Calcium and Cadmium Activate ESRRB to Mediate Cell Stemness and Pluripotency
Abstract
1. Introduction
2. Results
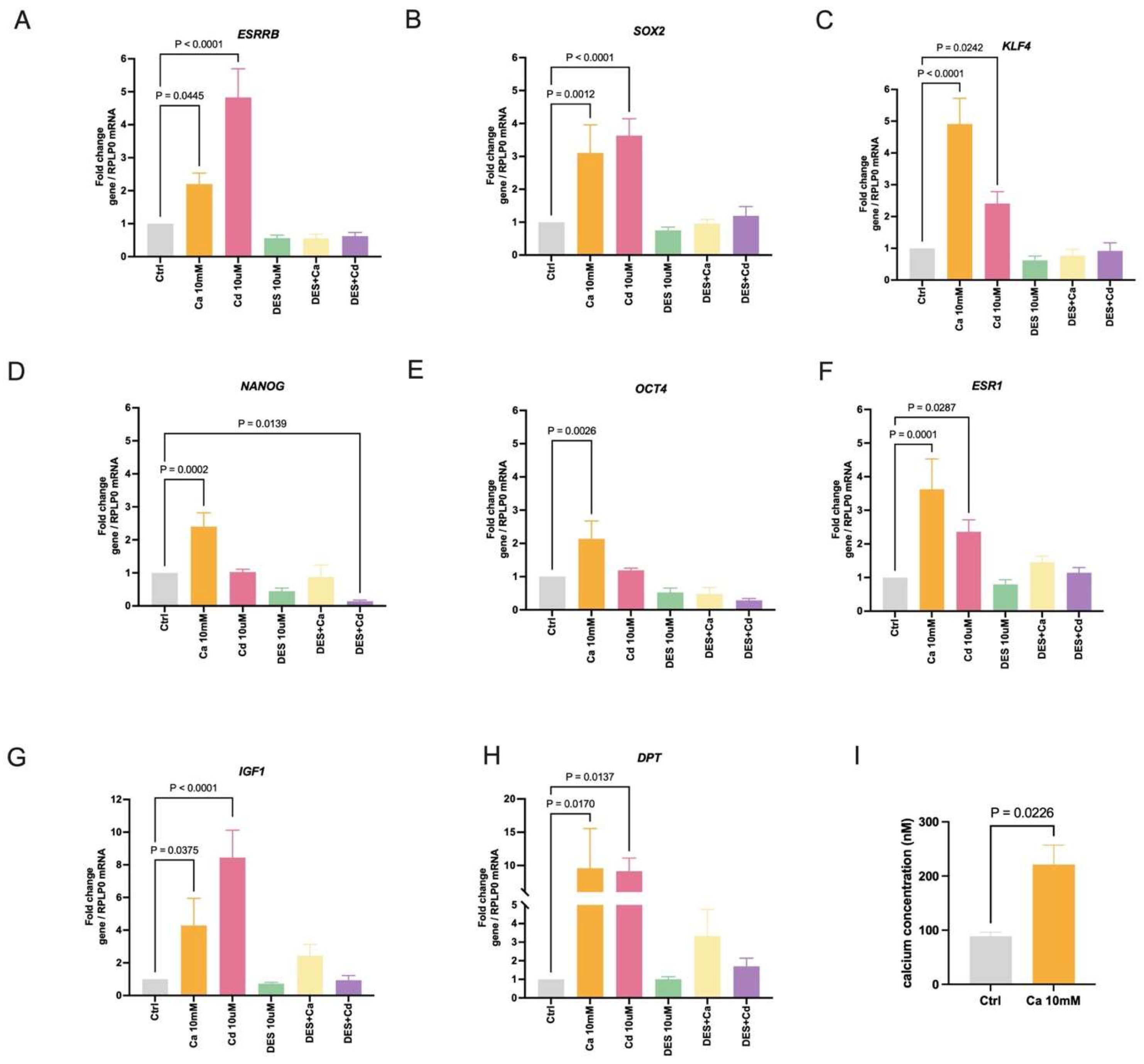
2.1. Calcium and Cadmium Induce ESRRB-Regulated Genes in HEK293T Cells
2.2. Calcium and Cadmium Induce ESRRB-Regulated Genes in MDA-MB-453 Cells
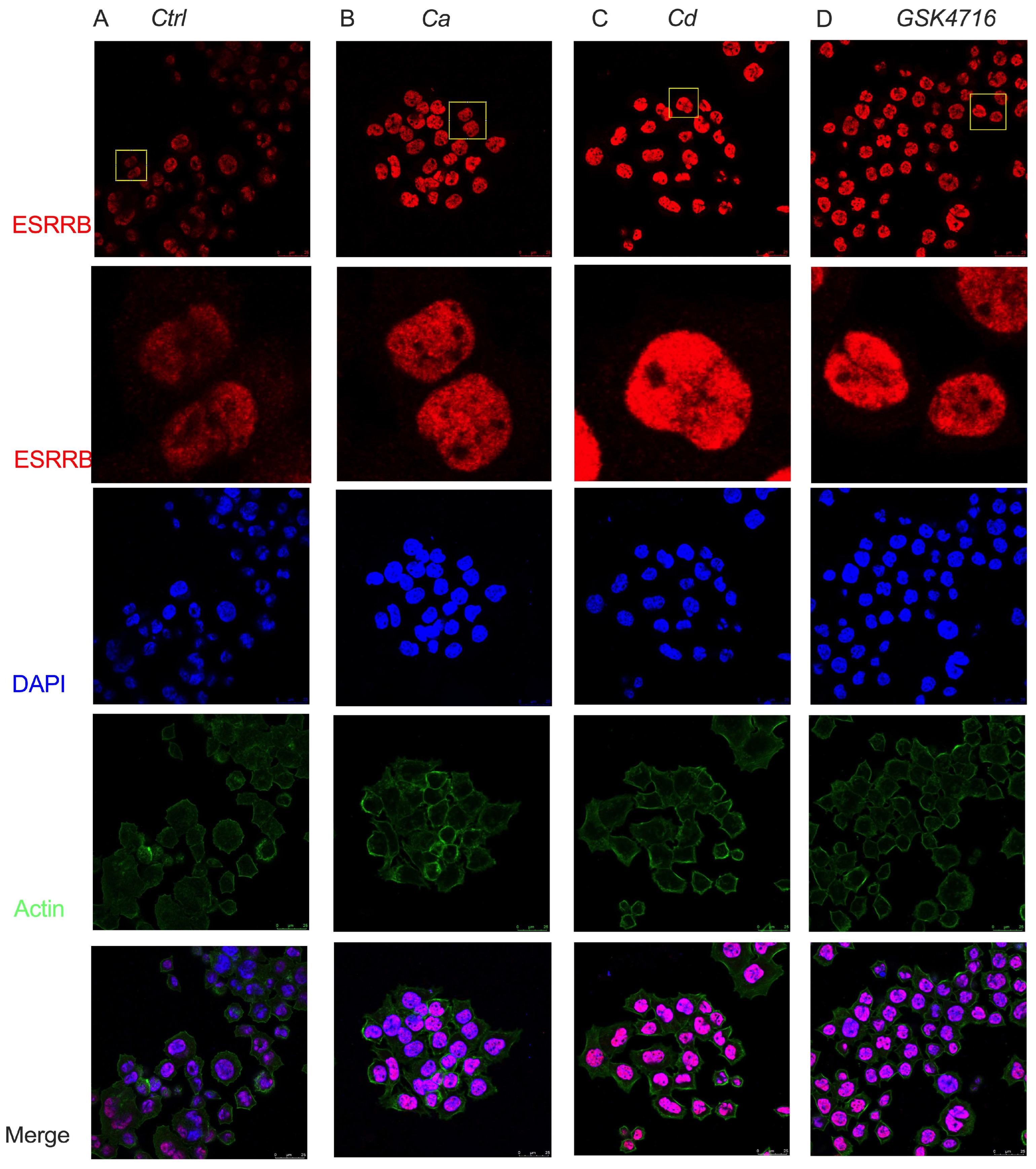
2.3. Calcium and Cadmium Enhance the Localization of ESRRB
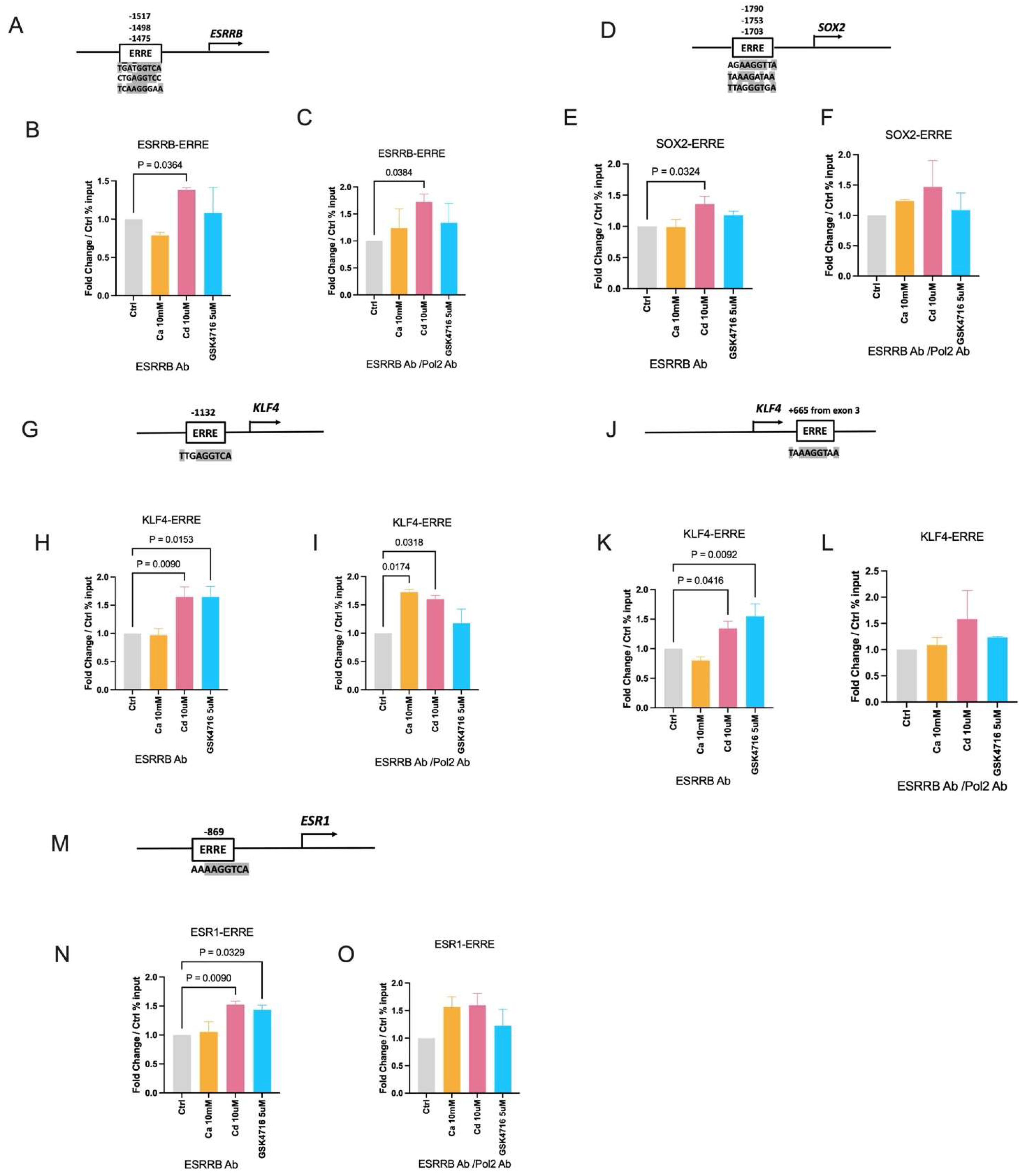
2.4. Effects of Calcium and Cadmium on the Binding of ESRRB to the Enhancers of ESRRB Target Genes
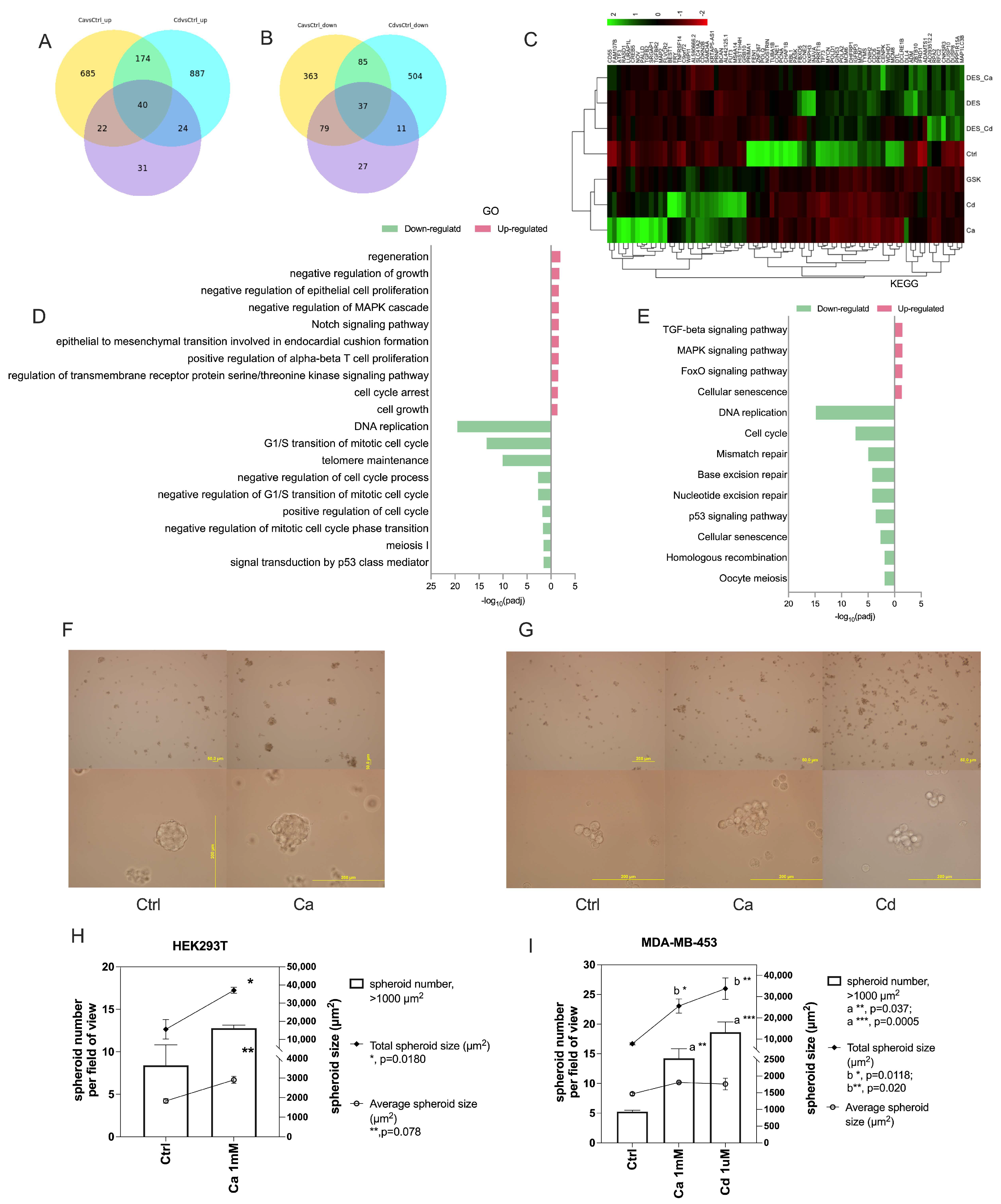
2.5. Calcium and Cadmium Mediate Cell Stemness/Pluripotency Through ESRRB
2.6. Calcium and Cadmium Activate ESRRB Through Amino Acids in the Ligand-Binding Domain
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Real-Time-qPCR
4.3. Intracellular Calcium Measurement
4.4. Immunofluorescence
4.5. Chromatin Immunoprecipitation (ChIP) and Re-ChIP
4.6. Spheroid Formation Assay
4.7. Chloramphenicol Acetyltransferase (CAT) Reporter Assay
4.8. Mutagenesis and Transfection Assays
4.9. RNA-Seq Analysis
4.10. Molecular Docking
4.11. Statistical Analysis
- STAR Methods
| Reagent/Resource | Reference or Source | Identifier or Catalog Number |
|---|---|---|
| Experimental Models | ||
| HEK-293T cells (H. sapiens) | ATCC | CVCL_0063 |
| MDA-MB-453 cells (H. sapiens) | Tissue Culture and Biobanking Shared Resource (Georgetown University) | CVCL_0418 |
| COS-1 cells (C. aethiops) | Tissue Culture and Biobanking Shared Resource (Georgetown University) | CVCL_0223 |
| Subcloning Efficiency DH5α Competent Cells | Invitrogen | 18265-017 |
| Recombinant DNA | ||
| pSG5-ERRbeta2 | Heckler & Riggins, (2015) [53] | Addgene plasmid #52186 |
| pCMV-GAL4 | Potter et al., (2010) [95] | Addgene plasmid #24345 |
| pEQ176 | Schleiss et al., (1991) [96] | Addgene plasmid #83943 |
| pG6(5′Pro) | Taube et al., (2002) [97] | Addgene plasmid #14659 |
| Antibodies | ||
| Human ERR beta/NR3B2 Antibody | R&D systems, Minneapolis, MN, USA | PP-H6705-00 |
| POLR2A Monoclonal Antibody (8WG16) | Thermo Fisher Scientific, Waltham, MA, USA | MA1-26249 |
| mouse IgG control | Abcam, Cambridge, UK | ab18413 RRID: AB_2631983 |
| goat anti-mouse IgG polyclonal antibody (594; Alexa Fluor) | Invitrogen, Waltham, MA, USA | RRID: AB_2313921 |
| Oligonucleotides and other sequence-based reagents | ||
| PCR Primers | This study | Supplemental Table S2 |
| EsRRB Taqman probe | Thermo Fisher Scientific, Waltham, MA, USA | Hs01584024_m1 |
| SOX2 Taqman probe | Thermo Fisher Scientific, Waltham, MA, USA | Hs01053049_s1 |
| KLF4 Taqman probe | Thermo Fisher Scientific, Waltham, MA, USA | Hs00358836_m1 |
| NANOG Taqman probe | Thermo Fisher Scientific, Waltham, MA, USA | Hs02387400_g1 |
| POU5F1 (OCT4) Taqman probe | Thermo Fisher Scientific, Waltham, MA, USA | Hs00999632_g1 |
| PRL Taqman probe | Thermo Fisher Scientific, Waltham, MA, USA | Hs00168730_m1 |
| SFRP2 Taqman probe | Thermo Fisher Scientific, Waltham, MA, USA | Hs00293258_m1 |
| DPT Taqman probe | Thermo Fisher Scientific, Waltham, MA, USA | Hs00355056_m1 |
| IGF1 Taqman probe | Thermo Fisher Scientific, Waltham, MA, USA | Hs01547656_m1 |
| ESR1 Taqman probe | Thermo Fisher Scientific, Waltham, MA, USA | Hs00174860_m1 |
| RPLP0 Taqman probe | Thermo Fisher Scientific, Waltham, MA, USA | Hs00420895_gH |
| Chemicals, Enzymes and other reagents | ||
| calcium chloride | Sigma-Aldrich, St. Louis, MO, USA | C4901 |
| cadmium chloride | Sigma-Aldrich, St. Louis, MO, USA | 202908 |
| Diethylstilbestrol (DES) | Sigma-Aldrich, St. Louis, MO, USA | D4628 |
| GSK4716 | Tocris Bioscience, Briston, UK | 3075 |
| CorningTM Improved Minimal Essential Medium (IMEM), phenol red-free | Corning, Corning, NY, USA | MT10026CV |
| fetal bovine serum (FBS) | Sigma-Aldrich, St. Louis, MO, USA | F0926 |
| Dulbecco’s Modified Eagle’s Medium (DMEM) | Corning, Corning, NY, USA | MT10013CV |
| lipoic-acid and phenol-red free IMEM | Crystalgen Commack, NY, USA | 226-072-12 |
| charcoal stripped calf serum (CCS) | Sigma-Aldrich, St. Louis, MO, USA | F6765 |
| Trizol | Life Technologies, Carlsbad, CA, USA | 15596026 |
| chloroform | Fisher Scientific, Waltham, MA, USA | J67241.K4 |
| isopropanol | Fisher Scientific, Waltham, MA, USA | T036181000 |
| Hank’s balanced salt solution (HBSS) | Gibco, Billings, MT, USA | 14175095 |
| Fluo-4 AM | Invitrogen, Waltham, MA, USA | F14217 |
| formaldehyde | Fisher Scientific, Waltham, MA, USA | 28908 |
| Triton X-100 | Sigma Aldrich, St. Louis, MO, USA | 93443 |
