Nakaseomyces glabratus (Candida glabrata) MLST Genotypes in Central Poland
Abstract
1. Introduction
2. Results
2.1. Genotyping
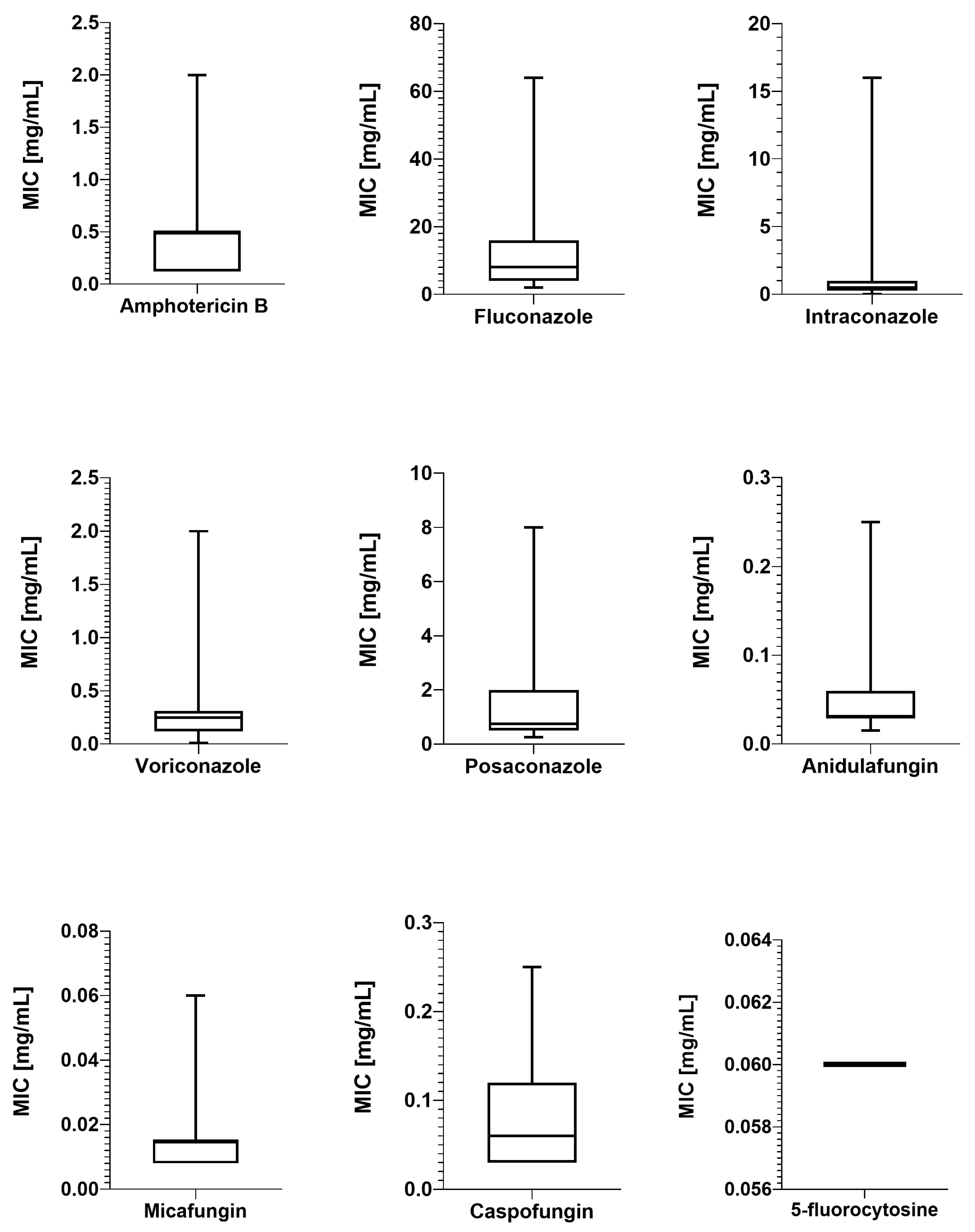
2.2. Antimicrobial Susceptibility Testing
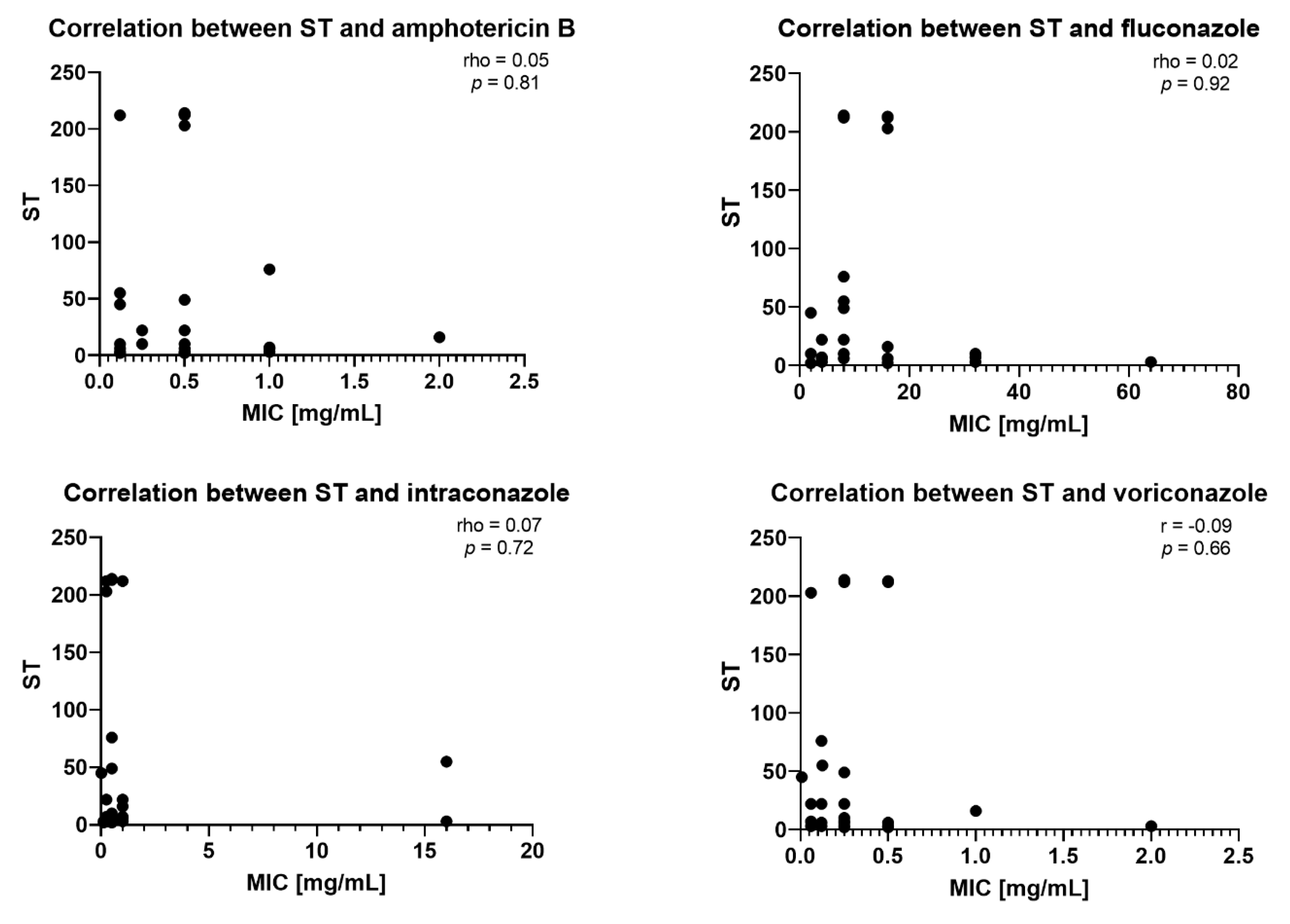
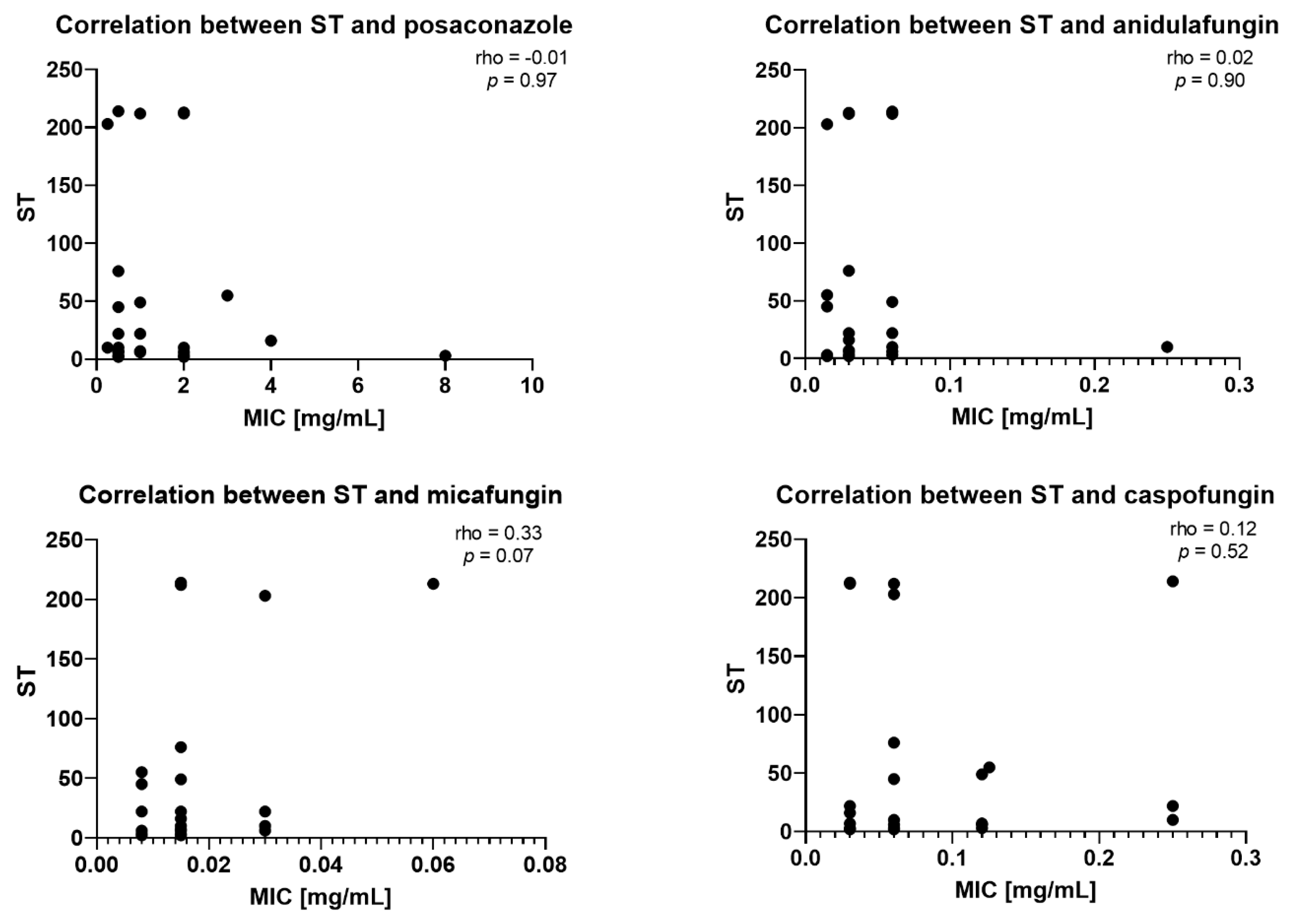
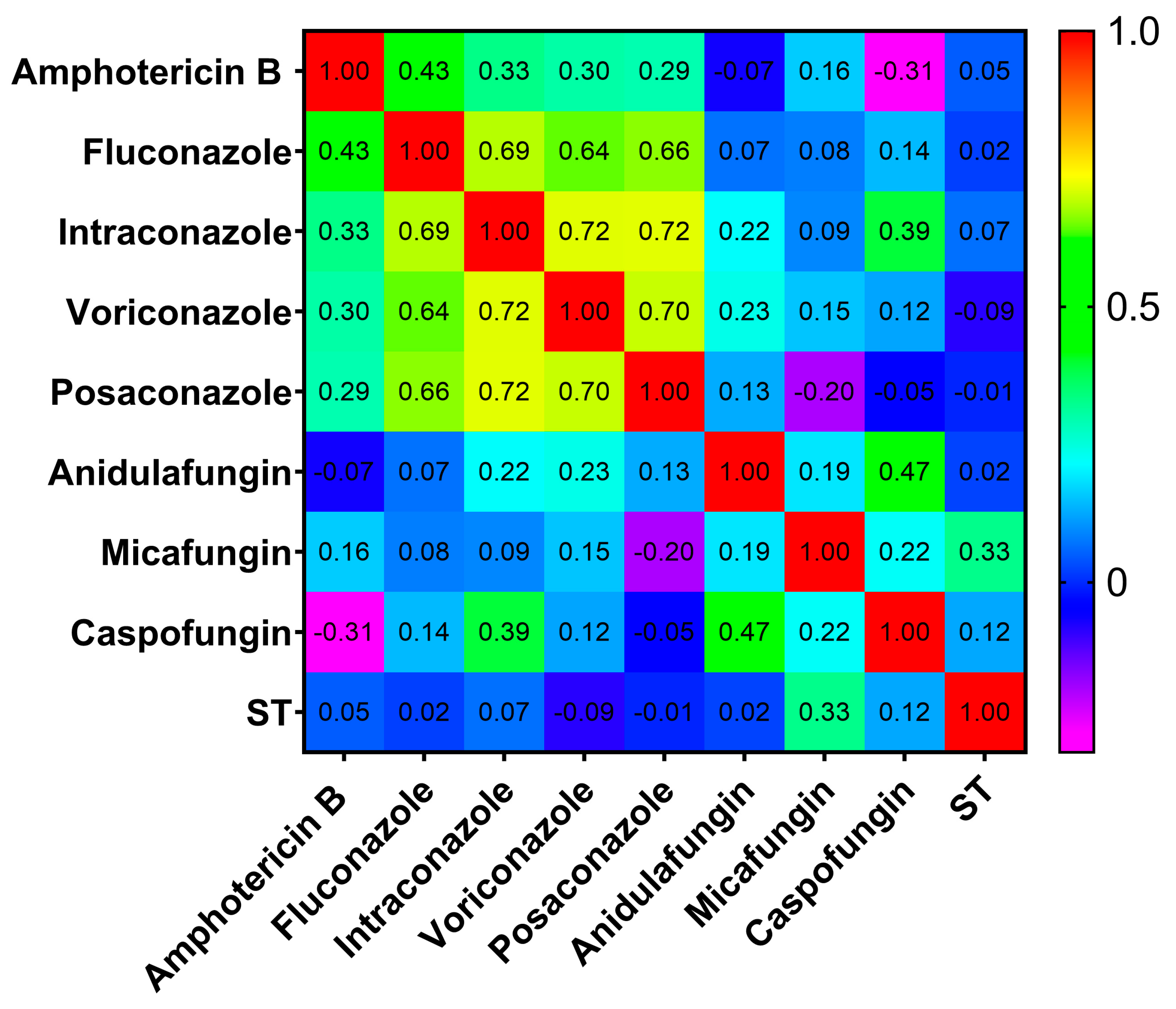
2.3. Correlation Between STs and MICs
3. Discussion
Comparison of Poland to the World STs
4. Materials and Methods
4.1. Clinical Strains and Identification
4.2. Genomic DNA Isolation
4.3. Genotyping by MLST
4.4. Sample Preparation Sequencing
4.5. Drug Susceptibility Testing
4.6. Statistical Analysis
4.7. Phylogenetic Analysis
5. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Chr | chromosome |
| MALDI ToF MS | matrix-assisted laser desorption/ionization time-of-flight mass spectrometry |
| ITS | internal transcribed spacer |
| MLST | multilocus sequence typing |
| ST | sequence type |
| STs | sequence types |
| FKS | 1,3-beta-glucan synthase |
| LEU2 | 3-isopropylmalate dehydrogenase |
| NMT1 | myristoyl-CoA protein N-myristoyltransferase |
| TRP1 | phosphoribosylanthranilate isomerase |
| UGP1 | UTP-glucose-1-phosphate uridylyltransferase |
| URA3 | orotidine-5′-phosphate decarboxylase |
| ID | identification number in the PubMLST database |
| AB | amphotericin B |
| FZ | fluconazole |
| IZ | itraconazole |
| VOR | voriconazole |
| PZ | posaconazole |
| AND | anidulafungin |
| MF | micafungin |
| CAS | caspofungin |
| FC | 5-fluorocytosine |
| MIC | minimal inhibitory concentration [mg/mL] |
| Interpr. | interpretation |
| S | susceptible |
| R | resistant |
| I | value between the S and the R breakpoint; |
| IE | insufficient evidence that the organism is a good target for therapy with the agent |
| NA | not available. |
| WT | wildtype |
| non-WT | non-wildtype |
| ECOFF | Epidemiological Cut-Off Value |
| EUCAST | The European Committee on Antimicrobial Susceptibility Testing |
| WGS | whole genome sequencing |
| NGS | next-generation sequencing |
References
- Lass-Flörl, C.; Kanj, S.S.; Govender, N.P.; Thompson GR3rd Ostrosky-Zeichner, L.; Govrins, M.A. Invasive candidiasis. Nat. Rev. Dis. Primers 2024, 10, 20. [Google Scholar] [CrossRef] [PubMed]
- Parambath, S.; Dao, A.; Kim, H.Y.; Zawahir, S.; Izquierdo, A.A.; Tacconelli, E.; Govender, N.; Oladele, R.; Colombo, A.; Sorrell, T.; et al. Candida albicans—A systematic review to inform the World Health Organization Fungal Priority Pathogens List. Med. Mycol. 2024, 62, myae045, Erratum in: Med. Mycol. 2024, 62, myae074. [Google Scholar] [CrossRef]
- Katsipoulaki, M.; Stappers, M.H.T.; Malavia-Jones, D.; Brunke, S.; Hube, B.; Gow, N.A.R. Candida albicans and Candida glabrata: Global priority pathogens. Microbiol. Mol. Biol. Rev. 2024, 88, e0002123. [Google Scholar] [CrossRef]
- Kumar, K.; Askari, F.; Sahu, M.S.; Kaur, R. Candida glabrata: A Lot More Than Meets the Eye. Microorganisms 2019, 7, 39. [Google Scholar] [CrossRef] [PubMed]
- Tortorano, A.M.; Prigitano, A.; Morroni, G.; Brescini, L.; Barchiesi, F. Candidemia: Evolution of Drug Resistance and Novel Therapeutic Approaches. Infect. Drug Resist. 2021, 14, 5543–5553. [Google Scholar] [CrossRef]
- Lockhart, S.R. Current epidemiology of Candida infection. Clin. Microbiol. Newsl. 2014, 36, 131–136. [Google Scholar] [CrossRef]
- Frías-De-León, M.G.; Hernández-Castro, R.; Conde-Cuevas, E.; García-Coronel, I.H.; Vázquez-Aceituno, V.A.; Soriano-Ursúa, M.A.; Farfán-García, E.D.; Ocharán-Hernández, E.; Rodríguez-Cerdeira, C.; Arenas, R.; et al. Candida glabrata Antifungal Resistance and Virulence Factors, a Perfect Pathogenic Combination. Pharmaceutics 2021, 13, 1529. [Google Scholar] [CrossRef]
- Deckers, C.; Bélik, F.; Khourssaji, M.; Plum, P.E.; Ausselet, N.; Bulpa, P.; Sonet, A.; Bihin, B.; Huang, T.D.; Denis, O.; et al. A decade of candidaemia: A comprehensive analysis of prognosis and risk factors at a Belgian tertiary hospital. Diagn. Microbiol. Infect. Dis. 2024, 110, 116493. [Google Scholar] [CrossRef] [PubMed]
- Healey, K.R.; Perlin, D.S. Fungal resistance to echinocandins and the MDR ohenomenon in Candida glabrata. J. Fungi 2018, 4, 105. [Google Scholar] [CrossRef]
- Pristov, K.E.; Ghannoum, M.A. Resistance of Candida to azoles and echinocandins worldwide. Clin. Microbiol. Infect. 2019, 25, 792–798. [Google Scholar] [CrossRef]
- Aldejohann, A.M.; Herz, M.; Martin, R.; Walther, G.; Kurzai, O. Emergence of resistant Candida glabrata in Germany. JAC Antimicrob. Resist. 2021, 3, dlab122. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Cerdeira, C.; Pinto-Almazán, R.; Saunte, D.M.L.; Hay, R.; Szepietowski, J.C.; Moreno-Coutiño, G.; Skerlev, M.; Prohic, A.; Martínez-Herrera, E. Virulence and resistance factors of Nakaseomyces glabratus (formerly known as Candida glabrata) in Europe: A systematic review. J. Eur. Acad. Dermatol. Venereol. 2025, 39, 377–388. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.K.; Wang, T. The conundrum of antifungal resistance: Emergence of Nakaseomyces glabratus (Candida glabrata) in Europe with global implications. J. Eur. Acad. Dermatol. Venereol. 2025, 39, 255–256. [Google Scholar] [CrossRef]
- Alexander, B.D.; Johnson, M.D.; Pfeiffer, C.D.; Jiménez-Ortigosa, C.; Catania, J.; Booker, R.; Castanheira, M.; Messer, S.A.; Perlin, D.S.; Pfaller, M.A. Increasing echinocandin resistance in Candida glabrata: Clinical failure correlates with presence of FKS mutations and elevated minimum inhibitory concentrations. Clin. Infect. Dis. 2013, 56, 1724–1732. [Google Scholar] [CrossRef]
- Katiyar, S.; Pfaller, M.; Edlind, T. Candida albicans and Candida glabrata clinical isolates exhibiting reduced echinocandin susceptibility. Antimicrob. Agents Chemother. 2006, 50, 2892–2894. [Google Scholar] [CrossRef]
- Ksiezopolska, E.; Schikora-Tamarit, M.À.; Beyer, R.; Nunez-Rodriguez, J.C.; Schüller, C.; Gabaldón, T. Narrow mutation-al signatures drive acquisition of multidrug resistance in the fungal pathogen Candida glabrata. Curr. Biol. 2021, 31, 5314–5326.e10. [Google Scholar] [CrossRef] [PubMed]
- Sanguinetti, M.; Posteraro, B.; Fiori, B.; Ranno, S.; Torelli, R.; Fadda, G. Mechanisms of azole resistance in clinical isolates of Candida glabrata collected during a hospital sur-vey of antifungal resistance. Antimicrob. Agents Chemother. 2005, 49, 668–679. [Google Scholar] [CrossRef]
- Ksiezopolska, E.; Schikora-Tamarit, M.À.; Carlos Nunez-Rodriguez, J.; Gabaldón, T. Long-term stability of acquired drug resistance and resistance associated mutations in the fungal pathogen Nakaseomyces glabratus (Candida glabrata). Front. Cell. Infect. Microbiol. 2024, 14, 1416509. [Google Scholar] [CrossRef]
- Alcoba-Flórez, J.; Méndez-Alvarez, S.; Cano, J.; Guarro, J.; Pérez-Roth, E.; del Pilar Arévalo, M. Phenotypic and molecular characterization of Candida nivariensis sp. nov., a possible new opportunistic fungus. J. Clin. Microbiol. 2005, 43, 4107–4111. [Google Scholar] [CrossRef]
- Miranda-Zapico, I.; Eraso, E.; Hernández-Almaraz, J.L.; López-Soria, L.M.; Carrillo-Muñoz, A.J.; Hernández-Molina, J.M.; Quindós, G. Prevalence and antifungal susceptibility patterns of new cryptic species inside the species complexes Candida parapsilosis and Candida glabrata among blood isolates from a Spanish tertiary hospital. J. Antimicrob. Chemother. 2011, 66, 2315–2322. [Google Scholar] [CrossRef][Green Version]
- Correia, A.; Sampaio, P.; James, S.; Pais, C. Candida bracarensis sp. nov., a novel anamorphic yeast species phenotypically similar to Candida glabrata. Int. J. Syst. Evol. Microbiol. 2006, 56 Pt 1, 313–317. [Google Scholar] [CrossRef] [PubMed]
- Schoch, C.L.; Ciufo, S.; Domrachev, M.; Hotton, C.L.; Kannan, S.; Khovanskaya, R.; Leipe, D.; Mcveigh, R.; O’Neill, K.; Robbertse, B.; et al. NCBI Taxonomy: A comprehensive update on curation, resources and tools. Database 2020, baaa062. [Google Scholar] [CrossRef]
- Angoulvant, A.; Guitard, J.; Hennequin, C. Old and new pathogenic Nakaseomyces species: Epidemiology, biology, identification, pathogenicity and antifungal resistance. FEMS Yeast Res. 2016, 16, fov114. [Google Scholar] [CrossRef] [PubMed]
- Borman, A.M.; Petch, R.; Linton, C.J.; Palmer, M.D.; Bridge, P.D.; Johnson, E.M. Candida nivariensis, an emerging pathogenic fungus with multidrug resistance to antifungal agents. J. Clin. Microbiol. 2008, 46, 933–938. [Google Scholar] [CrossRef] [PubMed]
- Takashima, M.; Sugita, T. Taxonomy of Pathogenic Yeasts Candida, Cryptococcus, Malassezia, and Trichosporon. Med. Mycol. J. 2022, 63, 119–132. [Google Scholar] [CrossRef]
- Koehler, P.; Stecher, M.; Cornely, O.A.; Koehler, D.; Vehreschild, M.J.G.T.; Bohlius, J.; Wisplinghoff, H.; Vehreschild, J.J. Morbidity and mortality of candidaemia in Europe: An epidemiologic meta-analysis. Clin. Microbiol. Infect. 2019, 25, 1200–1212. [Google Scholar] [CrossRef]
- Dai, Z.; Lan, X.; Cai, M.; Liao, Y.; Zhang, J.; Ye, N.; Lu, X.; Wang, J.; Xiao, Y.; Zhang, Y.; et al. Nineteen years retrospective analysis of epidemiology, antifungal resistance and a nomogram model for 30-day mortality in nosocomial candidemia patients. Front. Cell. Infect. Microbiol. 2025, 15, 1504866. [Google Scholar] [CrossRef]
- Acosta-Mosquera, Y.; Tapia, J.C.; Armas-González, R.; Cáceres-Valdiviezo, M.J.; Fernández-Cadena, J.C.; Andrade-Molina, D. Prevalence and Species Distribution of Candida Clinical Isolates in a Tertiary Care Hospital in Ecuador Tested from January 2019 to February 2020. J. Fungi 2024, 10, 304. [Google Scholar] [CrossRef]
- Sikora, M.; Kuthan, R.; Piskorska-Malolepsza, K.; Golas-Pradzynska, M.; Domański, D.; Augustynowicz-Kopeć, E.; Swoboda-Kopec, E. Prevalence and Antifungal Susceptibility of the Emerging Fungal Species, Candida nivariensis, Isolated in a Teaching Hospital in Poland. Pol. J. Microbiol. 2019, 68, 303–308. [Google Scholar] [CrossRef]
- Domański, D.; Sikora, M.A.; Kuthan, R.T.; Augustynowicz-Kopeć, E.; Swoboda-Kopec, E. New species within Candida parapsilosis and Candida glabrata. Med. Dośw. Mikrobiol. 2019, 71, 51–57. [Google Scholar] [CrossRef]
- Suárez-Urquiza, P.; Pemán, J.; Gordon, M.; Favier, P.; Muñoz-Brell, P.; López-Hontangas, J.L.; Ruiz-Gaitán, A. Predicting Fungemia in the ICU: Unveiling the value of weekly fungal surveillance and yeast colonisation monitoring. J. Fungi 2024, 10, 674. [Google Scholar] [CrossRef]
- Sprague, J.L.; Kasper, L.; Hube, B. From intestinal colonization to systemic infections: Candida albicans translocation and dissemination. Gut Microbes 2022, 14, 2154548. [Google Scholar] [CrossRef] [PubMed]
- Hassan, Y.; Chew, S.Y.; Than, L.T.L. Candida glabrata: Pathogenicity and resistance mechanisms for adaptation and survival. J. Fungi 2021, 7, 667. [Google Scholar] [CrossRef] [PubMed]
- Radcliffe, C.; Patel, K.K.; Azar, M.M.; Koff, A.; Belfield, K.D.; Peaper, D.R.; Topal, J.E.; Malinis, M. Rectal screening for azole non-susceptible Candida species in patients undergoing liver transplantation. Transpl. Infect. Dis. 2022, 24, e13811. [Google Scholar] [CrossRef]
- Abe, M.; Sekizuka, T.; Miyazaki, Y. Gastrointestinal anaerobes and Enterococcus faecalis promote Candida glabrata gastrointestinal colonization and organ dissemination. J. Infect. Chemother. 2025, 31, 102658. [Google Scholar] [CrossRef] [PubMed]
- Skrzypek, M.S.; Binkley, J.; Binkley, G.; Miyasato, S.R.; Simison, M.; Sherlock, G. The Candida Genome Database (CGD): Incorporation of Assembly 22, systematic identifiers and visualization of high throughput sequencing data. Nucleic Acids Res. 2017, 45, D592–D596. [Google Scholar] [CrossRef]
- Dodgson, A.R.; Pujol, C.; Denning, D.W.; Soll, D.R.; Fox, A.J. Multilocus sequence typing of Candida glabrata reveals geographically enriched clades. J. Clin. Microbiol. 2003, 41, 5709–5717. [Google Scholar] [CrossRef]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications [version 1; peer review: 2 approved]. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef]
- Hallen-Adams, H.E.; Kachman, S.D.; Kim, J.; Legge, R.M.; Martínez, I. Fungi inhabiting the healthy human gastrointestinal tract: A diverse and dynamic community. Fungal Ecol. 2015, 15, 9–17. [Google Scholar] [CrossRef]
- Jiang, S.; Chen, Y.; Han, S.; Lv, L.; Li, L. Next-generation sequencing applications for the study of fungal pathogens. Microorganisms 2022, 10, 1882. [Google Scholar] [CrossRef]
- Auchtung, T.A.; Fofanova, T.Y.; Stewart, C.J.; Nash, A.K.; Wong, M.C.; Gesell, J.R.; Auchtung, J.M.; Ajami, N.J.; Petrosino, J.F. Investigating colonization of the healthy adult gastrointestinal tract by fungi. mSphere 2018, 3, e00092-18. [Google Scholar] [CrossRef] [PubMed]
- Biswas, C.; Chen, S.C.; Halliday, C.; Kennedy, K.; Playford, E.G.; Marriott, D.J.; Slavin, M.A.; Sorrell, T.C.; Sintchenko, V. Identification of genetic markers of resistance to echinocandins, azoles and 5-fluorocytosine in Candida glabrata by next-generation sequencing: A feasibility study. Clin. Microbiol. Infect. 2017, 23, e7–e676. [Google Scholar] [CrossRef]
- Dunaiski, C.M.; Kock, M.M.; Chan, W.Y.; Ismail, A.; Peters, R.P.H. Molecular epidemiology and antimicrobial resistance of vaginal Candida glabrata isolates in Namibia. Med. Mycol. 2024, 62, myae009. [Google Scholar] [CrossRef]
- Biswas, C.; Marcelino, V.R.; Van Hal, S.; Halliday, C.; Martinez, E.; Wang, Q.; Kidd, S.; Kennedy, K.; Marriott, D.; Morrissey, C.O.; et al. Whole genome sequencing of Australian Candida glabrata isolates reveals genetic diversity and novel sequence types. Front. Microbiol. 2018, 9, 2946, Erratum in: Front. Microbiol. 2019, 10, 2218. [Google Scholar] [CrossRef] [PubMed]
- Szarvas, J.; Rebelo, A.R.; Bortolaia, V.; Leekitcharoenphon, P.; Schrøder Hansen, D.; Nielsen, H.L.; Nørskov-Lauritsen, N.; Kemp, M.; Røder, B.L.; Frimodt-Møller, N.; et al. Danish whole-genome-sequenced Candida albicans and Candida glabrata samples fit into globally prevalent clades. J. Fungi 2021, 7, 962. [Google Scholar] [CrossRef] [PubMed]
- McDermott, A. Drug-resistant fungi on the rise. Proc. Natl. Acad. Sci. USA 2022, 119, e2217948119. [Google Scholar] [CrossRef]
- Friedman, D.Z.P.; Schwartz, I.S. Emerging fungal infections: New patients, new patterns, and new pathogens. J. Fungi 2019, 5, 67. [Google Scholar] [CrossRef]
- Granada, M.; Cook, E.; Sherlock, G.; Rosenzweig, F. Microbe Profile: Candida glabrata—A master of deception. Microbiology 2024, 170. [Google Scholar] [CrossRef]
- Jiménez Rosales, R.; Ayuso Carrasco, C.A.B.; Ojeda Hinojosa, M. The first reported case of colonic infection caused by Candida glabrata. Rev. Esp. Enferm. Dig. 2019, 111, 648. [Google Scholar] [CrossRef]
- Mori, T.; Matsumura, M.; Oguri, T. Analysis by pulsed-field gel electrophoresis of Candida albicans that developed resistance during antifungal therapy. Nippon Ishinkin Gakkai Zasshi 1998, 39, 229–233. [Google Scholar] [CrossRef]
- Shin, J.H.; Shin, D.H.; Song, J.W.; Kee, S.J.; Suh, S.P.; Ryang, D.W. Electrophoretic karyotype analysis of sequential Candida parapsilosis isolates from patients with persistent or pecurrent fungemia. J. Clin. Microbiol. 2001, 39, 1258–1263. [Google Scholar] [CrossRef] [PubMed]
- Clancy, C.J.; Barchiesi, F.; Falconi DiFrancesco, L.; Morris, A.J.; Snydman, D.R.; Yu, V.L.; Scalise, G.; Nguyen, M.H. Clinical manifestations and molecular epidemiology of late recurrent candidemia, and implications for management. Eur. J. Clin. Microbiol. Infect. Dis. 2000, 19, 585–592. [Google Scholar] [CrossRef]
- Benoit, D.; Decruyenaere, J.; Vandewoude, K.; Roosens, C.; Hoste, E.; Poelaert, J.; Vermassen, F.; Colardyn, F. Management of candidal thrombophlebitis of the central veins: Case report and review. Clin. Infect. Dis. 1998, 26, 393–397. [Google Scholar] [CrossRef]
- Khan, S.; Cai, L.; Bilal, H.; Khan, M.N.; Fang, W.; Zhang, D.; Yao, F.; Wang, X.; Wang, Q.; Hou, B.; et al. An 11-Year retrospective analysis of candidiasis epidemiology, risk factors, and antifungal susceptibility in a tertiary care hospital in China. Sci. Rep. 2025, 15, 7240. [Google Scholar] [CrossRef]
- Chew, K.L.; Achik, R.; Osman, N.H.; Octavia, S.; Teo, J.W.P. Genomic epidemiology of human candidaemia isolates in a tertiary hospital. Microb. Genom. 2023, 9, mgen001047. [Google Scholar] [CrossRef]
- Pyrpasopoulou, A.; Zarras, C.; Mouloudi, E.; Vakalis, G.; Ftergioti, A.; Kouroupis, D.; Papathanasiou, A.I.; Iosifidis, E.; Goumperi, S.; Lampada, C.; et al. Changing epidemiology of Candida spp. causing bloodstream infections in a tertiary hospital in Northern Greece: Appearance of Candida auris. Pathogens 2025, 14, 161. [Google Scholar] [CrossRef] [PubMed]
- Pérez, J.C. Fungi of the human gut microbiota: Roles and significance. Int. J. Med. Microbiol. 2021, 311, 151490. [Google Scholar] [CrossRef]
- Pultz, N.J.; Stiefel, U.; Ghannoum, M.; Helfand, M.S.; Donskey, C.J. Effect of parenteral antibiotic administration on establishment of intestinal colonization by Candida glabrata in adult mice. Antimicrob. Agents Chemother. 2005, 49, 438–440. [Google Scholar] [CrossRef] [PubMed]
- Ben-Ami, R.; Olshtain-Pops, K.; Krieger, M.; Oren, I.; Bishara, J.; Dan, M.; Wiener-Well, Y.; Weinberger, M.; Zimhony, O.; Chowers, M.; et al. Antibiotic exposure as a risk factor for fluconazole-resistant Candida bloodstream infection. Antimicrob. Agents Chemother. 2012, 56, 2518–2523. [Google Scholar] [CrossRef]
- WHO Fungal Priority Pathogens List to Guide Research, Development and Public Health Action; World Health Organization: Geneva, Switzerland, 2022.
- Sikora, M.; Gołaś, M.; Piskorska, K.; Swoboda-Kopeć, E. Genetic relatedness analysis of yeast-like fungal strains isolated from patients with total parenteral nutrition support. Postępy Nauk Medycznych 2015, 4b, 4–9. [Google Scholar]
- Paluchowska, P.; Tokarczyk, M.; Bogusz, B.; Skiba, I.; Budak, A. Molecular epidemiology of Candida albicans and Candida glabrata strains isolated from intensive care unit patients in Poland. Mem. Inst. Oswaldo Cruz. 2014, 109, 436–441. [Google Scholar] [CrossRef] [PubMed]
- Małek, M.; Paluchowska, P.; Bogusz, B.; Budak, A. Molecular characterization of Candida isolates from intensive care unit patients, Krakow, Poland. Rev. Iberoam. Micol. 2017, 34, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Tokarczyk, M.; Bogusz, B.; Węglowska-Jędryka, M.; Trojanowska, D.; Drygaś, A.; Nowak, P.; Mudyna, W.; Budak, A. Molecular typing of Candida albicans and Candida glabrata isolates from patients hospitalized in an Intensive Care Unit. Mikologia Lekarska 2010, 17, 207–210. [Google Scholar]
- Mushi, M.F.; Gross, U.; Mshana, S.E.; Bader, O. High diversity of Candida glabrata in a tertiary hospital-Mwanza, Tanzania. Med. Mycol. 2019, 57, 914–917. [Google Scholar] [CrossRef]
- Lott, T.J.; Frade, J.P.; Lyon, G.M.; Iqbal, N.; Lockhart, S.R. Bloodstream and non-invasive isolates of Candida glabrata have similar population structures and fluconazole susceptibilities. Med. Mycol. 2012, 50, 136–142. [Google Scholar] [CrossRef][Green Version]
- Khalifa, H.O.; Arai, T.; Majima, H.; Watanabe, A.; Kamei, K. Genetic basis of azole and echinocandin resistance in clinical Candida glabrata in Japan. Antimicrob. Agents Chemother. 2020, 64, e00783-20. [Google Scholar] [CrossRef]
- Odds, F.C.; Hanson, M.F.; Davidson, A.D.; Jacobsen, M.D.; Wright, P.; Whyte, J.A.; Gow, N.A.R.; Jones, B.L. One year prospective survey of Candida bloodstream infections in Scotland. J. Med. Microbiol. 2007, 56 Pt 8, 1066–1075. [Google Scholar] [CrossRef] [PubMed]
- Helmstetter, N.; Chybowska, A.D.; Delaney, C.; Da Silva Dantas, A.; Gifford, H.; Wacker, T.; Munro, C.; Warris, A.; Jones, B.; Cuomo, C.A.; et al. Population genetics and microevolution of clinical Candida glabrata reveals recombinant sequence types and hyper-variation within mitochondrial genomes, virulence genes, and drug targets. Genetics 2022, 221, iyac031. [Google Scholar] [CrossRef]
- Canela, H.M.S.; Cardoso, B.; Frazão, M.R.; Falcão, J.P.; Vitali, L.H.; Martinez, R.; da Silva Ferreira, M.E. Genetic diversity assessed using PFGE, MLP and MLST in Candida spp. candidemia isolates obtained from a Brazilian hospital. Braz. J. Microbiol. 2021, 52, 503–516. [Google Scholar] [CrossRef]
- Carreté, L.; Ksiezopolska, E.; Pegueroles, C.; Gómez-Molero, E.; Saus, E.; Iraola-Guzmán, S.; Loska, D.; Bader, O.; Fairhead, C.; Gabaldón, T. Patterns of genomic variation in the opportunistic Pathogen Candida glabrata suggest the existence of mating and a secondary association with humans. Curr. Biol. 2018, 28, 15–27.e7, Erratum in: Curr Biol. 2022, 32, 3219. [Google Scholar] [CrossRef]
- Håvelsrud, O.E.; Gaustad, P. Draft Genome sequences of Candida glabrata isolates 1A, 1B, 2A, 2B, 3A, and 3B. Genome Announc. 2017, 5, e00328-16. [Google Scholar] [CrossRef]
- Bordallo-Cardona, M.Á.; Agnelli, C.; Gómez-Nuñez, A.; Sánchez-Carrillo, C.; Bouza, E.; Muñoz, P.; Escribano, P.; Guinea, J. MSH2 gene point mutations are not antifungal resistance markers in Candida glabrata. Antimicrob. Agents Chemother. 2018, 63, e01876-18. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.; Joseph, L.; Parker, J.E.; Asadzadeh, M.; Kelly, S.L.; Meis, J.F.; Khan, Z. ERG6 and ERG2 are major targets conferring reduced susceptibility to amphotericin B in Clinical Candida glabrata Isolates in Kuwait. Antimicrob. Agents Chemother. 2019, 63, e01900-18. [Google Scholar] [CrossRef] [PubMed]
- Sasso, M.; Normand, A.C.; Pantel, A.; Bourgeois, N.; Lachaud, L. Evaluation of the DiversiLab® automated repetitive sequence-based PCR system for the characterization of Candida albicans and Candida glabrata isolates. J. Mycol. Med. 2018, 28, 320–326. [Google Scholar] [CrossRef]
- Healey, K.R.; Jimenez Ortigosa, C.; Shor, E.; Perlin, D.S. Genetic drivers of multidrug resistance in Candida glabrata. Front. Microbiol. 2016, 7, 1995. [Google Scholar] [CrossRef]
- Jensen, R.H.; Johansen, H.K.; Søes, L.M.; Lemming, L.E.; Rosenvinge, F.S.; Nielsen, L.; Olesen, B.; Kristensen, L.; Dzajic, E.; Astvad, K.M.; et al. Posttreatment antifungal resistance among colonizing Candida isolates in candidemia patients: Results from a systematic multicenter study. Antimicrob. Agents Chemother. 2015, 60, 1500–1508. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, J.; Ben Abid, F.; Salah, H.; Sundararaju, S.; Al Ismail, K.; Wang, K.; Sara Matthew, L.; Taj-Aldeen, S.; Ibrahim, E.B.; et al. Population genomic analyses reveal high diversity, recombination and nosocomial transmission among Candida glabrata (Nakaseomyces glabrata) isolates causing invasive infections. Microb. Genom. 2024, 10, 001179. [Google Scholar] [CrossRef] [PubMed]
- Amanloo, S.; Shams-Ghahfarokhi, M.; Ghahri, M.; Razzaghi-Abyaneh, M. Genotyping of clinical isolates of Candida glabrata from Iran by multilocus sequence typing and determination of population structure and drug resistance profile. Med. Mycol. 2018, 56, 207–215. [Google Scholar] [CrossRef]
- Gong, Y.B.; Jin, B.; Qi, H.; Zhang, R.; Zhang, X.Y.; Yuan, P.; Zhao, T.X.; Geng, X.H.; Zhang, M.; Zheng, J.L. Multilocus sequence typing of Candida albicans isolates from the oral cavities of patients undergoing haemodialysis. Sci. Rep. 2018, 8, 16413. [Google Scholar] [CrossRef]
- Takakura, S.; Ichiyama, S.; Bain, J.M.; Davidson, A.D.; Jacobsen, M.D.; Shaw, D.J.; Gow, N.A.; Odds, F.C. Comparison of Candida albicans strain types among isolates from three countries. Int. J. Med. Microbiol. 2008, 298, 663–668. [Google Scholar] [CrossRef]
- Meng, Q.; Wang, H.; Xiao, W.; Mai, W.; Liu, Y.; Xiao, Y.; Sui, J.; He, X.; Yin, F.; Xu, J. Prevalence, drug resistance and genetic diversity of Candida glabrata in the reproductive tract of pregnant women in Hainan and comparison with global multilocus sequence data. Mycology 2025, 1, 1–18. [Google Scholar] [CrossRef]
- Okolo, M.L.O.; Alaba, Z.A.; Samson, S.O.; Omatola, C.A.; Idache, B.M.; Omatola, J.A.; Okolo, E.U. Microbiological and molecular investigation of Candida spp. infection among women accessing antenatal care at Prince Abubakar Audu University Teaching Hospital Anyigba, North-Central Nigeria. Microbes Infect. Dis. 2023, 4, 1435–1445. [Google Scholar] [CrossRef]
- Fernandes, Â.; Azevedo, N.; Valente, A.; Dias, M.; Gomes, A.; Nogueira-Silva, C.; Henriques, M.; Silva, S.; Gonçalves, B. Vulvovaginal candidiasis and asymptomatic vaginal colonization in Portugal: Epidemiology, risk factors and antifungal pattern. Med. Mycol. 2022, 60, myac029. [Google Scholar] [CrossRef]
- Martínez-Herrera, E.; Frías-De-León, M.G.; Hernández-Castro, R.; García-Salazar, E.; Arenas, R.; Ocharan-Hernández, E.; Rodríguez-Cerdeira, C. Antifungal Resistance in Clinical Isolates of Candida glabrata in Ibero-America. J. Fungi 2021, 8, 14. [Google Scholar] [CrossRef] [PubMed]
- Chesdachai, S.; Yetmar, Z.A.; Ranganath, N.; Everson, J.J.; Wengenack, N.L.; Abu Saleh, O.M. Antifungal Susceptibility Pattern of Candida glabrata from a Referral Center and Reference Laboratory: 2012-2022. J. Fungi 2023, 9, 821. [Google Scholar] [CrossRef] [PubMed]
- Zarrinfar, H.; Kord, Z.; Fata, A. High incidence of azole resistance among Candida albicans and C. glabrata isolates in Northeastern Iran. Curr. Med. Mycol. 2021, 7, 18–21. [Google Scholar] [CrossRef]
- Huang, S.J.; Lv, G.; Song, Y.H.; Zhao, J.T.; Liu, J.Y.; Wang, L.L.; Xiang, M.J. Antifungal susceptibility, mo-lecular epidemiology, and clinical risk factors of Candida glabrata in intensive care unit in a Chinese Tertiary Hospital. Front. Cell. Infect. Microbiol. 2024, 14, 1455145. [Google Scholar] [CrossRef]
- Asadzadeh, M.; Ahmad, S.; Al-Sweih, N.; Khan, Z. Molecular fingerprinting by multi-locus sequence typing identifies microevolution and nosocomial transmission of Candida glabrata in Kuwait. Front. Public Health 2023, 11, 1242622. [Google Scholar] [CrossRef]
- Zhang, G.; Chen, Y.; Chen, J.; Yao, D. Association of multilocus sequence typing, MSH2 gene mutations, and antifungal resistance in Candida glabrata: Implications for clinical outcomes in Chinese hospitals. Ann. Clin. Microbiol. Antimicrob. 2024, 23, 100. [Google Scholar] [CrossRef]
- Byun, S.A.; Won, E.J.; Kim, M.N.; Lee, W.G.; Lee, K.; Lee, H.S.; Uh, Y.; Healey, K.R.; Perlin, D.S.; Choi, M.J.; et al. Multilocus Sequence Typing (MLST) Genotypes of Candida glabrata Bloodstream Isolates in Korea: Association With Antifungal Resistance, Mutations in Mismatch Repair Gene (Msh2), and Clinical Outcomes. Front. Microbiol. 2018, 9, 1523. [Google Scholar] [CrossRef]
- Kurtzman, C.P.; Robnett, C.J. Phylogenetic relationships among yeasts of the ‘Saccharomyces complex’ determined from multigene sequence analyses. FEMS Yeast Res. 2003, 3, 417–432. [Google Scholar] [CrossRef] [PubMed]
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs for Antifungal Agents, Version 11.0, 2024. Available online: http://www.eucast.org/astoffungi/clinicalbreakpointsforantifungals/ (accessed on 15 March 2025).
- Kumar, S.; Stecher, G.; Suleski, M.; Sanderford, M.; Sharma, S.; Tamura, K. MEGA12: Molecular Evolutionary Genetic Analysis Version 12 for Adaptive and Green Computing. Mol. Biol. Evol. 2024, 41, msae263. [Google Scholar] [CrossRef] [PubMed]
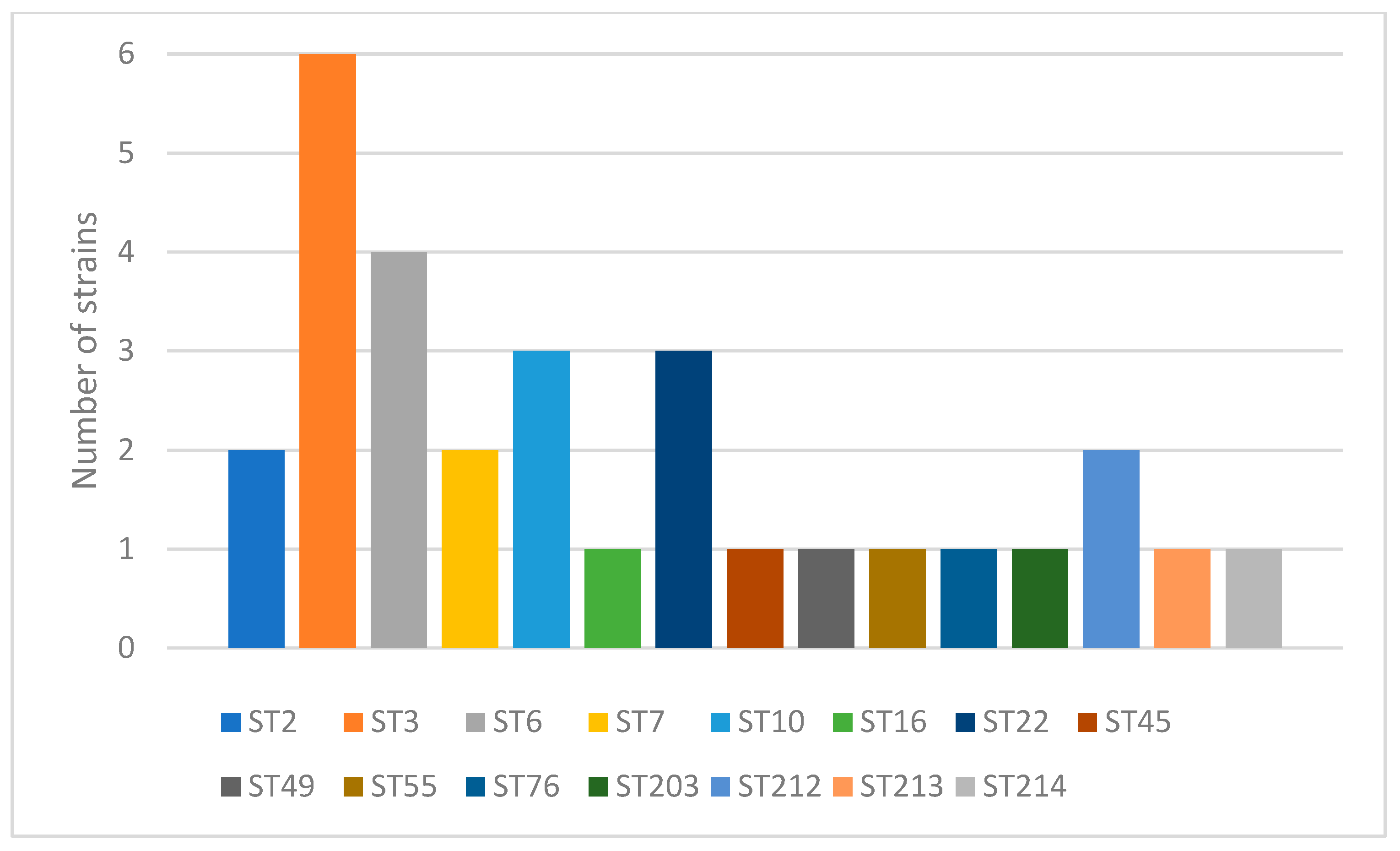
| ID in the PubMLST Database | Genotype | Sequence Type | |||||
|---|---|---|---|---|---|---|---|
| FKS | LEU2 | NMT1 | TRP1 | UGP1 | URA3 | ||
| 1936 | 1 | 2 | 2 | 1 | 1 | 1 | 2 |
| 1937 | 1 | 2 | 2 | 1 | 1 | 1 | 2 |
| 1914 | 5 | 7 | 8 | 7 | 3 | 6 | 3 |
| 1915 | 5 | 7 | 8 | 7 | 3 | 6 | 3 |
| 1924 | 5 | 7 | 8 | 7 | 3 | 6 | 3 |
| 1925 | 5 | 7 | 8 | 7 | 3 | 6 | 3 |
| 1926 | 5 | 7 | 8 | 7 | 3 | 6 | 3 |
| 1939 | 5 | 7 | 8 | 7 | 3 | 6 | 3 |
| 1919 | 2 | 5 | 7 | 5 | 1 | 2 | 6 |
| 1930 | 2 | 5 | 7 | 5 | 1 | 2 | 6 |
| 1931 | 2 | 5 | 7 | 5 | 1 | 2 | 6 |
| 1932 | 2 | 5 | 7 | 5 | 1 | 2 | 6 |
| 1917 | 3 | 4 | 4 | 3 | 3 | 4 | 7 |
| 1927 | 3 | 4 | 4 | 3 | 3 | 4 | 7 |
| 1928 | 8 | 4 | 3 | 5 | 1 | 2 | 10 |
| 1929 | 8 | 4 | 3 | 5 | 1 | 2 | 10 |
| 1933 | 8 | 4 | 3 | 5 | 1 | 2 | 10 |
| 1916 | 7 | 7 | 11 | 10 | 5 | 9 | 16 |
| 1918 | 7 | 5 | 6 | 12 | 1 | 8 | 22 |
| 1920 | 7 | 5 | 6 | 12 | 1 | 8 | 22 |
| 1922 | 7 | 5 | 6 | 12 | 1 | 8 | 22 |
| 1934 | 7 | 9 | 26 | 17 | 9 | 2 | 45 |
| 1921 | 6 | 6 | 29 | 2 | 3 | 4 | 49 |
| 1938 | 3 | 6 | 22 | 2 | 3 | 9 | 55 |
| 1923 | 10 | 14 | 12 | 11 | 3 | 2 | 76 |
| 1935 | 7 | 33 | 6 | 12 | 1 | 29 | 203 |
| 1383 | 1 | 23 | 2 | 1 | 1 | 1 | 212 |
| 1384 | 1 | 23 | 2 | 1 | 1 | 1 | 212 |
| 1385 | 20 | 13 | 21 | 9 | 3 | 2 | 213 |
| 1386 | 3 | 21 | 26 | 13 | 17 | 4 | 214 |
| FKS | LEU2 | NMT1 | TRP1 | UGP1 | URA3 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Allelic Type | Number of Incidence | Allelic Type | Number of Incidence | Allelic Type | Number of Incidence | Allelic Type | Number of Incidence | Allelic Type | Number of Incidence | Allelic Type | Number of Incidence |
| 1 | 4 | 2 | 2 | 2 | 4 | 1 | 4 | 1 | 16 | 1 | 4 |
| 2 | 4 | 4 | 5 | 3 | 3 | 2 | 2 | 3 | 12 | 2 | 10 |
| 3 | 4 | 5 | 7 | 4 | 2 | 3 | 2 | 5 | 1 | 4 | 4 |
| 5 | 6 | 6 | 2 | 6 | 4 | 5 | 7 | 9 | 1 | 6 | 6 |
| 6 | 1 | 7 | 7 | 7 | 4 | 7 | 6 | 17 | 1 | 8 | 3 |
| 7 | 6 | 9 | 1 | 8 | 6 | 9 | 1 | 9 | 2 | ||
| 8 | 3 | 13 | 1 | 11 | 1 | 10 | 1 | 29 | 1 | ||
| 10 | 1 | 14 | 1 | 12 | 1 | 11 | 1 | ||||
| 20 | 1 | 21 | 1 | 21 | 1 | 12 | 4 | ||||
| 23 | 2 | 22 | 1 | 13 | 1 | ||||||
| 33 | 1 | 26 | 2 | 17 | 1 | ||||||
| 29 | 1 | ||||||||||
| Primer Name | Primer Sequence | Target Gene | Concentration for PCR | Concentration for Sanger Sequencing | Primer Length [nt] | Product Size [bp] | Reference |
|---|---|---|---|---|---|---|---|
| FKS-F | GTCAAATGCCACAACAACAACCT | 1,3-beta-glucan synthase | 10 pM/µL | 1 pM/µL | 23 | 589 | [37] |
| FKS-R | AGCACTTCAGCAGCGTCTTCAG | 10 pM/µL | 1 pM/µL | 22 | |||
| LEU2-F | TTTCTTGTATCCTCCCATTGTTCA | 3-isopropylmalate dehydrogenase | 10 pM/µL | 1 pM/µL | 24 | 512 | |
| LEU2-R | ATAGGTAAAGGTGGGTTGTGTTGC | 10 pM/µL | 1 pM/µL | 24 | |||
| NMT1-F | GCCGGTGTGGTGTTGCCTGCTC | myristoyl-CoA, protein N-myristoyltransferasea | 10 pM/µL | 1 pM/µL | 22 | 607 | |
| NMT1-R | CGTTACTGCGGTGCTCGGTGTCG | 10 pM/µL | 1 pM/µL | 23 | |||
| TRP1-F | AATTGTTCCAGCGTTTTTGT | phosphoribosyl-anthranilate isomerase | 10 pM/µL | 1 pM/µL | 20 | 419 | |
| TRP1-R | GACCAGTCCAGCTCTTTCAC | 10 pM/µL | 1 pM/µL | 20 | |||
| UGP1-F | TTTCAACACCGACAAGGACACAGA | UTP-glucose-1-phosphate uridylyltransferase | 10 pM/µL | 1 pM/µL | 24 | 616 | |
| UGP-R | TCGGACTTCACTAGCAGCAAATCA | 10 pM/µL | 1 pM/µL | 24 | |||
| URA3-F | AGCGAATTGTTGAAGTTGGTTGA | orotidine-5′-phosphate decarboxylase | 10 pM/µL | 1 pM/µL | 23 | 602 | |
| URA3-R | AATTCGGTTGTAAGATGATGTTGC | 10 pM/µL | 1 pM/µL | 24 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuthan, R. Nakaseomyces glabratus (Candida glabrata) MLST Genotypes in Central Poland. Int. J. Mol. Sci. 2025, 26, 4407. https://doi.org/10.3390/ijms26094407
Kuthan R. Nakaseomyces glabratus (Candida glabrata) MLST Genotypes in Central Poland. International Journal of Molecular Sciences. 2025; 26(9):4407. https://doi.org/10.3390/ijms26094407
Chicago/Turabian StyleKuthan, Robert. 2025. "Nakaseomyces glabratus (Candida glabrata) MLST Genotypes in Central Poland" International Journal of Molecular Sciences 26, no. 9: 4407. https://doi.org/10.3390/ijms26094407
APA StyleKuthan, R. (2025). Nakaseomyces glabratus (Candida glabrata) MLST Genotypes in Central Poland. International Journal of Molecular Sciences, 26(9), 4407. https://doi.org/10.3390/ijms26094407






