exploratory RNA-seq studies but may limit the detection of subtle expression changes. Second, only male rats were included, restricting generalizability to females, who may exhibit distinct neuro-metabolic modulation to exercise training. Third, swimming exercise training may introduce stress-related transcriptional effects, and individual variability in exercise adaptation was not assessed. Fourth, the study relied exclusively on mRNA profiles without protein-level or functional assays, and pathway interpretation should therefore be considered hypothesis-generating rather than mechanistic. Future work incorporating region-specific assays, multimodal omics, and functional manipulation will be necessary to validate the cortical pathways identified here.
Despite these limitations, the analysis highlighted three neuron-related pathways—glutamatergic synapse, retrograde endocannabinoid signaling, and oxytocin signaling—that showed coordinated transcriptional patterns following exercise training. These findings provide a conceptual framework for exploring how exercise training may influence cortical signaling in diabetic–obese conditions. However, these interpretations remain speculative, and additional mechanistic studies will be required to determine whether the observed transcriptomic patterns translate to functional outcomes. Accordingly, these findings should be interpreted as hypothesis-generating and require future functional validation.
4. Materials and Methods
4.1. Animals
Male Obese Zucker Diabetic rats were used as a polygenic model of type 2 diabetes with obesity. Lean and obese Zucker rats were divided into three groups, sedentary Lean Zucker rats (LN), sedentary Obese Zucker Diabetic rats (OB), and Obese Zucker Diabetic rats after exercise training (OB-EX), with 10 rats per group. For RNA sequencing, four of the ten animals in each group were selected to represent transcriptomic profiles. All rats were housed under controlled temperature and lighting conditions with ad libitum access to standard chow and water.
4.2. Physiology and Metabolic Analysis
Systolic, diastolic, and mean arterial blood pressures were measured using an automated tail-cuff system (29SSP; IITC/Life Science Instruments, Woodland Hills, CA, USA), following procedures previously established in our laboratory for obese rat exercise studies. Rats were habituated to the apparatus for several sessions prior to data collection to minimize stress-induced variability, and at least three consecutive stable readings were averaged for each parameter.
Fasting blood glucose concentrations were obtained from tail vein blood samples using Roche Accu Soft test strips, based on protocols applied in earlier investigations of exercise training in obese or diabetic rats [
19]. All glucose measurements were collected after a 10–12 h overnight fast and prior to euthanasia.
Citrate synthase (CS) activity was analyzed by homogenizing soleus tissue in HES buffer and measuring enzymatic activity spectrophotometrically according to established methods from our previous studies. Soleus CS activity is presented as μmol/min/g tissue and served as an index of oxidative enzyme capacity after exercise training.
4.3. Exercise Training Protocol
The Obese Zucker Diabetic rats after exercise training (OB-EX) group underwent a swimming training program modified from earlier studies conducted in obese diabetic rats [
19]. Rats swam for 15 min per day, 5 days per week during the first 2 weeks. Training duration was increased to 20 min per day in week 3 and then maintained at 30 min per day from weeks 4 to 12. All exercise sessions were performed in a 60 × 90 × 50 cm tank filled to an appropriate depth and maintained at 35 °C to prevent hypothermia.
To minimize handling stress, animals were gently transferred into and out of the swimming tank and monitored continuously throughout each session. After swimming, rats were thoroughly dried using towels followed by a warm air dryer. All animals were euthanized 48 h after the final training session to avoid acute exercise effects on physiological or molecular measurements.
4.4. RNA Isolation
Total RNA was extracted from the cerebral cortex using a standard phenol–chloroform procedure based on the manufacturer’s guidelines and our laboratory’s established protocol for neural tissue processing. Approximately 30 mg of frozen cortical tissue was homogenized in 1 mL of TRIzol reagent (Invitrogen, Carlsbad, CA, USA) using a motorized homogenizer, followed by phase separation with chloroform and centrifugation at 12,000× g for 15 min at 4 °C. The aqueous phase was transferred to a fresh tube, and RNA was precipitated with isopropanol, washed with 75% ethanol, and resuspended in RNase-free water.
RNA concentration and purity were assessed by spectrophotometric measurement (NanoDrop 2000; Thermo Fisher Scientific, Waltham, MA, USA). Samples with A260/280 ratios between 1.9 and 2.1 were retained for sequencing. RNA integrity was further evaluated using microfluidic electrophoresis (Bioanalyzer 2100; Agilent Technologies, Santa Clara, CA, USA), and only samples with an RNA integrity number (RIN) ≥ 7.0 were included in downstream library preparation. RNA samples were stored at −80 °C until sequencing.
RNA-seq library preparation and sequencing were performed by Genomics, BioSci & Tech Co. (New Taipei City, Taiwan) using a standard TruSeq-based protocol on an Illumina NovaSeq platform (150 bp paired-end).