| Fisher BioReagents bovine serum albumin (BSA) | Fisher BioReagents, Waltham, MA, USA | BP9700100 |
| Phalloidin-iFluor Conjugate (488) | AAT Bioquest, Pleasanton, CA, USA | 23115 |
| 4′,6′-diamidine-2-phenylindole dihydrochloride (DAPI) | Sigma Aldrich, St. Louis, MO, USA | 10236276001 |
| PhosSTOP/PI cocktail | Sigma Aldrich, St. Louis, MO, USA | 4906837001 and 4693159001 |
| Nuclei lysis buffer | Sigma Aldrich, St. Louis, MO, USA | 20-163 |
| Chromatin Immunoprecipitation Dilution Buffer | Sigma Aldrich, St. Louis, MO, USA | 20-153 |
| Low Salt Immune Complex Wash Buffer | Sigma Aldrich, St. Louis, MO, USA | 20-154 |
| High Salt Immune Complex Wash Buffer | Sigma Aldrich, St. Louis, MO, USA | 20-155 |
| LiCl Immune Complex Wash Buffer | Sigma Aldrich, St. Louis, MO, USA | 20-156 |
| PCR purification kit (QIAquick PCR purification kit) | QIAGEN, Germantown, MD, USA | 28106 |
| SYBR green mastermix (PowerUP SYBR Green Master Mix) | Thermo Fishier Scientific, Waltham, MA, USA | A25780 |
| BspDI Restriction Enzyme | New England Biolabs, Ipswich, MA, USA | R0557 |
| NotI-HF Restriction Enzyme | New England Biolabs, Ipswich, MA, USA | R3189 |
| Taq 2X Master Mix | New England Biolabs, Ipswich, MA, USA | M0270L |
| T4 DNA Ligase | New England Biolabs, Ipswich, MA, USA | M0202 |
| Maxima H MA Synthesis Master Mix with dsDNase | Thermo scientific, Waltham, MA, USA | M1681 |
| PureLinkTM HiPure Plasmid Maxiprep Kit | Invitrogen, Waltham, MA, USA | K210007 |
| QuikChange Lightning Site-Directed Mutagenesis Kit | Agilent, Santa Clara, CA, USA | 210518 |
| Trans-IT LT-1 | MirusBio, Madison, WI, USA | MIR 2306 |
| SsoAdvanced Universal Probes Supermix | Bio-Rad, Hercules, CA, USA | 172-5281 |
| Software | ||
| GraphPad Prism version 10.2 | https://www.graphpad.com (accessed on 27 March 2024). | |
| ImageJ version 1.53p | https://imagej.nih.gov/ij/index.html (accessed on 16 June 2021) | |
| UCSC Genome Browser | https://genome.ucsc.edu/ (accessed on 20 August 2017) | |
| Cistrome Data Browser | http://cistrome.org/db/#/ (accessed on 11 September 2017) | |
| QuikChange Primer Design of Agilent | https://www.agilent.com/store/primerDesignProgram.jsp (accessed on 10 December 2017) | |
| Novo Magic | https://us-magic.novogene.com (accessed on 31 October 2023) | |
| Autodock Vina version 1.1.2 | https://vina.scripps.edu/ (accessed on 13 July 2023) [98,99,100] | |
| Pymol version 2.5.5 | https://www.pymol.org/ (accessed on 11 April 2023) | |
| Other | ||
| fluorimeter | Photon Technology International, Lawrenceville, NJ, USA | |
| Olympus IX71 microscope | Olympus, Tokyo, Japan | |
| SP8 laser confocal microscope | Leica, Wetzlar, Germany | |
| Novaseq 6000 | Illumina, San Diego, CA, USA |
Supplementary Materials
Author Contributions
Funding
Institutional Review Board statement
Informed Consent statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AF-1 or AF-2 | activation function-1 or -2 |
| CAT | chloramphenicol acetyltransferase |
| CCS | charcoal-stripped calf serum |
| ChIP | Chromatin Immunoprecipitation Assay |
| CSCs | cancer stem cells |
| DAPI | 4, 6-diamidino-2-phenylindole |
| DBD | DNA-binding domain |
| DEGs | differentially expressed genes |
| DES | diethylstilbestrol |
| DMEM | Dulbecco’s Modified Eagle’s Medium |
| DPT | dermatopontin, tyrosin-rich acidic matrix protein |
| ERRE | estrogen-related receptor response elements |
| ESR | estrogen receptor |
| ESR1 or ERα | estrogen receptor alpha |
| ESRRA | estrogen-related receptor alpha |
| ESRRB-∆10 | ESRRB splice variant delta 10 |
| ESRRB-sf | ESRRB splice variant short form |
| ESRRB | estrogen-related receptor beta |
| ESRRB2 | ESRRB splice variant beta 2 |
| ESRRG | estrogen-related receptor gama |
| ESRRs | estrogen-related receptors |
| FBS | fetal bovine serum |
| FoxO | forehead box protein, member of class O |
| GAL4 | galectin 4 |
| GO | Gene Ontology |
| HBSS | Hank’s balanced salt solution |
| HER2 | human epidermal growth factor 2 |
| IF | immunofluorescence |
| IGF1 | insulin-like growth factor 1 |
| IMEM | improved minimal essential medium |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| KLF4 | Kruppel-like factor 4 |
| LBD | ligand-binding domain |
| MAPK | mitogen-activated protein kinase |
| MaSCs | mammary stem cells |
| NANOG | Nanog homeobox |
| OCT4 | OCT3/4 or POU class 5 homeobox1 |
| PGR | progesterone receptor |
| Pol2 | RNA polymerase 2 |
| PRL | prolactin |
| Re-ChIP | subsequential Chromatin Immunoprecipitation Assay |
