Advances in the Evolutionary Mechanisms and Genomic Studies of Sexual Differentiation in Lauraceae Plants
Abstract
1. Introduction
2. Sex Determination Mechanisms in Angiosperms
2.1. Sex Chromosome Systems: Evolution from Autosomes to Sex-Determining Factors
- (1)
- Dosage-dependent sex determination: In Rumex and Humulus, sex is determined by the ratio of X chromosomes to autosomes (X/A). For example, a female phenotype arises when the X/A ratio ≥ 1, a male phenotype when ≤0.5, and intersex flowers develop at intermediate ratios [11].
- (2)
- (3)
- Studies in Silene latifolia and Phoenix dactylifera reveal a positive correlation between sex chromosome heterochromatization and recombination suppression, supporting the “degeneration–differentiation” model of sex chromosome evolution. According to this model, sex chromosomes originate from a pair of autosomes, with differentiation triggered by the emergence of a sex-determining gene. Chromosomal inversions near this gene suppress recombination, leading to progressive gene loss and accumulation of repetitive sequences. Heterochromatization, marked by transposon proliferation and stabilized by DNA methylation and other epigenetic modifications, reinforces recombination suppression, ultimately resulting in heteromorphic sex chromosomes (e.g., XY/ZW systems) [17].
2.2. Sex-Determining Genes: From Single-Gene Regulation to Multifactor Interaction Networks
2.2.1. Strict Dioecy Systems
2.2.2. Pseudo-Dioecy Systems
2.3. Epigenetic Regulation: A Multi-Layered Network Integrating Transposon Dynamics
2.3.1. DNA Methylation Synergizes with Metabolic–Hormonal Signaling
2.3.2. Spatiotemporal Specificity of Histone Modifications
2.3.3. Non-Coding RNAs Drive the Evolution of Sex Determination Systems
3. Floral Development and Morphological Diversity in Lauraceae
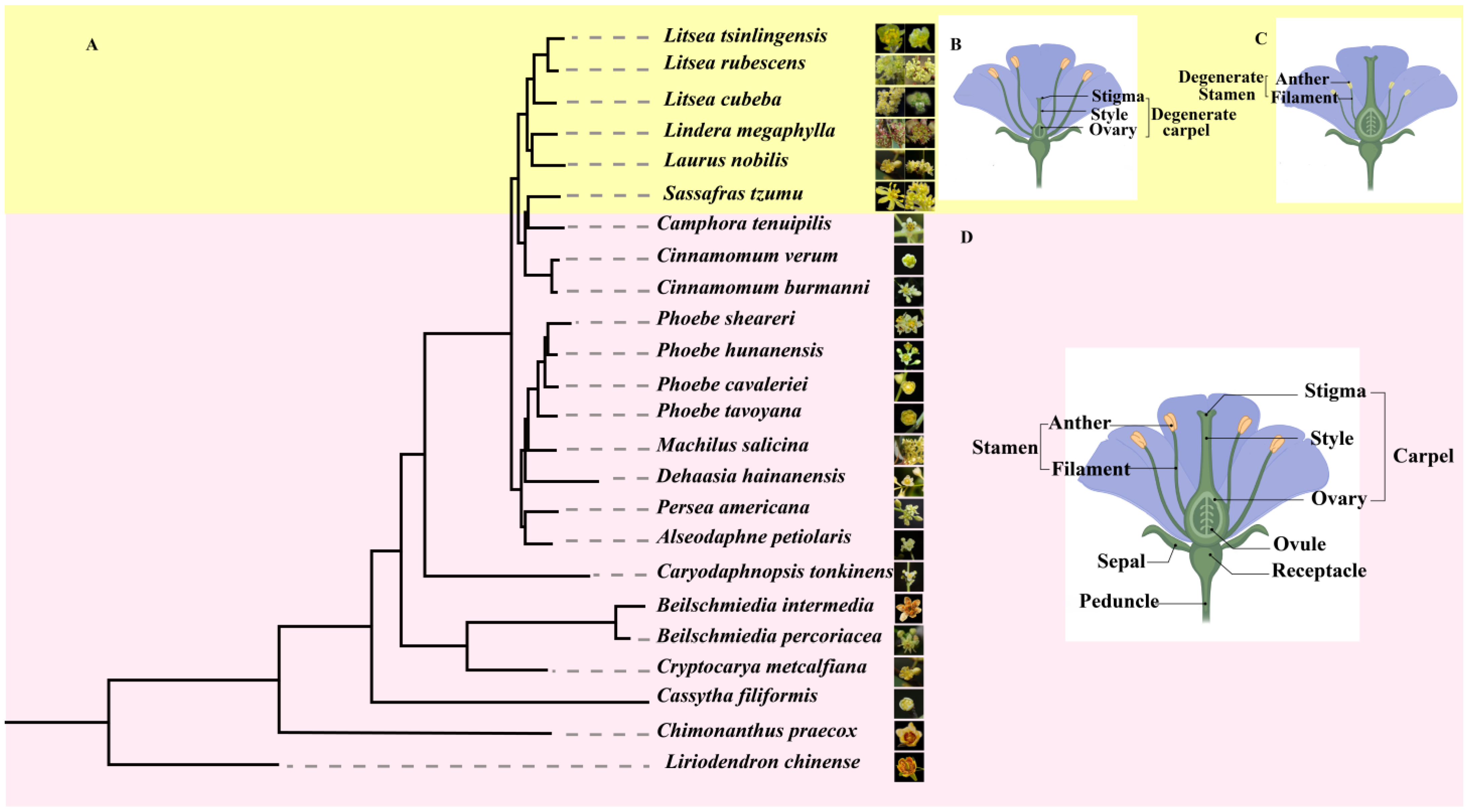
3.1. Evolution of Lauraceae Classification Systems and Floral MorphologicalMechanisms
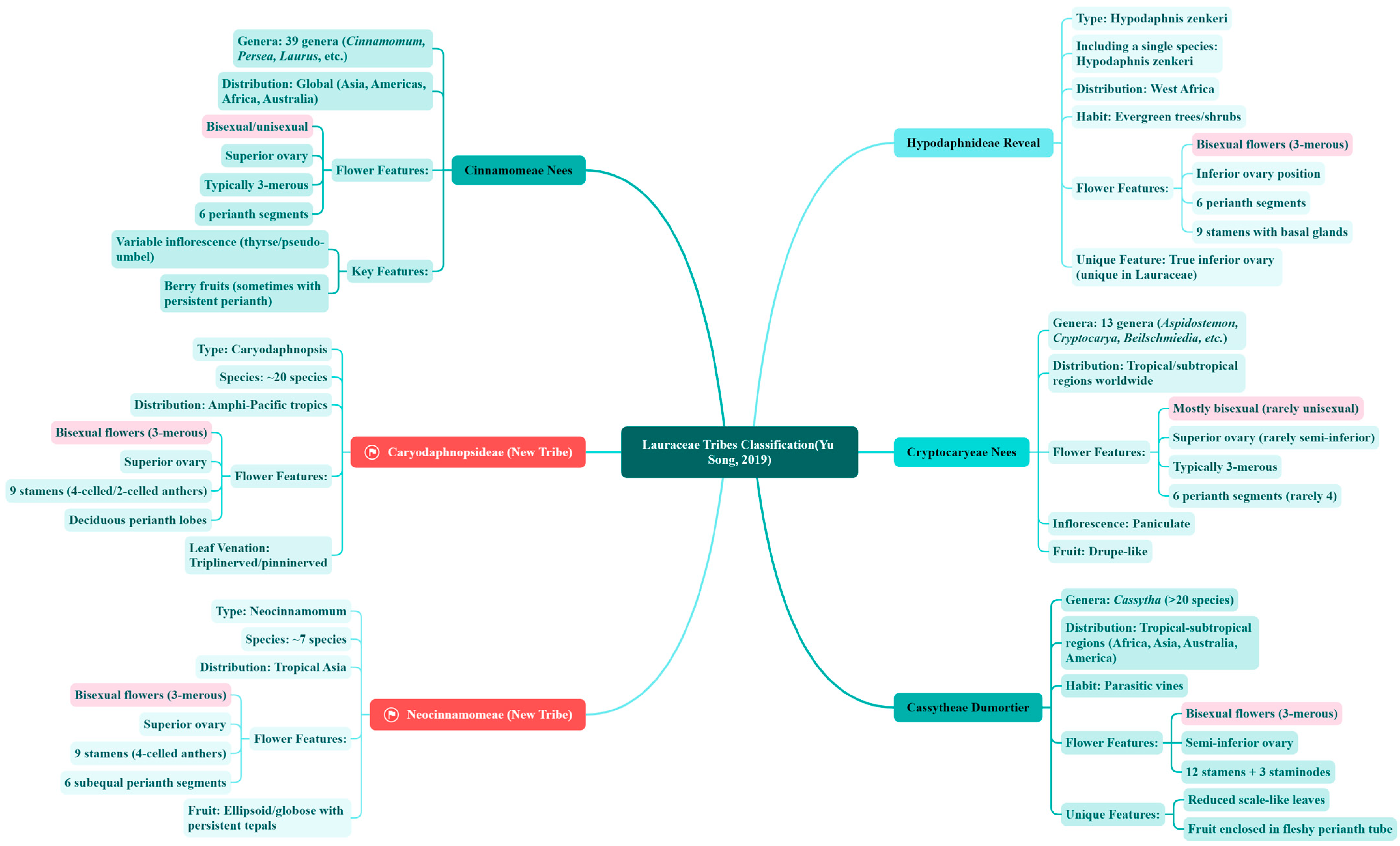
3.1.1. Classic Classification Systems and Morphological Basis
3.1.2. Molecular Phylogenetic Reconstruction of Taxonomic Frameworks
3.1.3. Whole-Genome Duplication (WGD) Events in Lauraceae
3.1.4. Conservatism and Diversity of Floral Organs in Lauraceae
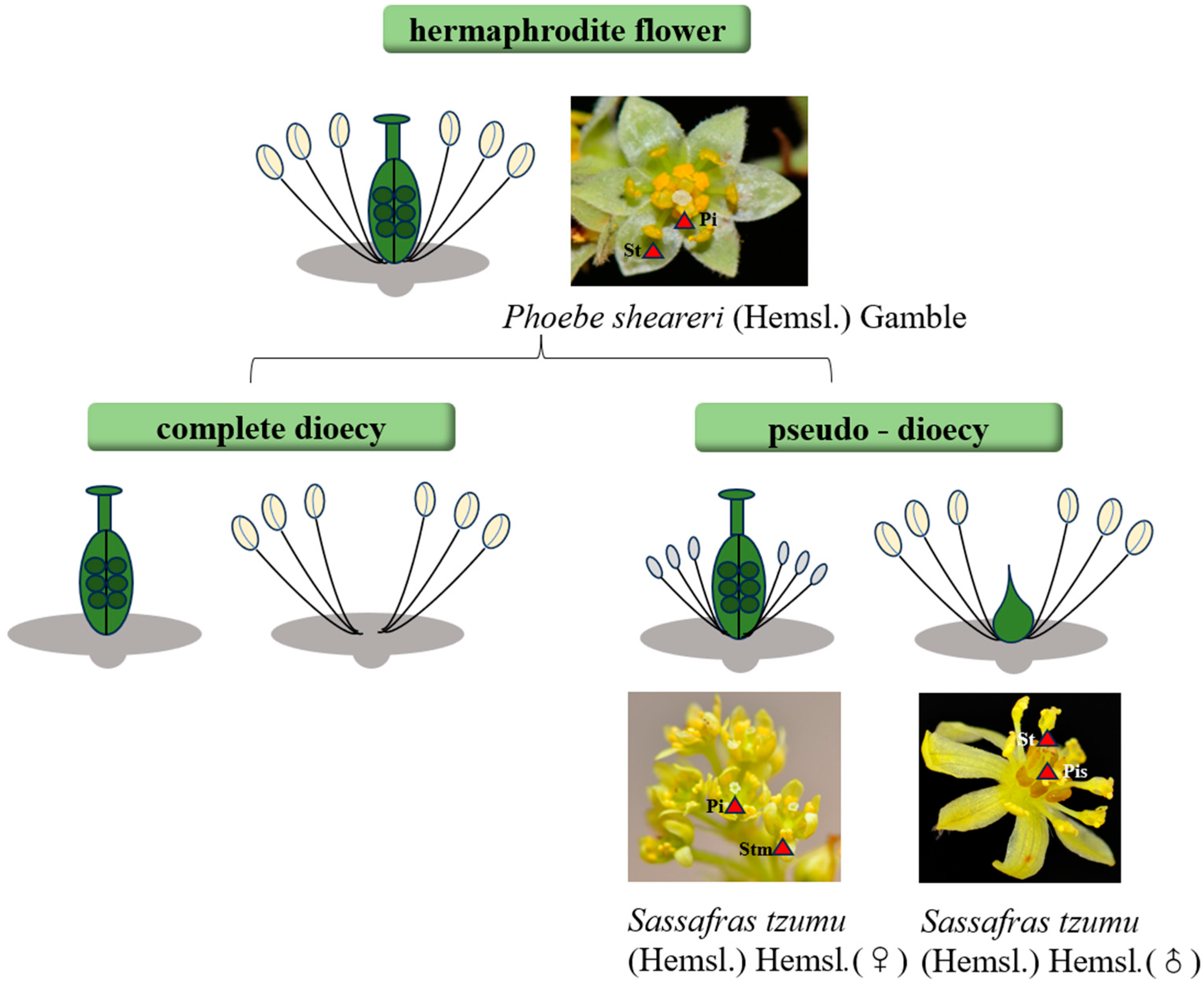
3.2. Sexual Differentiation and Evolutionary Trajectories in Lauraceae
3.2.1. Ancestral Reconstruction and Phylogeny
3.2.2. Transitional Lineages and Evolutionary “Intermediate States”
3.2.3. Genomic Insights into Sex Determination Mechanisms
3.2.4. Environmental Factors Drive Gender Differentiation in Lauraceae
3.2.5. Current Challenges in Lauraceae Sex Determination Research
4. Perspectives: New Frontiers in Multi-Omics Integration and Evolutionary Developmental Research
4.1. Multi-Omics-Driven Dissection of Sex Determination Networks
4.2. Spatiotemporal Specificity of Epigenetic Regulation
4.3. Environment–Gene Interactions and Ecological Adaptation
4.4. Comparative Genomics and Evolutionary Reconstruction
4.5. Application-Oriented Molecular Breeding
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- She, H.; Liu, Z.; Xu, Z.; Zhang, H.; Wu, J.; Cheng, F.; Wang, X.; Qian, W. Pan-genome analysis of 13 Spinacia accessions reveals structural variations associated with sex chromosome evolution and domestication traits in spinach. Plant Biotechnol. J. 2024, 22, 3102–3117. [Google Scholar] [CrossRef]
- Sather, D.N.; Jovanovic, M.; Golenberg, E.M. Functional analysis of B and C class floral organ genes in spinach demonstrates their role in sexual dimorphism. BMC Plant Biol. 2010, 10, 46. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Tan, F.-Q.; Chung, C.-H.; Slavkovic, F.; Devani, R.S.; Troadec, C.; Marcel, F.; Morin, H.; Camps, C.; Gomez Roldan, M.V. The control of carpel determinacy pathway leads to sex determination in cucurbits. Science 2022, 378, 543–549. [Google Scholar] [CrossRef] [PubMed]
- Rashid, D.; Devani, R.S.; Rodriguez-Granados, N.Y.; Abou-Choucha, F.; Troadec, C.; Morin, H.; Tan, F.Q.; Marcel, F.; Huang, H.Y.; Hanique, M.; et al. Ethylene produced in carpel primordia controls CmHB40 expression to inhibit stamen development. Nat. Plants 2023, 9, 1675–1687. [Google Scholar] [CrossRef] [PubMed]
- Akagi, T.; Pilkington, S.M.; Varkonyi-Gasic, E.; Henry, I.M.; Sugano, S.S.; Sonoda, M.; Firl, A.; McNeilage, M.A.; Douglas, M.J.; Wang, T.; et al. Two Y-chromosome-encoded genes determine sex in kiwifruit. Nat. Plants 2019, 5, 801–809. [Google Scholar] [CrossRef]
- Akagi, T.; Henry, I.M.; Ohtani, H.; Morimoto, T.; Beppu, K.; Kataoka, I.; Tao, R. A Y-Encoded Suppressor of Feminization Arose via Lineage-Specific Duplication of a Cytokinin Response Regulator in Kiwifruit. Plant Cell 2018, 30, 780–795. [Google Scholar] [CrossRef]
- Xu, Z.; Wang, Y.; Chen, Y.; Yin, H.; Wu, L.; Zhao, Y.; Wang, M.; Gao, M. A model of hormonal regulation of stamen abortion during pre-meiosis of Litsea cubeba. Genes 2019, 11, 48. [Google Scholar] [CrossRef]
- Akagi, T.; Henry, I.M.; Kawai, T.; Comai, L.; Tao, R. Epigenetic Regulation of the Sex Determination Gene MeGI in Polyploid Persimmon. Plant Cell 2016, 28, 2905–2915. [Google Scholar] [CrossRef]
- Harkess, A.; Zhou, J.; Xu, C.; Bowers, J.; Van der Hulst, R.; Ayyampalayam, S.; Mercati, F.; Riccardi, P.; McKain, M.; Kakrana, A. The asparagus genome sheds light on the origin and evolution of a young Y chromosome. Nat. Commun. 2017, 8, 1279. [Google Scholar] [CrossRef]
- Renner, S.S.; Ricklefs, R.E. Dioecy and Its Correlates in the Flowering Plants. Am. J. Bot. 1995, 82, 596–606. [Google Scholar] [CrossRef]
- Sun, W.H.; Xiang, S.; Zhang, Q.G.; Xiao, L.; Zhang, D.; Zhang, P.; Chen, D.Q.; Hao, Y.; Liu, D.K.; Ding, L.; et al. The camphor tree genome enhances the understanding of magnoliid evolution. J. Genet. Genom. Yi Chuan Xue Bao 2022, 49, 249–253. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-C.; Li, Z.; Zhao, Y.-X.; Gao, M.; Wang, J.-Y.; Liu, K.-W.; Wang, X.; Wu, L.-W.; Jiao, Y.-L.; Xu, Z.-L.; et al. The Litsea genome and the evolution of the laurel family. Nat. Commun. 2020, 11, 1675. [Google Scholar] [CrossRef]
- Ming, R.; Bendahmane, A.; Renner, S.S. Sex chromosomes in land plants. Annu. Rev. Plant Biol. 2011, 62, 485–514. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Chen, J.; Pan, C.; Ye, L.; Lu, G. Research Progress on the Evolution of Plant Sex Chromosomes and Sex Determination Genes. J. Integrative Plant Bio. 2016, 51, 841–848. [Google Scholar]
- Zhang, B.; Yao, X.; Chen, H.; Lu, L. High-quality chromosome-level genome assembly of Litsea coreana L. provides insights into Magnoliids evolution and flavonoid biosynthesis. Genomics 2022, 114, 110394. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, F.; Zhang, X.; Li, Z.; Zhao, Y.; Lohaus, R.; Chang, X.; Dong, W.; Ho, S.Y.W.; Liu, X.; et al. The water lily genome and the early evolution of flowering plants. Nature 2020, 577, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Gamble, T.; Geneva, A.J.; Glor, R.E.; Zarkower, D. Anolis sex chromosomes are derived from a single ancestral pair. Evol. Int. J. Org. Evol. 2014, 68, 1027–1041. [Google Scholar] [CrossRef]
- Chaw, S.-M.; Liu, Y.-C.; Wu, Y.-W.; Wang, H.-Y.; Lin, C.-Y.I.; Wu, C.-S.; Ke, H.-M.; Chang, L.-Y.; Hsu, C.-Y.; Yang, H.-T.J. Stout camphor tree genome fills gaps in understanding of flowering plant genome evolution. Nat. Plants 2019, 5, 63–73. [Google Scholar] [CrossRef]
- Jiang, R.; Chen, X.; Liao, X.; Peng, D.; Han, X.; Zhu, C.; Wang, P.; Hufnagel, D.E.; Wang, L.; Li, K.; et al. A Chromosome-Level Genome of the Camphor Tree and the Underlying Genetic and Climatic Factors for Its Top-Geoherbalism. Front. Plant Sci. 2022, 13, 827890. [Google Scholar] [CrossRef]
- Latrasse, D.; Rodriguez-Granados, N.Y.; Veluchamy, A.; Mariappan, K.G.; Bevilacqua, C.; Crapart, N.; Camps, C.; Sommard, V.; Raynaud, C.; Dogimont, C.; et al. The quest for epigenetic regulation underlying unisexual flower development in Cucumis melo. Epigenet. Chromatin 2017, 10, 22. [Google Scholar] [CrossRef]
- Hu, W.; Hu, S.; Li, S.; Zhou, Q.; Xie, Z.; Hao, X.; Wu, S.; Tian, L.; Li, D. AtSAMS regulates floral organ development by DNA methylation and ethylene signaling pathway. Plant Sci. Int. J. Exp. Plant Biol. 2023, 334, 111767. [Google Scholar] [CrossRef]
- Liu, J.; Chatham, L.; Aryal, R.; Yu, Q.; Ming, R. Differential methylation and expression of HUA1 ortholog in three sex types of papaya. Plant Sci. Int. J. Exp. Plant Biol. 2018, 272, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Von Linne, C. Species Plantarum; 1797. Available online: https://archive.org/details/mobot31753003413983/page/633/mode/2up (accessed on 20 December 2024).
- Kostermans, A.J.G.H. Lauraceae. REINWARDTIA 1957, 4, 193–256. [Google Scholar]
- van der Werff, H.; Richter, H.G. Toward an Improved Classification of Lauraceae. Ann. Mo. Bot. Gard. 1996, 83, 409–418. [Google Scholar] [CrossRef]
- Song, Y.; Yu, W.B.; Tan, Y.H.; Jin, J.J.; Wang, B.; Yang, J.B.; Liu, B.; Corlett, R.T.J. Plastid phylogenomics improve phylogenetic resolution in the Lauraceae. J. Syst. Evol. 2020, 58, 423–439. [Google Scholar] [CrossRef]
- Song, Y.; Yu, Q.-F.; Zhang, D.; Chen, L.-G.; Tan, Y.-H.; Zhu, W.; Su, H.-L.; Yao, X.; Liu, C.; Corlett, R.T. New insights into the phylogenetic relationships within the Lauraceae from mitogenomes. BMC Biol. 2024, 22, 241. [Google Scholar] [CrossRef] [PubMed]
- Editorial Committee of Flora Reipublicae Popularis Sinicae; Chinese Academy of Sciences. Flora Reipublicae Popularis Sinicae; Science Press: Beijing, China, 1984; Volume 31. [Google Scholar]
- Xue, X.; Hou, M.; Fang, Y. Scanning Electron Microscopy Observation of the Pollen of Several Common Lauraceae Plants. J. Anhui Agri. Sci. 2010, 38, 20945–20962. [Google Scholar] [CrossRef]
- Xiong, B.; Zhang, L.; Xie, L.; Li, L.; He, X.; Niu, Y.; Zhang, T.; Liao, S.; Dong, S.; Zhang, Z. Genome of Lindera glauca provides insights into the evolution of biosynthesis genes for aromatic compounds. iScience 2022, 25, 104761. [Google Scholar] [CrossRef]
- Wang, X.-D.; Xu, C.-Y.; Zheng, Y.-J.; Wu, Y.-F.; Zhang, Y.-T.; Zhang, T.; Xiong, Z.-Y.; Yang, H.-K.; Li, J.; Fu, C. Chromosome-level genome assembly and resequencing of camphor tree (Cinnamomum camphora) provides insight into phylogeny and diversification of terpenoid and triglyceride biosynthesis of Cinnamomum. Hortic. Res. 2022, 9, uhac216. [Google Scholar] [CrossRef]
- Shen, T.; Qi, H.; Luan, X.; Xu, W.; Yu, F.; Zhong, Y.; Xu, M. The chromosome-level genome sequence of the camphor tree provides insights into Lauraceae evolution and terpene biosynthesis. Plant Biotechnol. J. 2022, 20, 244–246. [Google Scholar] [CrossRef]
- Li, F.; Huang, S.; Mei, Y.; Wu, B.; Hou, Z.; Zhan, P.; Hou, Z.; Huang, W.; Zhao, J.; Wang, J.; et al. Genome assembly provided new insights into the Cinnamomum burmannii evolution and D-borneol biosynthesis differences between chemotypes. Ind. Crop. Prod. 2022, 186, 115181. [Google Scholar] [CrossRef]
- Chen, S.P.; Sun, W.H.; Xiong, Y.F.; Jiang, Y.T.; Liu, X.D.; Liao, X.Y.; Zhang, D.Y.; Jiang, S.Z.; Li, Y.; Liu, B.; et al. The Phoebe genome sheds light on the evolution of magnoliids. Hortic. Res. 2020, 7, 146. [Google Scholar] [CrossRef]
- Rendón-Anaya, M.; Ibarra-Laclette, E.; Méndez-Bravo, A.; Lan, T.; Zheng, C.; Carretero-Paulet, L.; Perez-Torres, C.A.; Chacón-López, A.; Hernandez-Guzmán, G.; Chang, T.H.; et al. The avocado genome informs deep angiosperm phylogeny, highlights introgressive hybridization, and reveals pathogen-influenced gene space adaptation. Proc. Natl. Acad. Sci. USA 2019, 116, 17081–17089. [Google Scholar] [CrossRef] [PubMed]
- Nath, O.; Fletcher, S.J.; Hayward, A.; Shaw, L.M.; Masouleh, A.K.; Furtado, A.; Henry, R.J.; Mitter, N. A haplotype resolved chromosomal level avocado genome allows analysis of novel avocado genes. Hortic. Res. 2022, 9, uhac157. [Google Scholar] [CrossRef] [PubMed]
- Tao, L.; Guo, S.; Xiong, Z.; Zhang, R.; Sun, W. Chromosome-level genome assembly of the threatened resource plant Cinnamomum chago. Sci. Data 2024, 11, 447. [Google Scholar] [CrossRef]
- Beech, E.; Rivers, M.C.; Oldfield, S.; Smith, P.P. GlobalTreeSearch: The first complete global database of tree species and country distributions. J. Sustain. For. 2017, 36, 454–489. [Google Scholar] [CrossRef]
- Stevens, P.F.; Kubitzki, K.; Rohwer, J.G.; Bittrich, V. The Families and Genera of Vascular Plants. Vol. 2. Flowering Plants. Dicotyledons: Magnoliid, Hamamelid and Caryophyllid Families. Taxon 1994, 43, 517. [Google Scholar] [CrossRef]
- Xu, Z. Mining and Identification of Salicylic Acid-Responsive Genes during Staminode Development in Litsea cubeba (Lour.) Pers. Ph.D. Theisis, Research Institute of Subtropical Forestry, Chinese Academy of Forestry, Hangzhou, China, 2020. [Google Scholar]
- Liu, C.; Chen, H.; Er, H.L.; Soo, H.M.; Kumar, P.P.; Han, J.H.; Liou, Y.C.; Yu, H. Direct interaction of AGL24 and SOC1 integrates flowering signals in Arabidopsis. Development 2008, 135, 1481–1491. [Google Scholar] [CrossRef]
- Zhu, S.S.; Comes, H.P.; Tamaki, I.; Cao, Y.N.; Sakaguchi, S.; Yap, Z.Y.; Ding, Y.Q.; Qiu, Y.X. Patterns of genotype variation and demographic history in Lindera glauca (Lauraceae), an apomict-containing dioecious forest tree. J. Biogeogr. 2020, 47, 2002–2016. [Google Scholar] [CrossRef]
- Baker, H.G.J.E. Self-compatibility and establishment after ‘long-distance’ dispersal. Evolution 1955, 9, 347–349. [Google Scholar]
- Zhou, G.; Yin, H.; Chen, F.; Wang, Y.; Gao, Q.; Yang, F.; He, C.; Zhang, L.; Wan, Y. The genome of Areca catechu provides insights into sex determination of monoecious plants. New Phytol. 2022, 236, 2327–2343. [Google Scholar] [CrossRef]
- He, L.; Wang, Y.; Wang, Y.; Zhang, R.-G.; Wang, Y.; Hörandl, E.; Ma, T.; Mao, Y.-F.; Mank, J.E.; Ming, R. Allopolyploidization from two dioecious ancestors leads to recurrent evolution of sex chromosomes. Nat. Commun. 2024, 15, 6893. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.S.; Shen, D.; Zhang, W.; Zhang, X.; Qiu, Y.; Wang, H.; Dou, X.; Li, S.; Wu, Y.; Song, J.; et al. Temperature and photoperiod changes affect cucumber sex expression by different epigenetic regulations. BMC Plant Biol. 2018, 18, 268. [Google Scholar] [CrossRef]
- He, L.; Fan, Y.; Zhang, Z.; Wei, X.; Yu, J. Identifying Genes Associated with Female Flower Development of Phellodendron amurense Rupr. Using a Transcriptomics Approach. Genes 2023, 14, 661. [Google Scholar] [CrossRef] [PubMed]
- Xia, Z.; He, Y.; Korpelainen, H.; Niinemets, Ü.; Li, C. Allelochemicals and soil microorganisms jointly mediate sex-specific belowground interactions in dioecious Populus cathayana. New Phytol. 2023, 240, 1519–1533. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Wang, D.; Li, Y.; Zhang, Z.; Tong, S.; Li, M.; Zhang, X.; Zhang, L.; Ren, L.; Ma, X.; et al. A General Model to Explain Repeated Turnovers of Sex Determination in the Salicaceae. Mol. Biol. Evol. 2021, 38, 968–980. [Google Scholar] [CrossRef]
- Huang, H.; Li, J. Flower fossils of Lauraceae in the geological timeand its phylogenetic evolutionary significance. Guihaia 2018, 38, 210–219. [Google Scholar]
| Degenerative Stage | Species | Patterns of Control | References |
|---|---|---|---|
| Stage 0 | Spinacia oleracea | XY system with Y-linked insertion duplications suppressing recombination | [1,2] |
| Stage 1 | Cucumis melo | Regulation of ethylene synthesis pathway and epigenetic regulation (DNA methylation); The key factor in sex determination: CRC | [3,4] |
| Stage 1 | Actinidia spp. | The SyGI gene inhibits carpel development. The FrBy acts for the maintenance of male functions | [5,6] |
| Stage2 | Litsea cubeba | Hormonal regulation | [7] |
| Stage 3 | Diospyros lotus | Epigenetic regulation (Srna) | [8] |
| Stage 3 | Asparagus officinalis | The two-mutation model | [9] |
| Number | Species | Genus/Tribe | Genome Size (Mb) | Chromosome Number (2n) | Sequencing Technology | Assembly Level | Research Focus | References |
|---|---|---|---|---|---|---|---|---|
| 1 | Litsea cubeba | Laureae | 1325.7 | 24 | PacBio CLR + Hi-C | Chromosome | Association between Monoterpene Synthesis and Sex Evolution | [16] |
| 2 | Litsea coreana | Laureae | 1139.5 | 24 | Illumina + PacBio CCS + Hi-C | Chromosome | Flavonoid Metabolism and Stress Resistance Mechanisms | [15] |
| 3 | Lindera glauca | Laureae | 2092.2 | 24 | Illumina + Nanopore + Hi-C | Chromosome | Parthenogenesis and Heterozygous Genomic Features | [30] |
| 4 | Cinnamomum kanehirae | Cinn amomeae | 730.7 | 24 | Illumina + PacBio CLR + Chicago + Hi-C | Chromosome | Terpenoids and Fatty Acid Biosynthesis Pathways | [18] |
| 5 | Cinnamomum camphora | Cinn amomeae | 755.4 | 24 | PacBio CCS + Hi-C | Chromosome | Molecular Basis of Chemotypic Diversity in Terpenoids | [19] |
| 6 | Cinnamomum camphora | Cinn amomeae | 723.1 | 24 | Illumina + PacBio CCS + Hi-C | Chromosome | Phylogenetics and Key Genes in Essential Oil Biosynthesis | [31] |
| 7 | Cinnamomum camphora | Cinn amomeae | 719.9 | 24 | Illumina + PacBio CCS + Hi-C | Chromosome | Genome Resequencing and Chemotype Evolution | [11] |
| 8 | Cinnamomum camphora | Cinn amomeae | 785.0 | 24 | PacBio CCS + Hi-C | Evolution and Terpenoid Biosynthesis | [32] | |
| 9 | Cinnamomum burmanni | Cinn amomeae | 1177.6 | 24 | Illumina + PacBio CLR + Hi-C | Chromosome | Terpenoid Synthesis and Mining of Disease-Resistance Genes | [33] |
| 10 | Phoebe bournei | Perseeae | 989.2 | 24 | PacBio CLR | Scaffold | Wood Properties and Secondary Metabolic Pathways | [34] |
| 11 | Phoebe bournei | Perseeae | 941.8 | 24 | PacBio CLR; BioNano and Hi-C | Chromosome | Terpenoid biosynthesis, WGD evolutionary mechanisms, and disease resistance applications | [15] |
| 12 | Persea americana | Perseeae | 912.6 | 24 | PacBio CLR | Chromosome | Paleopolyploidization and the Origin of Sex Chromosomes | [35] |
| 13 | Persea america-na | Perseeae | 913.0 | 24 | Illumina and PacBio CCS | Chromosome | Evolutionary context, metabolic pathways, and fruit traits | [36] |
| 14 | Cinnamomum chago | Cinn amomeae | 785.0 | 24 | PacBio CCS + Hi-C | Chromosome | Conservation of Endangered Resources and Terpenoid Synthesis | [37] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, S.; Wang, Y.; Chen, Y.; Zhao, Y.; Gao, M. Advances in the Evolutionary Mechanisms and Genomic Studies of Sexual Differentiation in Lauraceae Plants. Int. J. Mol. Sci. 2025, 26, 4335. https://doi.org/10.3390/ijms26094335
Wang S, Wang Y, Chen Y, Zhao Y, Gao M. Advances in the Evolutionary Mechanisms and Genomic Studies of Sexual Differentiation in Lauraceae Plants. International Journal of Molecular Sciences. 2025; 26(9):4335. https://doi.org/10.3390/ijms26094335
Chicago/Turabian StyleWang, Siqi, Yangdong Wang, Yicun Chen, Yunxiao Zhao, and Ming Gao. 2025. "Advances in the Evolutionary Mechanisms and Genomic Studies of Sexual Differentiation in Lauraceae Plants" International Journal of Molecular Sciences 26, no. 9: 4335. https://doi.org/10.3390/ijms26094335
APA StyleWang, S., Wang, Y., Chen, Y., Zhao, Y., & Gao, M. (2025). Advances in the Evolutionary Mechanisms and Genomic Studies of Sexual Differentiation in Lauraceae Plants. International Journal of Molecular Sciences, 26(9), 4335. https://doi.org/10.3390/ijms26094335