4.5. RNA-Seq Data Analysis and Functional Enrichment
Differential gene expression was defined based on FPKM-derived expression values using a significance threshold of
p < 0.05 without a fold-change cutoff, consistent with exploratory transcriptomic analyses. Functional enrichment analysis was performed using Metascape (
https://metascape.org, accessed on 21 December 2023) with default settings (minimum overlap = 3,
p < 0.01) [
20]. Pathway clustering was conducted using GSOAP (
https://gsoap.csb.pitt.edu, accessed on 21 December 2023) based on gene overlap-derived similarity between pathways [
21]. Protein–protein interaction networks were generated using the STRING (version 11.5) database with a medium-confidence interaction threshold (interaction score ≥ 0.7) [
22]. These analytical parameters are reported to ensure transparency and reproducibility, as requested by reviewer comments.
4.6. Additional Software and Visualization
Visualization of gene expression patterns and network structures was performed using the ggpubr package (
https://cran.r-project.org/package=ggpubr, accessed on 21 December 2023) and related R-based tools [
23]. These analyses supported pathway-level interpretation of transcriptional changes as hypothesis-generating in silico findings. BioRender (
https://www.biorender.com, accessed on 21 December 2023) was used to generate the schematic illustration shown in
Figure 1.
4.7. Ethics Statement
All animal experimental procedures were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of China Medical University, Taichung, Taiwan (Approval No. CMUIACUC-104-183-N), approved on 30 December 2014. All procedures complied with institutional and national guidelines for the care and use of laboratory animals.
5. Conclusions
Exercise training produced clear metabolic improvements in obese diabetic rats, including reductions in fasting glucose and systolic blood pressure. In parallel with these physiological effects, transcriptomic analysis revealed broad exercise-associated changes in cortical gene expression. Pathway enrichment consistently highlighted three neuron-related pathways—glutamatergic synapse, retrograde endocannabinoid signaling, and oxytocin signaling—as prominent components of the cortical response to exercise. The shared and interconnected nature of these pathways indicates that exercise is associated with coordinated, network-level transcriptional patterns rather than changes confined to isolated signaling routes.
Although RNA-seq data cannot determine directionality or functional consequences, several observed patterns aligned with gene groups involved in excitatory, neuromodulatory, and Ca2+-related signaling processes. These observations should be interpreted as hypothesis-generating, as no functional or correlational analyses were performed to establish mechanistic relationships. Accordingly, future studies will be required to clarify the functional relevance of these pathways in the diabetic–obese cortex.
Overall, this work provides a conceptual framework suggesting that exercise may reshape cortical signaling networks under metabolic dysfunctions. The findings offer a foundation for future investigations aimed at defining how exercise influences neuronal regulatory processes in diabetic–obese conditions.
Author Contributions
Conceptualization, Y.-Y.C., M.A.W. and S.-D.L.; Methodology, Y.-Y.C., M.A.W. and S.-D.L.; Software, M.A.W.; Validation, Y.-Y.C., M.A.W. and S.-D.L.; Formal Analysis, M.A.W.; Investigation, Y.-Y.C. and M.A.W.; Resources, S.-D.L.; Data Curation, M.A.W.; Writing—Original Draft Preparation, Y.-Y.C. and M.A.W.; Writing—Review & Editing, Y.-Y.C., M.A.W. and S.-D.L.; Visualization, M.A.W.; Supervision, S.-D.L.; Project Administration, S.-D.L.; Funding Acquisition, Y.-Y.C. and S.-D.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
All animal experimental procedures were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of China Medical University, Taichung, Taiwan (Approval No. CMUIACUC-104-183-N), approved on 30 December 2014.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. (Raw sequencing files are available upon reasonable request due to ongoing related research activities).
Acknowledgments
Figures were created with BioRender.com.
Conflicts of Interest
The authors declare that there are no conflicts of interest.