| real time-qPCR | real-time polymerase chain reaction |
| RMSF | Root mean square fluctuation |
| RPLP0 | ribosomal protein P0 |
| RXRα | Retinoid X receptor-alpha |
| SFRP2 | secreted frizzle protein 2 |
| SOX2 | sex-determining region Y-box 2 |
| TFs | transcription factors |
| TGFβ | tumor growth factor-beta |
| TNBC | triple negative breast cancer |
| TSS | transcription start site |
References
- Dunn, S.-J.; Martello, G.; Yordanov, B.; Emmott, S.; Smith, A.G. Defining an Essential Transcription Factor Program for Naïve Pluripotency. Science 2014, 344, 1156–1160. [Google Scholar] [CrossRef]
- Festuccia, N.; Owens, N.; Navarro, P. Esrrb, an Estrogen-Related Receptor Involved in Early Development, Pluripotency, and Reprogramming. Febs Lett. 2018, 592, 852–877. [Google Scholar] [CrossRef] [PubMed]
- Martello, G.; Sugimoto, T.; Diamanti, E.; Joshi, A.; Hannah, R.; Ohtsuka, S.; Göttgens, B.; Niwa, H.; Smith, A. Esrrb Is a Pivotal Target of the Gsk3/Tcf3 Axis Regulating Embryonic Stem Cell Self-Renewal. Cell Stem Cell 2012, 11, 491–504, Erratum in Cell Stem Cell 2013, 12, 630. [Google Scholar] [CrossRef] [PubMed]
- Papp, B.; Plath, K. Pluripotency Re-Centered around Esrrb. Embo J. 2012, 31, 4255–4257. [Google Scholar] [CrossRef]
- Young, R.A. Control of the Embryonic Stem Cell State. Cell 2011, 144, 940–954. [Google Scholar] [CrossRef]
- Zhang, M.; Leitch, H.G.; Tang, W.W.C.; Festuccia, N.; Hall-Ponsele, E.; Nichols, J.; Surani, M.A.; Smith, A.; Chambers, I. Esrrb Complementation Rescues Development of Nanog-Null Germ Cells. Cell Rep. 2018, 22, 332–339. [Google Scholar] [CrossRef] [PubMed]
- Adachi, K.; Kopp, W.; Wu, G.; Heising, S.; Greber, B.; Stehling, M.; Araúzo-Bravo, M.J.; Boerno, S.T.; Timmermann, B.; Vingron, M.; et al. Esrrb Unlocks Silenced Enhancers for Reprogramming to Naive Pluripotency. Cell Stem Cell 2018, 23, 266–275.e6, Correction in Cell Stem Cell 2018, 23, 900–904. [Google Scholar] [CrossRef]
- Feng, B.; Jiang, J.; Kraus, P.; Ng, J.H.; Heng, J.C.D.; Chan, Y.S.; Yaw, L.P.; Zhang, W.; Loh, Y.H.; Han, J.; et al. Reprogramming of Fibroblasts into Induced Pluripotent Stem Cells with Orphan Nuclear Receptor Esrrb. Nat. Cell Biol. 2009, 11, 197–203. [Google Scholar] [CrossRef]
- Festuccia, N.; Dubois, A.; Vandormael-Pournin, S.; Gallego Tejeda, E.; Mouren, A.; Bessonnard, S.; Mueller, F.; Proux, C.; Cohen-Tannoudji, M.; Navarro, P. Mitotic Binding of Esrrb Marks Key Regulatory Regions of the Pluripotency Network. Nat. Cell Biol. 2016, 18, 1139–1148. [Google Scholar] [CrossRef]
- Okamura, E.; Tam, O.H.; Posfai, E.; Li, L.; Cockburn, K.; Lee, C.Q.E.; Garner, J.; Rossant, J. Esrrb Function Is Required for Proper Primordial Germ Cell Development in Presomite Stage Mouse Embryos. Dev. Biol. 2019, 455, 382–392. [Google Scholar] [CrossRef]
- Chervova, A.; Molliex, A.; Baymaz, H.I.; Coux, R.-X.; Papadopoulou, T.; Mueller, F.; Hercul, E.; Fournier, D.; Dubois, A.; Gaiani, N.; et al. Mitotic Bookmarking Redundancy by Nuclear Receptors in Pluripotent Cells. Nat. Struct. Mol. Biol. 2024, 31, 513–522. [Google Scholar] [CrossRef]
- Weikum, E.R.; Liu, X.; Ortlund, E.A. The Nuclear Receptor Superfamily: A Structural Perspective. Protein Sci. 2018, 27, 1876–1892. [Google Scholar] [CrossRef] [PubMed]
- Greschik, H.; Wurtz, J.-M.; Sanglier, S.; Bourguet, W.; van Dorsselaer, A.; Moras, D.; Renaud, J.-P. Structural and Functional Evidence for Ligand-Independent Transcriptional Activation by the Estrogen-Related Receptor 3. Mol. Cell 2002, 9, 303–313. [Google Scholar] [CrossRef]
- Wärnmark, A.; Treuter, E.; Wright, A.P.H.; Gustafsson, J.-Å. Activation Functions 1 and 2 of Nuclear Receptors: Molecular Strategies for Transcriptional Activation. Mol. Endocrinol. 2003, 17, 1901–1909. [Google Scholar] [CrossRef] [PubMed]
- Bianco, S.; Sailland, J.; Vanacker, J.-M. ERRs and Cancers: Effects on Metabolism and on Proliferation and Migration Capacities. J. Steroid Biochem. Mol. Biol. 2012, 130, 180–185. [Google Scholar] [CrossRef] [PubMed]
- Huss, J.M.; Garbacz, W.G.; Xie, W. Constitutive Activities of Estrogen-Related Receptors: Transcriptional Regulation of Metabolism by the ERR Pathways in Health and Disease. Biochim. Biophys. Acta Mol. Basis Dis. 2015, 1852, 1912–1927. [Google Scholar] [CrossRef]
- Kumar, P.; Mendelson, C.R. Estrogen-Related Receptor γ (ERRγ) Mediates Oxygen-Dependent Induction of Aromatase (CYP19) Gene Expression during Human Trophoblast Differentiation. Mol. Endocrinol. 2011, 25, 1513–1526. [Google Scholar] [CrossRef]
- Takacs, M.; Petoukhov, M.V.; Atkinson, R.A.; Roblin, P.; Ogi, F.X.; Demeler, B.; Potier, N.; Chebaro, Y.; Dejaegere, A.; Svergun, D.I.; et al. The Asymmetric Binding of PGC-1α to the ERRα and ERRγ Nuclear Receptor Homodimers Involves a Similar Recognition Mechanism. PLoS ONE 2013, 8, e67810. [Google Scholar] [CrossRef]
- Vanacker, J.-M.; Bonnelye, E.; Chopin-Delannoy, S.; Delmarre, C.; Cavaillès, V.; Laudet, V. Transcriptional Activities of the Orphan Nuclear Receptor ERRα (Estrogen Receptor-Related Receptor-α). Mol. Endocrinol. 1999, 13, 764–773. [Google Scholar] [CrossRef]
- Kida, Y.S.; Kawamura, T.; Wei, Z.; Sogo, T.; Jacinto, S.; Shigeno, A.; Kushige, H.; Yoshihara, E.; Liddle, C.; Ecker, J.R.; et al. ERRs Mediate a Metabolic Switch Required for Somatic Cell Reprogramming to Pluripotency. Cell Stem Cell 2015, 16, 547–555. [Google Scholar] [CrossRef]
- Deblois, G.; Giguère, V. Functional and Physiological Genomics of Estrogen-Related Receptors (ERRs) in Health and Disease. Biochim. Biophys. Acta Mol. Basis Dis. 2011, 1812, 1032–1040. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Fan, X.; Shi, Y.; Zhang, C.; Sun, D.; Qin, K.; Qin, W.; Zhou, W.; Chen, X. Next-Generation Unnatural Monosaccharides Reveal That ESRRB O-GlcNAcylation Regulates Pluripotency of Mouse Embryonic Stem Cells. Nat. Commun. 2019, 10, 4065. [Google Scholar] [CrossRef]
- Heard, D.J.; Norby, P.L.; Holloway, J.; Vissing, H. Human ERRγ, a Third Member of the Estrogen Receptor-Related Receptor (ERR) Subfamily of Orphan Nuclear Receptors: Tissue-Specific Isoforms Are Expressed during Development and in the Adult. Mol. Endocrinol. 2000, 14, 382–392. [Google Scholar] [CrossRef]
- Singh, T.D.; Song, J.; Kim, J.; Chin, J.; Ji, H.D.; Lee, J.-E.; Lee, S.B.; Yoon, H.; Yu, J.H.; Kim, S.K.; et al. A Novel Orally Active Inverse Agonist of Estrogen-Related Receptor Gamma (ERRγ), DN200434, A Booster of NIS in Anaplastic Thyroid Cancer. Clin. Cancer Res. 2019, 25, 5069–5081. [Google Scholar] [CrossRef]
- Tremblay, G.B.; Kunath, T.; Bergeron, D.; Lapointe, L.; Champigny, C.; Bader, J.A.; Rossant, J.; Giguère, V. Diethylstilbestrol Regulates Trophoblast Stem Cell Differentiation as a Ligand of Orphan Nuclear Receptor ERRβ. Genes Dev. 2001, 15, 833–838. [Google Scholar] [CrossRef]
- Yu, D.D.; Huss, J.M.; Li, H.; Forman, B.M. Identification of Novel Inverse Agonists of Estrogen-Related Receptors ERRγ and ERRβ. Bioorganic Med. Chem. 2017, 25, 1585–1599. [Google Scholar] [CrossRef]
- Casaburi, I.; Chimento, A.; De Luca, A.; Nocito, M.; Sculco, S.; Avena, P.; Trotta, F.; Rago, V.; Sirianni, R.; Pezzi, V. Cholesterol as an Endogenous ERRα Agonist: A New Perspective to Cancer Treatment. Front. Endocrinol. 2018, 9, 525. [Google Scholar] [CrossRef]
- Wei, W.; Schwaid, A.G.; Wang, X.; Wang, X.; Chen, S.; Chu, Q.; Saghatelian, A.; Wan, Y. Ligand Activation of ERRα by Cholesterol Mediates Statin and Bisphosphonate Effects. Cell Metab. 2016, 23, 479–491. [Google Scholar] [CrossRef] [PubMed]
- Ghanbari, F.; Hebert-Losier, A.; Barry, J.; Poirier, D.; Giguere, V.; Mader, S.; Philip, A. Isolation and Functional Characterization of a Novel Endogenous Inverse Agonist of Estrogen Related Receptors (ERRs) from Human Pregnancy Urine. J. Steroid Biochem. Mol. Biol. 2019, 191, 105352. [Google Scholar] [CrossRef]
- Lu, Y.; Li, J.; Cheng, J.; Lubahn, D.B. Messenger RNA Profile Analysis Deciphers New Esrrb Responsive Genes in Prostate Cancer Cells. BMC Mol. Biol. 2015, 16, 21. [Google Scholar] [CrossRef] [PubMed]
- Di Micco, S.; Renga, B.; Carino, A.; D’Auria, M.V.; Zampella, A.; Riccio, R.; Fiorucci, S.; Bifulco, G. Structural Insights into Estrogen Related Receptor-β Modulation: 4-Methylenesterols from Theonella swinhoei Sponge as the First Example of Marine Natural Antagonists. Steroids 2014, 80, 51–63. [Google Scholar] [CrossRef]
- Wang, Q.; Huang, N.; Psaltis, J.B.; Gahtani, R.M.; Yan, G.; Lu, D.; Cahalan, S.R.; Shi, X.; Copeland, R.L.; Haddad, B.R.; et al. Activation of Estrogen-Related Receptor γ by Calcium and Cadmium. Front. Endocrinol. 2024, 15, 1400022. [Google Scholar] [CrossRef] [PubMed]
- Apáti, Á.; Berecz, T.; Sarkadi, B. Calcium Signaling in Human Pluripotent Stem Cells. Cell Calcium 2016, 59, 117–123. [Google Scholar] [CrossRef]
- Clapham, D.E. Calcium Signaling. Cell 2007, 131, 1047–1058. [Google Scholar] [CrossRef]
- Feher, J. 9.7—Calcium and Phosphorus Homeostasis I: The Calcitropic Hormones. In Quantitative Human Physiology; Feher, J., Ed.; Academic Press: Boston, MA, USA, 2012; pp. 828–835. ISBN 978-0-12-382163-8. [Google Scholar]
- Lu, H.; Chen, I.; Shimoda, L.A.; Park, Y.; Zhang, C.; Tran, L.; Zhang, H.; Semenza, G.L. Chemotherapy-Induced Ca2+ Release Stimulates Breast Cancer Stem Cell Enrichment. Cell Rep. 2017, 18, 1946–1957, Correction in Cell Rep. 2021, 34, 108605. [Google Scholar] [CrossRef]
- Terrié, E.; Coronas, V.; Constantin, B. Role of the Calcium Toolkit in Cancer Stem Cells. Cell Calcium 2019, 80, 141–151. [Google Scholar] [CrossRef]
- Tonelli, F.M.P.; Santos, A.K.; Gomes, D.A.; da Silva, S.L.; Gomes, K.N.; Ladeira, L.O.; Resende, R.R. Stem Cells and Calcium Signaling. Adv. Exp. Med. Biol. 2012, 740, 891–916. [Google Scholar] [CrossRef]
- Yu, E.; Sharma, S. Physiology, Calcium. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2024. [Google Scholar]
- Divekar, S.D.; Storchan, G.B.; Sperle, K.; Veselik, D.J.; Johnson, E.; Dakshanamurthy, S.; Lajiminmuhip, Y.N.; Nakles, R.E.; Huang, L.; Martin, M.B. The Role of Calcium in the Activation of Estrogen Receptor-Alpha. Cancer Res. 2011, 71, 1658–1668. [Google Scholar] [CrossRef]
- Johnson, M.D.; Kenney, N.; Stoica, A.; Hilakivi-clarke, L.; Singh, B.; Chepko, G.; Clarke, R.; Sholler, P.F.; Lirio, A.A.; Foss, C.; et al. Cadmium Mimics the in Vivo Effects of Estrogen in The. Nat. Med. 2003, 9, 1081–1084. [Google Scholar] [CrossRef] [PubMed]
- Stoica, A.; Pentecost, E.; Martin, M.B. Effects of Arsenite on Estrogen Receptor-α Expression and Activity in MCF-7 Breast Cancer Cells1. Endocrinology 2000, 141, 3595–3602. [Google Scholar] [CrossRef] [PubMed]
- Davis, J.; Khan, G.; Martin, M.; Hilakivi-Clarke, L. Effects of Maternal Dietary Exposure to Cadmium During Pregnancy on Mammary Cancer Risk Among Female Offspring. J. Carcinog. 2013, 12, 11. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Parodi, D.A.; Greenfield, M.; Evans, C.; Chichura, A.; Alpaugh, A.; Williams, J.; Cyrus, K.C.; Martin, M.B. Alteration of Mammary Gland Development and Gene Expression by In Utero Exposure to Cadmium. Int. J. Mol. Sci. 2017, 18, 1939. [Google Scholar] [CrossRef]
- Ju, H.; Arumugam, P.; Lee, J.; Song, J.M. Impact of Environmental Pollutant Cadmium on the Establishment of a Cancer Stem Cell Population in Breast and Hepatic Cancer. ACS Omega 2017, 2, 563–572. [Google Scholar] [CrossRef]
- Lu, Y.; Li, J.; Cheng, J.; Lubahn, D.B. Genes Targeted by the Hedgehog-Signaling Pathway Can Be Regulated by Estrogen Related Receptor β. BMC Mol. Biol. 2015, 16, 19. [Google Scholar] [CrossRef]
- Carrillo-López, N.; Álvarez-Hernández, D.; González-Suárez, I.; Román-García, P.; Valdivielso, J.M.; Fernández-Martín, J.L.; Cannata-Andía, J.B. Simultaneous Changes in the Calcium-Sensing Receptor and the Vitamin D Receptor under the Influence of Calcium and Calcitriol. Nephrol. Dial. Transplant. 2008, 23, 3479–3484. [Google Scholar] [CrossRef]
- Pochampally, R.R.; Ylostalo, J.; Penfornis, P.; Matz, R.R.; Smith, J.R.; Prockop, D.J. Histamine Receptor H1 and Dermatopontin: New Downstream Targets of the Vitamin D Receptor. J. Bone Miner. Res. 2007, 22, 1338–1349. [Google Scholar] [CrossRef]
- Mangelsdorf, D.J.; Thummel, C.; Beato, M.; Herrlich, P.; Schütz, G.; Umesono, K.; Blumberg, B.; Kastner, P.; Mark, M.; Chambon, P.; et al. The Nuclear Receptor Superfamily: The Second Decade. Cell 1995, 83, 835–839. [Google Scholar] [CrossRef]
- Kulhari, H.; Pooja, D.; Shrivastava, S.; Kuncha, M.; Naidu, V.G.M.; Bansal, V.; Sistla, R.; Adams, D.J. Trastuzumab-Grafted PAMAM Dendrimers for the Selective Delivery of Anticancer Drugs to HER2-Positive Breast Cancer. Sci. Rep. 2016, 6, 23179. [Google Scholar] [CrossRef] [PubMed]
- Mishra, R.; Yuan, L.; Patel, H.; Karve, A.S.; Zhu, H.; White, A.; Alanazi, S.; Desai, P.; Merino, E.J.; Garrett, J.T. Phosphoinositide 3-Kinase (PI3K) Reactive Oxygen Species (ROS)-Activated Prodrug in Combination with Anthracycline Impairs PI3K Signaling, Increases DNA Damage Response and Reduces Breast Cancer Cell Growth. Int. J. Mol. Sci. 2021, 22, 2088. [Google Scholar] [CrossRef]
- Divekar, S.D.; Tiek, D.M.; Fernandez, A.; Riggins, R.B. Estrogen-Related Receptor β (ERRβ)—Renaissance Receptor or Receptor Renaissance? Nucl. Recept. Signal. 2016, 14, e002. [Google Scholar] [CrossRef] [PubMed]
- Heckler, M.M.; Riggins, R.B. ERRβ Splice Variants Differentially Regulate Cell Cycle Progression. Cell Cycle 2015, 14, 31–45. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Liu, Z.; Wu, J.; Liu, J.; Hyder, S.M.; Antoniou, E.; Lubahn, D.B. Identification and Characterization of Two Novel Splicing Isoforms of Human Estrogen-Related Receptor β. J. Clin. Endocr. Metab. 2006, 91, 569–579. [Google Scholar] [CrossRef]
- Rulíšek, L.; Vondrášek, J. Coordination Geometries of Selected Transition Metal Ions (Co2+, Ni2+, Cu2+, Zn2+, Cd2+, and Hg2+) in Metalloproteins. J. Inorg. Biochem. 1998, 71, 115–127. [Google Scholar] [CrossRef]
- Klatt Shaw, D.; Gunther, D.; Jurynec, M.J.; Chagovetz, A.A.; Ritchie, E.; Grunwald, D.J. Intracellular Calcium Mobilization Is Required for Sonic Hedgehog Signaling. Dev. Cell 2018, 45, 512–525.e5. [Google Scholar] [CrossRef]
- Mamaeva, O.A.; Kim, J.; Feng, G.; McDonald, J.M. Calcium/Calmodulin-Dependent Kinase II Regulates Notch-1 Signaling in Prostate Cancer Cells. J. Cell. Biochem. 2009, 106, 25–32. [Google Scholar] [CrossRef]
- Yao, B.; Zhang, S.; Wei, Y.; Tian, S.; Lu, Z.; Jin, L.; He, Y.; Xie, W.; Li, Y. Structural Insights into the Specificity of Ligand Binding and Coactivator Assembly by Estrogen-Related Receptor β. J. Mol. Biol. 2020, 432, 5460–5472. [Google Scholar] [CrossRef]
- Godt, J.; Scheidig, F.; Grosse-Siestrup, C.; Esche, V.; Brandenburg, P.; Reich, A.; Groneberg, D.A. The Toxicity of Cadmium and Resulting Hazards for Human Health. J. Occup. Med. Toxicol. 2006, 1, 22. [Google Scholar] [CrossRef]
- Friedman, R. Structural and Computational Insights into the Versatility of Cadmium Binding to Proteins. Dalton Trans. 2014, 43, 2878–2887. [Google Scholar] [CrossRef] [PubMed]
- Stoica, A.; Katzenellenbogen, B.S.; Martin, M.B. Activation of Estrogen Receptor-α by the Heavy Metal Cadmium. Mol. Endocrinol. 2000, 14, 545–553. [Google Scholar] [CrossRef]
- Maret, W.; Moulis, J.-M. The Bioinorganic Chemistry of Cadmium in the Context of Its Toxicity. In Cadmium: From Toxicity to Essentiality; Sigel, A., Sigel, H., Sigel, R.K., Eds.; Springer: Dordrecht, The Netherlands, 2013; pp. 1–29. ISBN 978-94-007-5179-8. [Google Scholar]
- Bourguet, W.; Ruff, M.; Chambon, P.; Gronemeyer, H.; Moras, D. Crystal Structure of the Ligand-Binding Domain of the Human Nuclear Receptor RXR-α. Nature 1995, 375, 377–382. [Google Scholar] [CrossRef] [PubMed]
- Brzozowski, A.M.; Pike, A.C.W.; Dauter, Z.; Hubbard, R.E.; Bonn, T.; Engström, O.; Öhman, L.; Greene, G.L.; Gustafsson, J.-Å.; Carlquist, M. Molecular Basis of Agonism and Antagonism in the Oestrogen Receptor. Nature 1997, 389, 753–758. [Google Scholar] [CrossRef]
- Renaud, J.-P.; Rochel, N.; Ruff, M.; Vivat, V.; Chambon, P.; Gronemeyer, H.; Moras, D. Crystal Structure of the RAR-γ Ligand-Binding Domain Bound to All-Trans Retinoic Acid. Nature 1995, 378, 681–689. [Google Scholar] [CrossRef]
- Shiau, A.K.; Barstad, D.; Loria, P.M.; Cheng, L.; Kushner, P.J.; Agard, D.A.; Greene, G.L. The Structural Basis of Estrogen Receptor/Coactivator Recognition and the Antagonism of This Interaction by Tamoxifen. Cell 1998, 95, 927–937. [Google Scholar] [CrossRef]
- Tanenbaum, D.M.; Wang, Y.; Williams, S.P.; Sigler, P.B. Crystallographic Comparison of the Estrogen and Progesterone Receptor’s Ligand Binding Domains. Proc. Natl. Acad. Sci. USA 1998, 95, 5998–6003. [Google Scholar] [CrossRef] [PubMed]
- Wagner, R.L.; Apriletti, J.W.; McGrath, M.E.; West, B.L.; Baxter, J.D.; Fletterick, R.J. A Structural Role for Hormone in the Thyroid Hormone Receptor. Nature 1995, 378, 690–697. [Google Scholar] [CrossRef]
- Wurtz, J.-M.; Bourguet, W.; Renaud, J.-P.; Vivat, V.; Chambon, P.; Moras, D.; Gronemeyer, H. A Canonical Structure for the Ligand-Binding Domain of Nuclear Receptors. Nat. Struct. Mol. Biol. 1996, 3, 87–94, Erratum in Nat. Struct. Mol. Biol. 1996, 3, 206. [Google Scholar] [CrossRef] [PubMed]
- Egea, P.F.; Mitschler, A.; Rochel, N.; Ruff, M.; Chambon, P.; Moras, D. Crystal Structure of the Human RXRα Ligand-binding Domain Bound to Its Natural Ligand: 9-cis Retinoic Acid. EMBO J. 2000, 19, 2592–2601. [Google Scholar] [CrossRef] [PubMed]
- Hamdaoui, L.; El Feki, H.; Ben Amor, M.; Oudadesse, H.; Badraoui, R.; Khalil, N.; Brahmi, F.; Jilani, S.; Aloufi, B.; Ben Amara, I.; et al. Physicochemical Exploration and Computational Analysis of Bone after Subchronic Exposure to Kalach 360 SL in Female Wistar Rats. Toxics 2025, 13, 456. [Google Scholar] [CrossRef]
- Harding, M.M. The Architecture of Metal Coordination Groups in Proteins. Acta Crystallogr. Sect. D 2004, 60, 849–859. [Google Scholar] [CrossRef]
- Kuwahara, K.; Angkawidjaja, C.; Matsumura, H.; Koga, Y.; Takano, K.; Kanaya, S. Importance of the Ca2+-Binding Sites in the N-Catalytic Domain of a Family I.3 Lipase for Activity and Stability. Protein Eng. Des. Sel. 2008, 21, 737–744. [Google Scholar] [CrossRef]
- DeOme, K.B.; Faulkin, L.J.J.; Bern, H.A.; Blair, P.B. Development of Mammary Tumors from Hyperplastic Alveolar Nodules Transplanted into Gland-Free Mammary Fat Pads of Female C3H Mice. Cancer Res. 1959, 19, 515–520. [Google Scholar] [CrossRef]
- Fu, N.Y.; Nolan, E.; Lindeman, G.J.; Visvader, J.E. Stem Cells and the Differentiation Hierarchy in Mammary Gland Development. Physiol. Rev. 2019, 100, 489–523. [Google Scholar] [CrossRef]
- Rangel, M.C.; Bertolette, D.; Castro, N.P.; Klauzinska, M.; Cuttitta, F.; Salomon, D.S. Developmental Signaling Pathways Regulating Mammary Stem Cells and Contributing to the Etiology of Triple-Negative Breast Cancer. Breast Cancer Res. Treat. 2016, 156, 211–226. [Google Scholar] [CrossRef]
- Visvader, J.E.; Stingl, J. Mammary Stem Cells and the Differentiation Hierarchy: Current Status and Perspectives. Genes Dev. 2014, 28, 1143–1158. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Zhang, R.; Shen, Q.; Zhu, Z.; Zhang, J.; Wu, X.; Zhao, W.; Li, N.; Yang, F.; Wei, H.; et al. ESRRB Facilitates the Conversion of Trophoblast-Like Stem Cells From Induced Pluripotent Stem Cells by Directly Regulating CDX2. Front. Cell Dev. Biol. 2021, 9, 712224. [Google Scholar] [CrossRef]
- Fernandez, A.I.; Geng, X.; Chaldekas, K.; Harris, B.; Duttargi, A.; Berry, V.L.; Berry, D.L.; Mahajan, A.; Cavalli, L.R.; Győrffy, B.; et al. The Orphan Nuclear Receptor Estrogen-Related Receptor Beta (ERRβ) in Triple-Negative Breast Cancer. Breast Cancer Res. Treat. 2020, 179, 585–604. [Google Scholar] [CrossRef]
- Naik, S.K.; Lam, E.W.-F.; Parija, M.; Prakash, S.; Jiramongkol, Y.; Adhya, A.K.; Parida, D.K.; Mishra, S.K. NEDDylation Negatively Regulates ERRβ Expression to Promote Breast Cancer Tumorigenesis and Progression. Cell Death Dis. 2020, 11, 703. [Google Scholar] [CrossRef]
- Sengupta, D.; Bhargava, D.K.; Dixit, A.; Sahoo, B.S.; Biswas, S.; Biswas, G.; Mishra, S.K. ERRβ Signalling through FST and BCAS2 Inhibits Cellular Proliferation in Breast Cancer Cells. Br. J. Cancer 2014, 110, 2144–2158. [Google Scholar] [CrossRef]
- Yu, S.; Wong, Y.C.; Wang, X.H.; Ling, M.T.; Ng, C.F.; Chen, S.; Chan, F.L. Orphan Nuclear Receptor Estrogen-Related Receptor-β Suppresses in Vitro and in Vivo Growth of Prostate Cancer Cells via p21WAF1/CIP1 Induction and as a Potential Therapeutic Target in Prostate Cancer. Oncogene 2008, 27, 3313–3328. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.-S.; Zheng, P.-S. ESRRB Inhibits the TGFβ Signaling Pathway to Drive Cell Proliferation in Cervical Cancer. Cancer Res. 2023, 83, 3095–3114. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.; Lin, Y. P53 Switches off Pluripotency on Differentiation. Stem Cell Res. Ther. 2017, 8, 44. [Google Scholar] [CrossRef] [PubMed]
- Aurilio, G.; Disalvatore, D.; Pruneri, G.; Bagnardi, V.; Viale, G.; Curigliano, G.; Adamoli, L.; Munzone, E.; Sciandivasci, A.; De Vita, F.; et al. A Meta-Analysis of Oestrogen Receptor, Progesterone Receptor and Human Epidermal Growth Factor Receptor 2 Discordance between Primary Breast Cancer and Metastases. Eur. J. Cancer 2014, 50, 277–289. [Google Scholar] [CrossRef]
- McAnena, P.F.; McGuire, A.; Ramli, A.; Curran, C.; Malone, C.; McLaughlin, R.; Barry, K.; Brown, J.A.L.; Kerin, M.J. Breast Cancer Subtype Discordance: Impact on Post-Recurrence Survival and Potential Treatment Options. BMC Cancer 2018, 18, 203, Erratum in BMC Cancer 2018, 18, 282. [Google Scholar] [CrossRef]
- Yeung, C.; Hilton, J.; Clemons, M.; Mazzarello, S.; Hutton, B.; Haggar, F.; Addison, C.L.; Kuchuk, I.; Zhu, X.; Gelmon, K.; et al. Estrogen, Progesterone, and HER2/Neu Receptor Discordance between Primary and Metastatic Breast Tumours—A Review. Cancer Metastasis Rev. 2016, 35, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Psaltis, J.B.; Wang, Q.; Yan, G.; Gahtani, R.; Huang, N.; Haddad, B.R.; Martin, M.B. Cadmium Activation of Wild-Type and Constitutively Active Estrogen Receptor Alpha. Front. Endocrinol. 2024, 15, 1380047. [Google Scholar] [CrossRef]
- Lu, D.D.; Huang, N.; Li, S.-W.A.; Fang, J.R.; Lai, C.-H.; Wang, J.-K.; Chan, K.-S.; Johnson, M.D.; Lin, C.-Y. HAI-1 Is Required for the Novel Role of FGFBP1 in Maintenance of Cell Morphology and F-Actin Rearrangement in Human Keratinocytes. Human. Cell 2023, 36, 1403–1415. [Google Scholar] [CrossRef]
- O’Geen, H.; Echipare, L.; Farnham, P.J. Using ChIP-Seq Technology to Generate High-Resolution Profiles of Histone Modifications. Methods Mol. Biol. 2011, 791, 265–286. [Google Scholar] [CrossRef] [PubMed]
- Furlan-Magaril, M.; Rincón-Arano, H.; Recillas-Targa, F. Sequential Chromatin Immunoprecipitation Protocol: ChIP-reChIP. Methods Mol. Biol. 2009, 543, 253–266. [Google Scholar] [CrossRef]
- Chronis, C.; Fiziev, P.; Papp, B.; Butz, S.; Bonora, G.; Sabri, S.; Ernst, J.; Plath, K. Cooperative Binding of Transcription Factors Orchestrates Reprogramming. Cell 2017, 168, 442–459.e20. [Google Scholar] [CrossRef]
- Chen, X.; Xu, H.; Yuan, P.; Fang, F.; Huss, M.; Vega, V.B.; Wong, E.; Orlov, Y.L.; Zhang, W.; Jiang, J.; et al. Integration of External Signaling Pathways with the Core Transcriptional Network in Embryonic Stem Cells. Cell 2008, 133, 1106–1117. [Google Scholar] [CrossRef]
- Xie, L.; Torigoe, S.E.; Xiao, J.; Mai, D.H.; Li, L.; Davis, F.P.; Dong, P.; Marie-Nelly, H.; Grimm, J.; Lavis, L.; et al. A Dynamic Interplay of Enhancer Elements Regulates Klf4 Expression in Naïve Pluripotency. Genes Dev. 2017, 31, 1795–1808. [Google Scholar] [CrossRef]
- Potter, C.J.; Tasic, B.; Russler, E.V.; Liang, L.; Luo, L. The Q System: A Repressible Binary System for Transgene Expression, Lineage Tracing, and Mosaic Analysis. Cell 2010, 141, 536–548. [Google Scholar] [CrossRef]
- Schleiss, M.R.; Degnin, C.R.; Geballe, A.P. Translational Control of Human Cytomegalovirus Gp48 Expression. J. Virol. 1991, 65, 6782–6789. [Google Scholar] [CrossRef]
- Taube, R.; Lin, X.; Irwin, D.; Fujinaga, K.; Peterlin, B.M. Interaction between P-TEFb and the C-Terminal Domain of RNA Polymerase II Activates Transcriptional Elongation from Sites Upstream or Downstream of Target Genes. Mol. Cell. Biol. 2002, 22, 321–331. [Google Scholar] [CrossRef] [PubMed]
- How to Perform Docking in a Specific Binding Site Using AutoDock Vina? Available online: https://bioinformaticsreview.com/20161214/how-to-perform-docking-in-a-specific-binding-site-using-autodock-vina/ (accessed on 13 July 2023).
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]


| No. | Protein | Metal | H2O | Affinity (Kcal/mol) | RMSD | Amino Acid in Interaction | Chains | Helix |
|---|---|---|---|---|---|---|---|---|
| 1 | 6LN4 (Apo) | Ca/Cd | Yes | −1.5 | 0 | S278, E282 | H4, H5 | |
| 2 | 6LN4 (Apo) | Ca/Cd | Yes | −1.3 | 14.67 | G270, D271, S274, Y356 | H4 | |
| 3 | 6LN4 (Apo) | Ca/Cd | No | −1.3 | 9.499 | G270, D271, S274, Y356 | H4 | |
| 4 | 6LN4 (Apo) | Ca/Cd | No | −1.2 | 7.908 | S274, S278 | H4, H4–5 | |
| 5 | 6LN4 (Apo) | Cd | Yes | −1.1 | 3.956 | Q277, S278 | H4, H4–5 | |
| 6 | 6LIT (Holo) | Ca | Yes | −1.5 | 30.673 | S278 | dimer-A | H4–5 |
| 7 | 6LIT (Holo) | Ca/Cd | Yes | −1.5 | 0–7.292 | S278 | monomer | H4–5 |
| 8 | 6LIT (Holo) | Ca/Cd | Yes | −1.5 | 29.55–30.966 | Q277, S278 | dimer-A/B | H4, H4–5 |
| 9 | 6LIT (Holo) | Ca/Cd | Yes | −1.5 | 1.349–10.131 | Q277, S278 | monomer | H4, H4–5 |
| 10 | 6LIT (Holo) | Ca/Cd | Yes | −1.4 | 24.96–27.243 | S278, S352 | dimer-A | H4–5, H8–9 |
| 11 | 6LIT (Holo) | Ca/Cd | Yes | −1.4 | 6.203–7.296 | S278, S352 | monomer | H4–5, H8–9 |
| 12 | 6LIT (Holo) | Ca | Yes | −1.5 | 27.288 | Q401 | dimer-B | H10 + H11 |
| 13 | 6LIT (Holo) | Cd | No | −1.3 | 22.767 | E427, L429, E430 | monomer | H12 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Shi, X.; Yan, G.; Zhao, N.C.; Wang, Q.; Lu, D.; Lawler, D.; Gahtani, R.M.; Byrne, C.; Haddad, B.R.; Copeland, R.L.; et al. Calcium and Cadmium Activate ESRRB to Mediate Cell Stemness and Pluripotency. Int. J. Mol. Sci. 2026, 27, 231. https://doi.org/10.3390/ijms27010231
Shi X, Yan G, Zhao NC, Wang Q, Lu D, Lawler D, Gahtani RM, Byrne C, Haddad BR, Copeland RL, et al. Calcium and Cadmium Activate ESRRB to Mediate Cell Stemness and Pluripotency. International Journal of Molecular Sciences. 2026; 27(1):231. https://doi.org/10.3390/ijms27010231
Chicago/Turabian StyleShi, Xu, Gai Yan, Nicole C. Zhao, Qiaochu Wang, Dajun Lu, Destiny Lawler, Reem M. Gahtani, Celia Byrne, Bassem R. Haddad, Robert L. Copeland, and et al. 2026. "Calcium and Cadmium Activate ESRRB to Mediate Cell Stemness and Pluripotency" International Journal of Molecular Sciences 27, no. 1: 231. https://doi.org/10.3390/ijms27010231
APA StyleShi, X., Yan, G., Zhao, N. C., Wang, Q., Lu, D., Lawler, D., Gahtani, R. M., Byrne, C., Haddad, B. R., Copeland, R. L., & Martin, M. B. (2026). Calcium and Cadmium Activate ESRRB to Mediate Cell Stemness and Pluripotency. International Journal of Molecular Sciences, 27(1), 231. https://doi.org/10.3390/ijms27010231