References
- Huber, J.D.; VanGilder, R.L.; Houser, K.A. Streptozotocin-induced diabetes progressively increases blood-brain barrier permeability in specific brain regions in rats. Am. J. Physiol. Heart Circ. Physiol. 2006, 291, H2660–H2668. [Google Scholar] [CrossRef]
- Meng, Y.; Wang, W.; Kang, J.; Wang, X.; Sun, L. Role of the PI3K/AKT signalling pathway in apoptotic cell death in the cerebral cortex of streptozotocin-induced diabetic rats. Exp. Ther. Med. 2017, 13, 2417–2422. [Google Scholar] [CrossRef]
- Wahl, P.; Mathes, S.; Kohler, K.; Achtzehn, S.; Bloch, W.; Mester, J. Acute metabolic, hormonal, and psychological responses to different endurance training protocols. Horm. Metab. Res. 2013, 45, 827–833. [Google Scholar] [CrossRef]
- Bae, J.H.; Choi, H.J.; Cho, K.I.K.; Kim, L.K.; Kwon, J.S.; Cho, Y.M. Glucagon-Like Peptide-1 Receptor Agonist Differentially Affects Brain Activation in Response to Visual Food Cues in Lean and Obese Individuals with Type 2 Diabetes Mellitus. Diabetes Metab. J. 2020, 44, 248–259. [Google Scholar] [CrossRef]
- Christiansen, L.B.; Toftager, M.; Boyle, E.; Kristensen, P.L.; Troelsen, J. Effect of a school environment intervention on adolescent adiposity and physical fitness. Scand. J. Med. Sci. Sports 2013, 23, e381–e389. [Google Scholar] [CrossRef] [PubMed]
- Kirk-Sanchez, N.J.; McGough, E.L. Physical exercise and cognitive performance in the elderly: Current perspectives. Clin. Interv. Aging 2014, 9, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Shi, H.; Guo, Q.; Gan, Y.; Zhang, Y.; Jia, J.; Zhang, L.; Zhou, Y. Aerobic Exercise Prevents Chronic Inflammation and Insulin Resistance in Skeletal Muscle of High-Fat Diet Mice. Nutrients 2022, 14, 3730. [Google Scholar] [CrossRef]
- Kim, Y.; Park, H. Does Regular Exercise without Weight Loss Reduce Insulin Resistance in Children and Adolescents? Int. J. Endocrinol. 2013, 2013, 402592. [Google Scholar] [CrossRef] [PubMed]
- Lumb, A. Diabetes and exercise. Clin. Med. 2014, 14, 673–676. [Google Scholar] [CrossRef]
- Wang, D.; Jiang, D.M.; Yu, R.R.; Zhang, L.L.; Liu, Y.Z.; Chen, J.X.; Chen, H.C.; Liu, Y.P. The Effect of Aerobic Exercise on the Oxidative Capacity of Skeletal Muscle Mitochondria in Mice with Impaired Glucose Tolerance. J. Diabetes Res. 2022, 2022, 3780156. [Google Scholar] [CrossRef]
- Yoon, G.; Cho, K.A.; Song, J.; Kim, Y.K. Transcriptomic Analysis of High Fat Diet Fed Mouse Brain Cortex. Front. Genet. 2019, 10, 83. [Google Scholar] [CrossRef]
- Crispoltoni, L.; Stabile, A.M.; Pistilli, A.; Venturelli, M.; Cerulli, G.; Fonte, C.; Smania, N.; Schena, F.; Rende, M. Changes in Plasma β-NGF and Its Receptors Expression on Peripheral Blood Monocytes During Alzheimer’s Disease Progression. J. Alzheimer’s Dis. 2017, 55, 1005–1017. [Google Scholar] [CrossRef]
- Valle-Bautista, R.; Marquez-Valadez, B.; Fragoso-Cabrera, A.D.; Garcia-Lopez, G.; Diaz, N.F.; Herrera-Lopez, G.; Griego, E.; Galvan, E.J.; Arias-Montano, J.A.; Molina-Hernandez, A. Impaired Cortical Cytoarchitecture and Reduced Excitability of Deep-Layer Neurons in the Offspring of Diabetic Rats. Front. Cell Dev. Biol. 2020, 8, 564561. [Google Scholar] [CrossRef] [PubMed]
- Kuru, O.; Senturk, U.K.; Demir, N.; Yesilkaya, A.; Erguler, G.; Erkilic, M. Effect of exercise on blood pressure in rats with chronic NOS inhibition. Eur. J. Appl. Physiol. 2002, 87, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Rossi Dare, L.; Garcia, A.; Alves, N.; Ventura Dias, D.; de Souza, M.A.; Mello-Carpes, P.B. Physical and cognitive training are able to prevent recognition memory deficits related to amyloid beta neurotoxicity. Behav. Brain Res. 2019, 365, 190–197. [Google Scholar] [CrossRef]
- Wiegers, E.C.; Rooijackers, H.M.; van Asten, J.J.A.; Tack, C.J.; Heerschap, A.; de Galan, B.E.; van der Graaf, M. Elevated brain glutamate levels in type 1 diabetes: Correlations with glycaemic control and age of disease onset but not with hypoglycaemia awareness status. Diabetologia 2019, 62, 1065–1073. [Google Scholar] [CrossRef]
- Matei, D.; Trofin, D.; Iordan, D.A.; Onu, I.; Condurache, I.; Ionite, C.; Buculei, I. The Endocannabinoid System and Physical Exercise. Int. J. Mol. Sci. 2023, 24, 1989. [Google Scholar] [CrossRef]
- Luo, J.; Zhu, H.Q.; Gou, B.; Zheng, Y.L. Mechanisms of exercise for diabetic neuropathic pain. Front. Aging Neurosci. 2022, 14, 975453. [Google Scholar] [CrossRef]
- Whitaker, K.W.; Totoki, K.; Reyes, T.M. Metabolic adaptations to early life protein restriction differ by offspring sex and post-weaning diet in the mouse. Nutr. Metab. Cardiovasc. Dis. 2012, 22, 1067–1074. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Serra, A.; Saarimaki, L.A.; Fratello, M.; Marwah, V.S.; Greco, D. BMDx: A graphical Shiny application to perform Benchmark Dose analysis for transcriptomics data. Bioinformatics 2020, 36, 2932–2933. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef] [PubMed]
- Kassambara, A. ggpubr: ‘ggplot2’-Based Publication Ready Plots. 2023. Available online: https://cran.r-project.org/package=ggpubr (accessed on 21 December 2023).
| Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |