Selective Detection of Fungal and Bacterial Glycans with Galactofuranose (Galf) Residues by Surface-Enhanced Raman Scattering and Machine Learning Methods
Abstract
1. Introduction
2. Results and Discussion
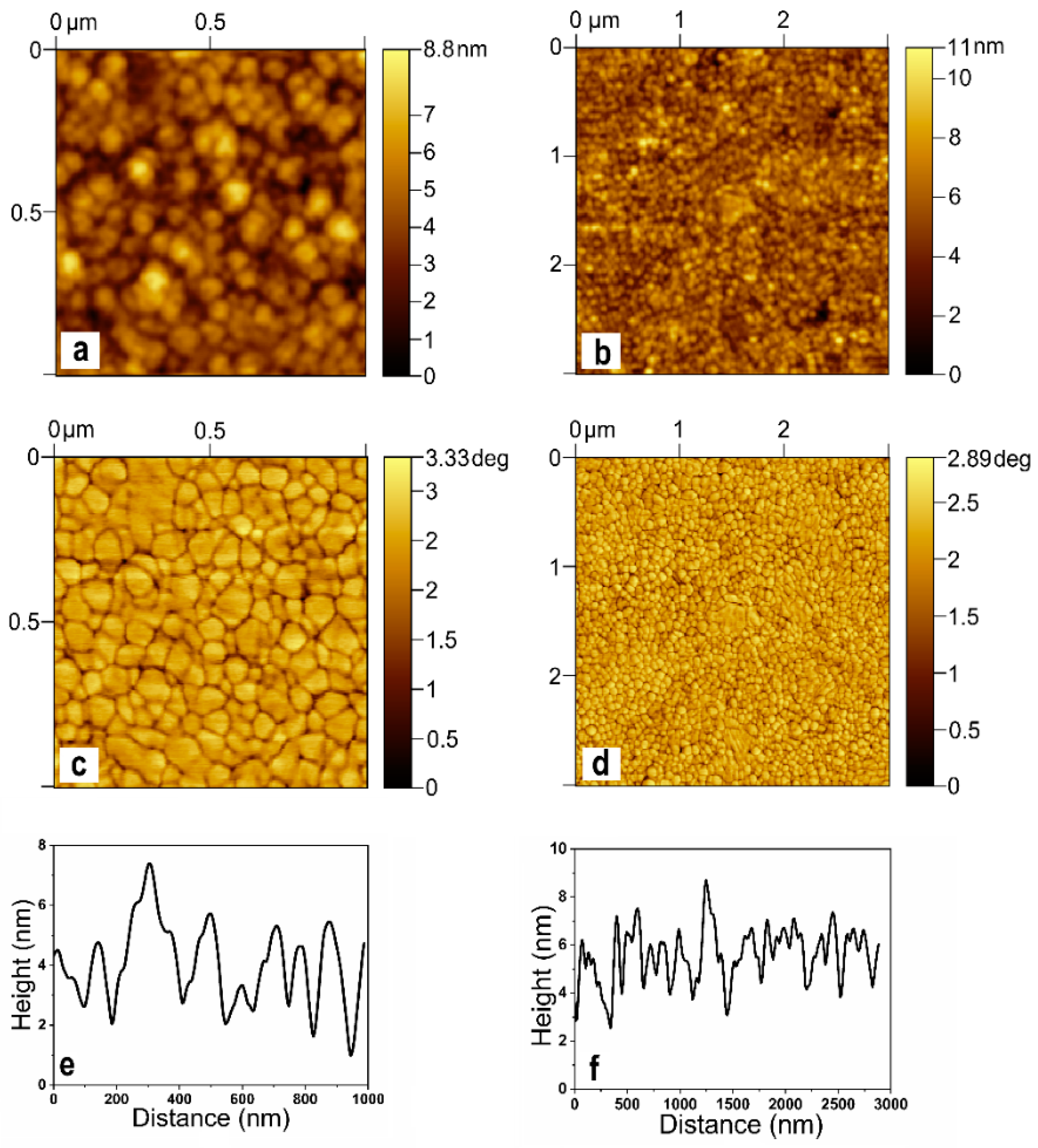
2.1. AFM of the SERS Substrate
2.2. Analysis of the Main Vibration Bands of the Analyte Spectra
2.3. Mathematica Treatment Results
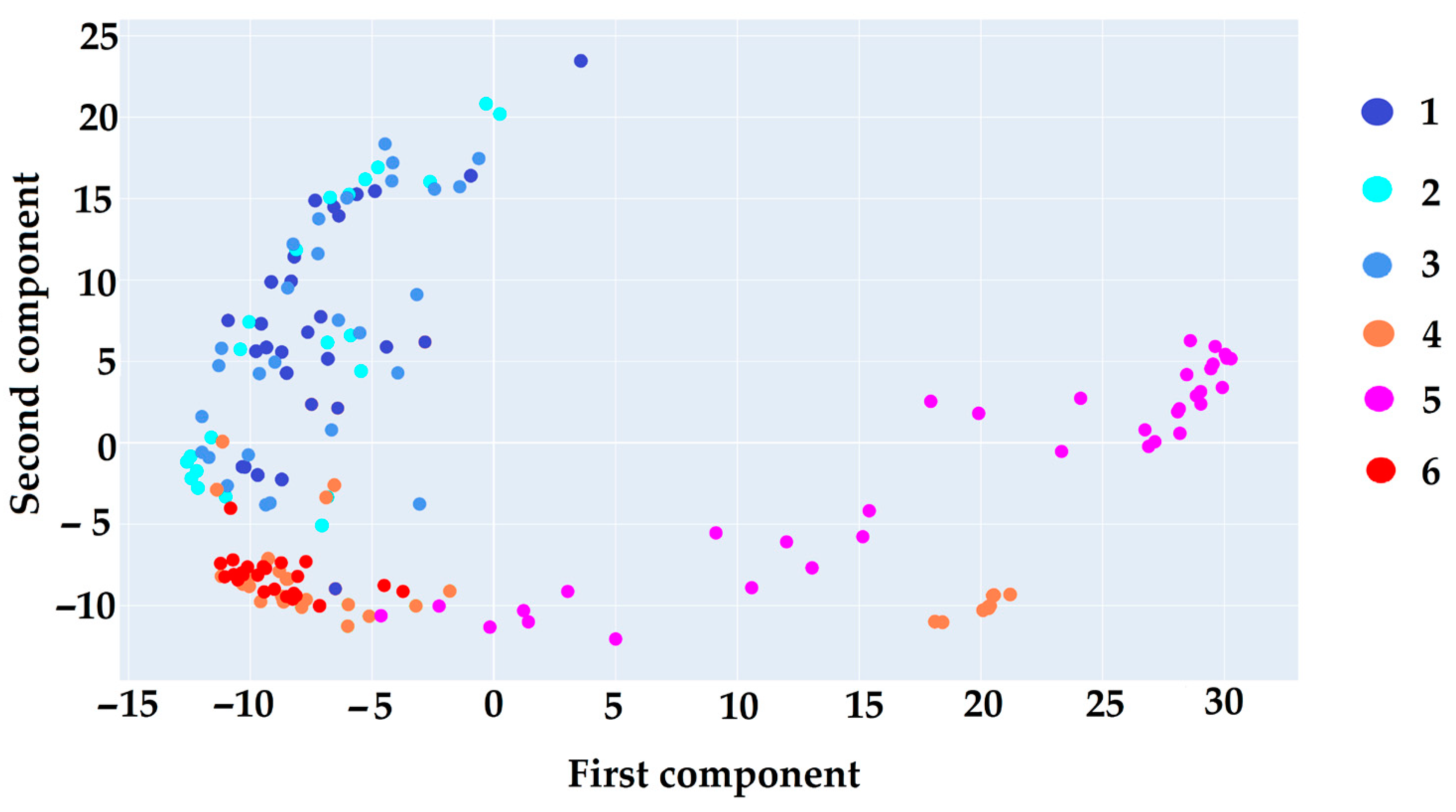
2.3.1. PCA
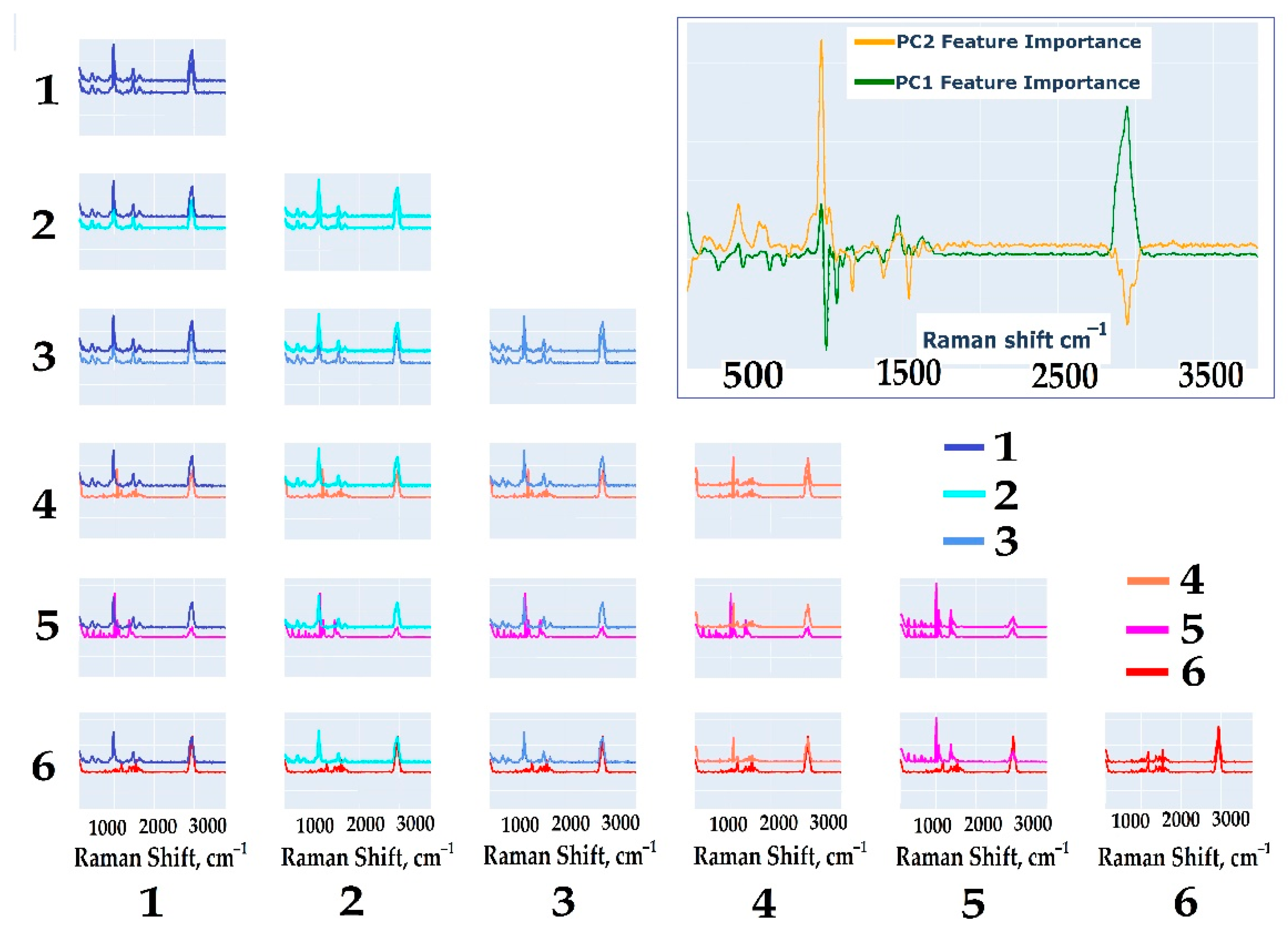
2.3.2. Determination of the Vibration Bands Responsible for the Separation of Spectra
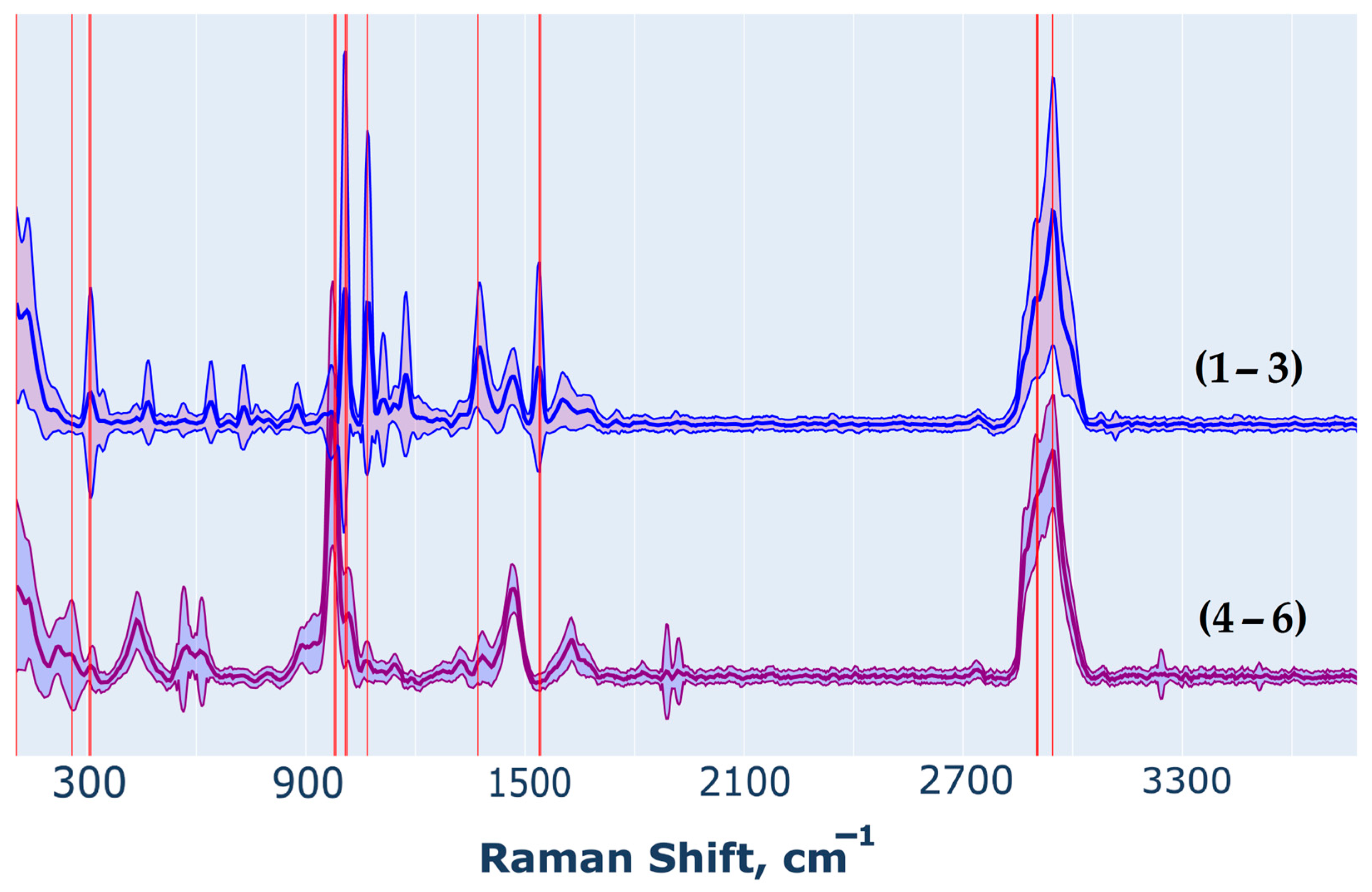
- Oligosaccharide groups without the Galf residue (1–3) exhibit similar spectral profiles. In pairwise comparisons, the differences between these groups are minimal and centered around shared peaks, indicating the presence of stable and comparable spectral fragments.
- Oligosaccharide groups containing the Galf residue (4–6) display distinct spectral differences. Their vibrational bands vary significantly when compared to those of oligosaccharide groups without Galf, indicating the strong influence of Galf on the spectral profile. This distinctive spectral behavior allows these groups to be categorized separately, highlighting their unique properties.
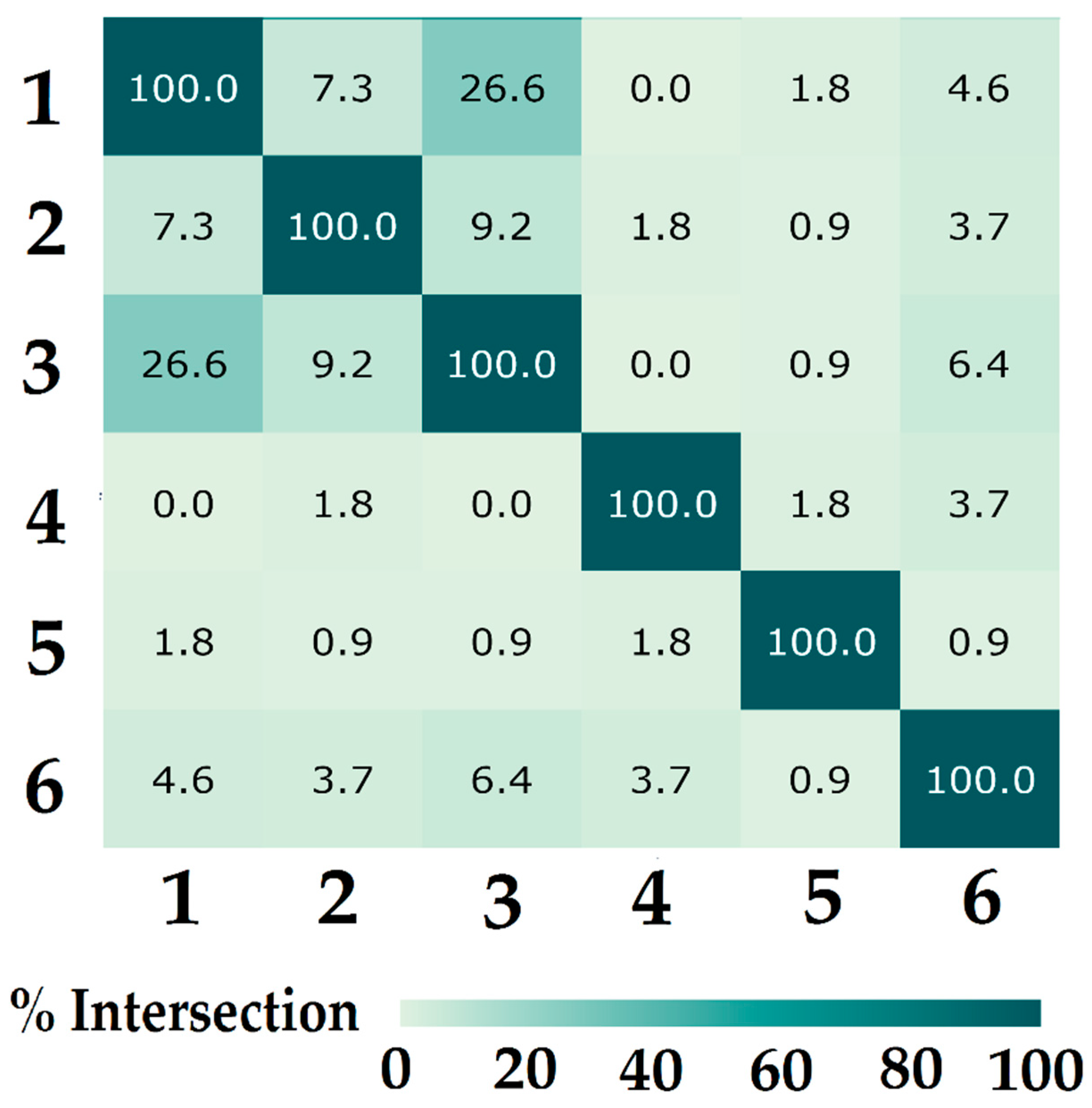
2.3.3. Confidence Interval Estimation
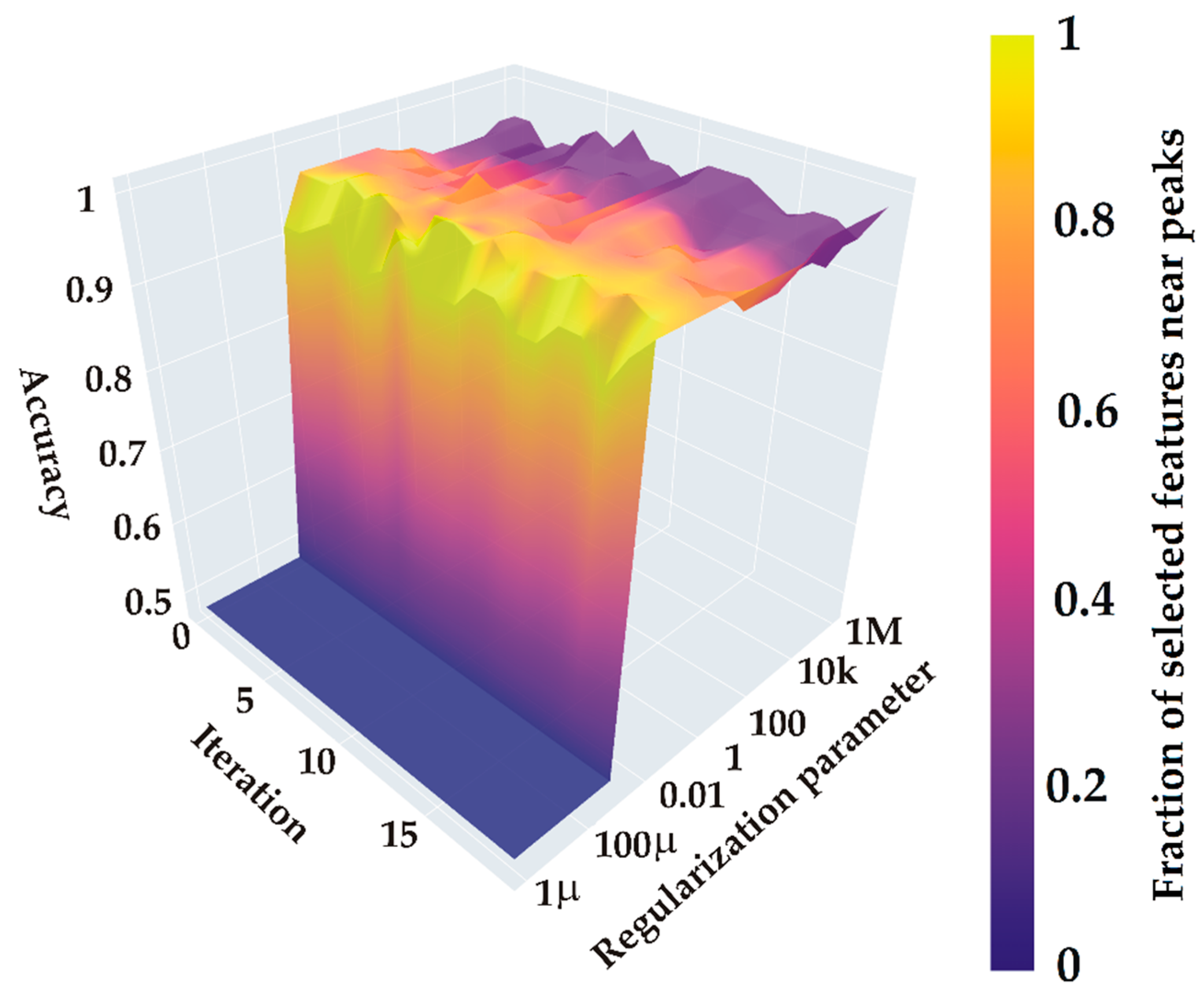
2.3.4. Computational Experiment
- Training the logistic regression model on a training subset;
- Assessing classification accuracy on the test subset;
- Recording the count of non-zero coefficients to measure feature selection;
- Mapping selected features relative to key spectral peaks identified by PCA and confidence interval analysis.
2.3.5. Model Estimation
2.3.6. Classification Results
3. Materials and Methods
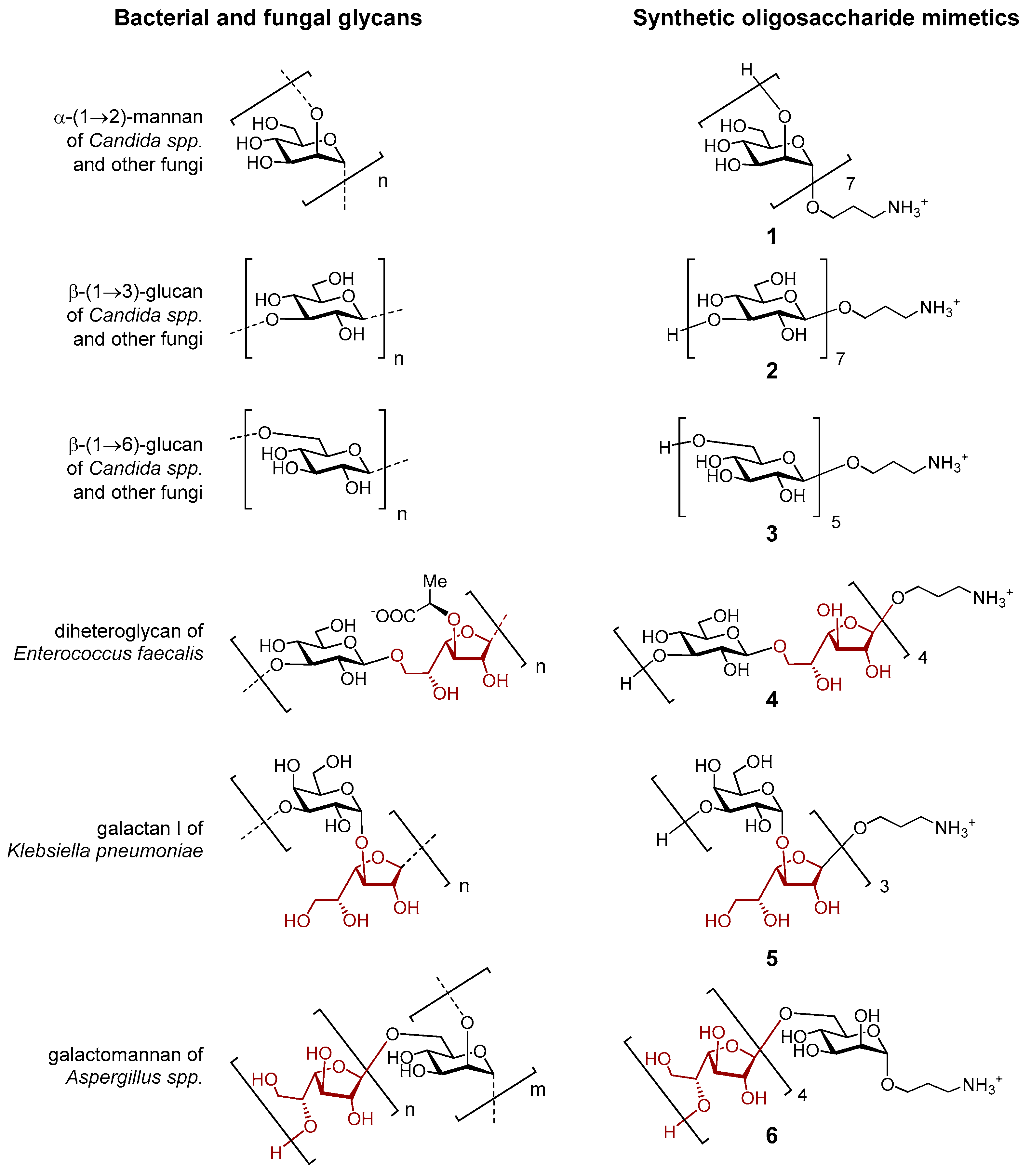
3.1. Research Objects
3.2. SERS Substrate Fabrication
3.3. AFM Studies of Substrate Surface
3.4. Analyte Preparation and Application
3.5. Spectra Measurements
3.6. Pre-Processing of Spectral Data
3.6.1. Outliers Removing
3.6.2. Spectra Filtering
3.6.3. Baseline Correction
3.6.4. Spectra Normalization
3.7. Analyte Discerning Differences Investigation and Their Logistic Regression Classification
3.7.1. PCA
3.7.2. Estimation of Overlap of Intensity Distributions at Peaks
3.7.3. Bootstrapping Procedure
- Generating Bootstrap Samples: Random sampling with replacement is conducted on the original intensity measurements at pi, producing 10,000 bootstrap samples;
- Calculating Statistics: The mean intensity is computed for each bootstrap sample;
- Building the Empirical Distribution: The distribution of mean values forms an empirical distribution of the mean intensity for each group at pi;
- Deriving Confidence Intervals: From the empirical distribution, confidence interval bounds are determined at a specified confidence level (95%).
- -
- tα/2,n−1 is the critical t—value at α/2 for n − 1 degrees of freedom.
- -
- Sx and Sy are the standard deviations for groups and, respectively.
- -
- and are the mean values for groups and.
- -
- n is the sample size.
3.7.4. Statistical Significance Threshold
- -
- r is the correlation coefficient between the standard deviations of intensities for two groups.
- -
- , where Sx and Sy are standard deviations for each group.
3.7.5. Application of the Method
3.7.6. Class Similarity Assessment
3.7.7. LR with L1 Regularization
- x—sample feature vector;
- w—vector of weight coefficients;
- b—free term.
- —class label for i-th sample ;
- —regularization coefficient;
- —number of features.
3.7.8. Classification Metrics
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Erwig, L.P.; Gow, N.A.R. Interactions of Fungal Pathogens with Phagocytes. Nat. Rev. Microbiol. 2016, 14, 163–176. [Google Scholar] [CrossRef] [PubMed]
- Mostowy, R.J.; Holt, K.E. Diversity-Generating Machines: Genetics of Bacterial Sugar-Coating. Trends Microbiol. 2018, 26, 1008–1021. [Google Scholar] [CrossRef]
- Ahamefula Osibe, D.; Lei, S.; Wang, B.; Jin, C.; Fang, W. Cell Wall Polysaccharides from Pathogenic Fungi for Diagnosis of Fungal Infectious Disease. Mycoses 2020, 63, 644–652. [Google Scholar] [CrossRef]
- Theel, E.S.; Doern, C.D. Point-Counterpoint: β-D-Glucan Testing Is Important for Diagnosis of Invasive Fungal Infections. J. Clin. Microbiol. 2013, 51, 3478–3483. [Google Scholar] [CrossRef]
- Chumpitazi, B.F.F.; Lebeau, B.; Faure-Cognet, O.; Hamidfar-Roy, R.; Timsit, J.-F.; Pavese, P.; Thiebaut-Bertrand, A.; Quesada, J.-L.; Pelloux, H.; Pinel, C. Characteristic and Clinical Relevance of Candida Mannan Test in the Diagnosis of Probable Invasive Candidiasis. Med. Mycol. 2014, 52, 462–471. [Google Scholar] [CrossRef]
- de Heer, K.; Gerritsen, M.G.; Visser, C.E.; Leeflang, M.M. Galactomannan Detection in Broncho-Alveolar Lavage Fluid for Invasive Aspergillosis in Immunocompromised Patients. Cochrane Database Syst. Rev. 2019, 2019, CD012399. [Google Scholar] [CrossRef]
- Huang, H.-R.; Fan, L.-C.; Rajbanshi, B.; Xu, J.-F. Evaluation of a New Cryptococcal Antigen Lateral Flow Immunoassay in Serum, Cerebrospinal Fluid and Urine for the Diagnosis of Cryptococcosis: A Meta-Analysis and Systematic Review. PLoS ONE 2015, 10, e0127117. [Google Scholar] [CrossRef]
- Latgé, J.P.; Kobayashi, H.; Debeaupuis, J.P.; Diaquin, M.; Sarfati, J.; Wieruszeski, J.M.; Parra, E.; Bouchara, J.P.; Fournet, B. Chemical and Immunological Characterization of the Extracellular Galactomannan of Aspergillus fumigatus. Infect. Immun. 1994, 62, 5424–5433. [Google Scholar] [CrossRef]
- Dorokhova, V.S.; Komarova, B.S.; Previato, J.O.; Mendonça Previato, L.; Krylov, V.B.; Nifantiev, N.E. Synthesis of Branched and Linear Galactooligosaccharides Related to Glucuronoxylomannogalactan of Cryptococcus Neoformans. Front. Chem. 2024, 12, 1501766. [Google Scholar] [CrossRef]
- Ikuta, K.; Shibata, N.; Blake, J.S.; Dahl, M.V.; Nelson, R.D.; Hisamichi, K.; Kobayashi, H.; Suzuki, S.; Okawa, Y. NMR Study of the Galactomannans of Trichophyton mentagrophytes and Trichophyton rubrum. Biochem. J. 1997, 323, 297–305. [Google Scholar] [CrossRef]
- Almeida, I.C.; Neville, D.C.A.; Mehlert, A.; Treumann, A.; Ferguson, A.J.; Previato, J.O.; Travassos, L.R. Structure of the N-Linked Oligosaccharide of the Main Diagnostic Antigen of the Pathogenic Fungus Paracoccidiodes brasiliensis. Glycobiology 1996, 6, 507–515. [Google Scholar] [CrossRef] [PubMed]
- Shibata, N.; Saitoh, T.; Tadokoro, Y.; Okawa, Y. The Cell Wall Galactomannan Antigen from Malassezia Furfur and Malassezia Pachydermatis Contains β-1,6-Linked Linear Galactofuranosyl Residues and Its Detection Has Diagnostic Potential. Microbiology 2009, 155, 3420–3429. [Google Scholar] [CrossRef]
- Denning, D.W. Global Incidence and Mortality of Severe Fungal Disease. Lancet Infect. Dis. 2024, 24, e428–e438. [Google Scholar] [CrossRef]
- Sendid, B.; Poissy, J.; François, N.; Mery, A.; Courtecuisse, S.; Krzewinski, F.; Jawhara, S.; Guerardel, Y.; Poulain, D. Preliminary Evidence for a Serum Disaccharide Signature of Invasive Candida Albicans Infection Detected by MALDI Mass Spectrometry. Clin. Microbiol. Infect. 2015, 21, 88.e1–88.e6. [Google Scholar] [CrossRef] [PubMed]
- Cornu, M.; Sendid, B.; Mery, A.; François, N.; Mikulska, M.; Letscher-Bru, V.; De Carolis, E.; Damonti, L.; Titecat, M.; Bochud, P.-Y.; et al. Evaluation of Mass Spectrometry-Based Detection of Panfungal Serum Disaccharide for Diagnosis of Invasive Fungal Infections: Results from a Collaborative Study Involving Six European Clinical Centers. J. Clin. Microbiol. 2019, 57, e01867-18. [Google Scholar] [CrossRef]
- Mery, A.; Jawhara, S.; François, N.; Cornu, M.; Poissy, J.; Martinez-Esparza, M.; Poulain, D.; Sendid, B.; Guerardel, Y. Identification of Fungal Trehalose for the Diagnosis of Invasive Candidiasis by Mass Spectrometry. Biochim. Biophys. Acta Gen. Subj. 2022, 1866, 130083. [Google Scholar] [CrossRef]
- Agrawal, P.K. NMR Spectroscopy in the Structural Elucidation of Oligosaccharides and Glycosides. Phytochemistry 1992, 31, 3307–3330. [Google Scholar] [CrossRef] [PubMed]
- Lahaye, M. NMR Spectroscopic Characterisation of Oligosaccharides from Two Ulva Rigida Ulvan Samples (Ulvales, Chlorophyta) Degraded by a Lyase. Carbohydr. Res. 1998, 314, 1–12. [Google Scholar] [CrossRef]
- Neely, L.A.; Audeh, M.; Phung, N.A.; Min, M.; Suchocki, A.; Plourde, D.; Blanco, M.; Demas, V.; Skewis, L.R.; Anagnostou, T.; et al. T2 Magnetic Resonance Enables Nanoparticle-Mediated Rapid Detection of Candidemia in Whole Blood. Sci. Transl. Med. 2013, 5, 182ra54. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Wolk, D.M.; Lowery, T.J. T2MR and T2Candida: Novel Technology for the Rapid Diagnosis of Candidemia and Invasive Candidiasis. Future Microbiol. 2016, 11, 103–117. [Google Scholar] [CrossRef]
- Sunamura, E.; Iwasaki, M.; Shiina, S.; Kitahara, S.; Yotani, T.; Manabe, M.; Miyazaki, O. A Novel Enzyme Immunoassay for the Measurement of Plasma (1→3)-β-D-Glucan Levels. J. Immunol. Methods 2020, 487, 112872. [Google Scholar] [CrossRef] [PubMed]
- Caceres, D.H.; Chiller, T.; Lindsley, M.D. Immunodiagnostic Assays for the Investigation of Fungal Outbreaks. Mycopathologia 2020, 185, 867–880. [Google Scholar] [CrossRef]
- Rehagel, C.; Akineden, Ö.; Geisen, R.; Cramer, B.; Plötz, M.; Usleber, E. Development and Application of a Competitive Enzyme Immunoassay for the Detection of Penitrem A in Fungal Mycelium: Evidence for Frequent Occurrence of Multiple Indole-Containing Mycotoxins in Mouldy Foods. Food Control 2022, 132, 108558. [Google Scholar] [CrossRef]
- Ou, K.-L.; Hsu, T.-C.; Liu, Y.-C.; Yang, K.-H.; Tsai, H.-Y. Silver Overlayer-Modified Surface-Enhanced Raman Scattering-Active Gold Substrates for Potential Applications in Trace Detection of Biochemical Species. Anal. Chim. Acta 2014, 806, 188–196. [Google Scholar] [CrossRef] [PubMed]
- Sanger, K.; Durucan, O.; Wu, K.; Thilsted, A.H.; Heiskanen, A.; Rindzevicius, T.; Schmidt, M.S.; Zór, K.; Boisen, A. Large-Scale, Lithography-Free Production of Transparent Nanostructured Surface for Dual-Functional Electrochemical and SERS Sensing. ACS Sens. 2017, 2, 1869–1875. [Google Scholar] [CrossRef]
- Fang, L.; Pan, X.-T.; Liu, K.; Jiang, D.; Ye, D.; Ji, L.-N.; Wang, K.; Xia, X.-H. Surface-Roughened SERS-Active Single Silver Nanowire for Simultaneous Detection of Intracellular and Extracellular PHs. ACS Appl. Mater. Interfaces 2023, 15, 20677–20685. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, B.L.; Patwardhan, A.J.; Ngola, S.M.; Chan, S.; Sundararajan, N. Experimental and Statistical Analysis Methods for Peptide Detection Using Surface-enhanced Raman Spectroscopy. J. Raman Spectrosc. 2008, 39, 380–388. [Google Scholar] [CrossRef]
- Kahraman, M.; Wachsmann-Hogiu, S. Label-Free and Direct Protein Detection on 3D Plasmonic Nanovoid Structures Using Surface-Enhanced Raman Scattering. Anal. Chim. Acta 2015, 856, 74–81. [Google Scholar] [CrossRef]
- Boginskaya, I.; Gainutdinova, A.; Gusev, A.; Mailyan, K.; Mikhailitsyn, A.; Sedova, M.; Vdovichenko, A.; Ryzhikov, I.; Chvalun, S.; Lagarkov, A. Detection of Organic Substances by a SERS Method Using a Special Ag-Poly(Chloro-P-Xylylene)-Ag Sandwich Substrate. Coatings 2020, 10, 799. [Google Scholar] [CrossRef]
- Boginskaya, I.; Gainutdinova, A.; Gusev, A.; Mailyan, K.; Mikhailitsyn, A.; Sedova, M.; Vdovichenko, A.; Glushchenkov, A.; Dorofeenko, A.; Ryzhikov, I. The Poly(Chloro-p-Xylylene)-Ag Metal-Polymer Nanocomposites Obtained by Controlled Vapor-Phase Synthesis for SERS Effect Realisation. Coatings 2021, 11, 1171. [Google Scholar] [CrossRef]
- Das, G.; Gentile, F.; Coluccio, M.L.; Perri, A.M.; Nicastri, A.; Mecarini, F.; Cojoc, G.; Candeloro, P.; Liberale, C.; De Angelis, F.; et al. Principal Component Analysis Based Methodology to Distinguish Protein SERS Spectra. J. Mol. Struct. 2011, 993, 500–505. [Google Scholar] [CrossRef]
- Puliga, F.; Zuffi, V.; Zambonelli, A.; Francioso, O.; Sanchez-Cortes, S. Spectroscopic Analysis of Mushrooms by Surface-Enhanced Raman Scattering (SERS). Chem. Biol. Technol. Agric. 2022, 9, 100. [Google Scholar] [CrossRef]
- Xia, J.; Li, W.; Sun, M.; Wang, H. Application of SERS in the Detection of Fungi, Bacteria and Viruses. Nanomaterials 2022, 12, 3572. [Google Scholar] [CrossRef]
- Dina, N.E.; Gherman, A.M.R.; Chiş, V.; Sârbu, C.; Wieser, A.; Bauer, D.; Haisch, C. Characterization of Clinically Relevant Fungi via SERS Fingerprinting Assisted by Novel Chemometric Models. Anal. Chem. 2018, 90, 2484–2492. [Google Scholar] [CrossRef] [PubMed]
- Santos, C.M.B.; da Silva, S.W.; Saldanha, C.A.; Santos, M.F.M.A.; Garcia, M.P.; Azevedo, R.B.; Morais, P.C. SERS as a Valuable Tool for Detection and Treatment Follow-up of Fungal Infection in Mice Lungs: Use of Amphotericin B and Its Nanoencapsulation onto Magnetic Nanoparticles. J. Raman Spectrosc. 2013, 44, 695–702. [Google Scholar] [CrossRef]
- Udupa, R.; Peralam Yegneswaran, P.; Lukose, J.; Chidangil, S. Utilization of Raman Spectroscopy for Identification and Characterization of Fungal Pathogens. Fungal Biol. Rev. 2024, 47, 100339. [Google Scholar] [CrossRef]
- Oh, M.-K.; Shin, Y.-S.; Lee, C.-L.; De, R.; Kang, H.; Yu, N.E.; Kim, B.H.; Kim, J.H.; Yang, J.-K. Morphological and SERS Properties of Silver Nanorod Array Films Fabricated by Oblique Thermal Evaporation at Various Substrate Temperatures. Nanoscale Res. Lett. 2015, 10, 259. [Google Scholar] [CrossRef]
- Gromov, D.G.; Dubkov, S.V.; Savitskiy, A.I.; Shaman, Y.P.; Polokhin, A.A.; Belogorokhov, I.A.; Trifonov, A.Y. Optimization of Nanostructures Based on Au, Ag, Au Ag Nanoparticles Formed by Thermal Evaporation in Vacuum for SERS Applications. Appl. Surf. Sci. 2019, 489, 701–707. [Google Scholar] [CrossRef]
- Khalil, I.; Chou, C.-M.; Tsai, K.-L.; Hsu, S.; Yehye, W.A.; Hsiao, V.K.S. Gold Nanofilm-Coated Porous Silicon as Surface-Enhanced Raman Scattering Substrate. Appl. Sci. 2019, 9, 4806. [Google Scholar] [CrossRef]
- Wu, H.-Y.; Lin, H.-C.; Hung, G.-Y.; Tu, C.-S.; Liu, T.-Y.; Hong, C.-H.; Yu, G.; Hsu, J.-C. High Sensitivity SERS Substrate of a Few Nanometers Single-Layer Silver Thickness Fabricated by DC Magnetron Sputtering Technology. Nanomaterials 2022, 12, 2742. [Google Scholar] [CrossRef]
- Jinachandran, A.; Kokulnathan, T.; Wang, T.-J.; Kumar, K.M.A.; Kumar, J.; Panneerselvam, R. Silver Nanopopcorns Decorated on Flexible Membrane for SERS Detection of Nitrofurazone. Microchim. Acta 2024, 191, 347. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Jia, T.; Li, X.; Yang, J.; Li, Z.; Shi, G.; Zhang, X.; Wang, Z. Fabrication of Nano/Microstructures for SERS Substrates Using an Electrochemical Method. Beilstein J. Nanotechnol. 2020, 11, 1568–1576. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.-Y.; Chien, C.-H. Facile Fabrication of Micro/Nano Hierarchical SERS Sensor via Anisotropic Etching and Electrochemical Treatment for Malachite Green Detection. Appl. Sci. 2019, 9, 5237. [Google Scholar] [CrossRef]
- Ma, C.; Trujillo, M.J.; Camden, J.P. Nanoporous Silver Film Fabricated by Oxygen Plasma: A Facile Approach for SERS Substrates. ACS Appl. Mater. Interfaces 2016, 8, 23978–23984. [Google Scholar] [CrossRef]
- Sojinrin, T.; Conde, J.; Liu, K.; Curtin, J.; Byrne, H.J.; Cui, D.; Tian, F. Plasmonic Gold Nanoparticles for Detection of Fungi and Human Cutaneous Fungal Infections. Anal. Bioanal. Chem. 2017, 409, 4647–4658. [Google Scholar] [CrossRef]
- Ramirez-Perez, J.C.; Reis, T.A.; Olivera, C.L.P.; Rizzutto, M.A. Impact of Silver Nanoparticles Size on SERS for Detection and Identification of Filamentous Fungi. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022, 272, 120980. [Google Scholar] [CrossRef]
- Boginskaya, I.; Sedova, M.; Baburin, A.; Afanas’ev, K.; Zverev, A.; Echeistov, V.; Ryzhkov, V.; Rodionov, I.; Tonanaiskii, B.; Ryzhikov, I.; et al. SERS-Active Substrates Nanoengineering Based on e-Beam Evaporated Self-Assembled Silver Films. Appl. Sci. 2019, 9, 3988. [Google Scholar] [CrossRef]
- Han, R.; Ketkaew, R.; Luber, S. A Concise Review on Recent Developments of Machine Learning for the Prediction of Vibrational Spectra. J. Phys. Chem. A 2022, 126, 801–812. [Google Scholar] [CrossRef]
- Luo, R.; Popp, J.; Bocklitz, T. Deep Learning for Raman Spectroscopy: A Review. Analytica 2022, 3, 287–301. [Google Scholar] [CrossRef]
- Beeram, R.; Banerjee, D.; Narlagiri, L.M.; Soma, V.R. Machine Learning for Rapid Quantification of Trace Analyte Molecules Using SERS and Flexible Plasmonic Paper Substrates. Anal. Methods 2022, 14, 1788–1796. [Google Scholar] [CrossRef]
- Lin, S.; Fang, X.; Fang, G.; Liu, F.; Dong, H.; Zhao, H.; Zhang, J.; Dong, B. Ultrasensitive Detection and Distinction of Pollutants Based on SERS Assisted by Machine Learning Algorithms. Sens. Actuators B Chem. 2023, 384, 133651. [Google Scholar] [CrossRef]
- Barucci, A.; D’Andrea, C.; Farnesi, E.; Banchelli, M.; Amicucci, C.; de Angelis, M.; Hwang, B.; Matteini, P. Label-Free SERS Detection of Proteins Based on Machine Learning Classification of Chemo-Structural Determinants. Analyst 2021, 146, 674–682. [Google Scholar] [CrossRef]
- Eiamchai, P.; Juntagran, C.; Somboonsaksri, P.; Waiwijit, U.; Eisiri, J.; Samarnjit, J.; Kaewseekhao, B.; Limwichean, S.; Horprathum, M.; Reechaipichitkul, W.; et al. Determination of Latent Tuberculosis Infection from Plasma Samples via Label-Free SERS Sensors and Machine Learning. Biosens. Bioelectron. 2024, 250, 116063. [Google Scholar] [CrossRef]
- Slipchenko, E.A.; Boginskaya, I.A.; Safiullin, R.R.; Ryzhikov, I.A.; Sedova, M.V.; Afanasev, K.N.; Nechaeva, N.L.; Kurochkin, I.N.; Merzlikin, A.M.; Lagarkov, A.N. SERS Sensor for Human Glycated Albumin Direct Assay Based on Machine Learning Methods. Chemosensors 2022, 10, 520. [Google Scholar] [CrossRef]
- Boginskaya, I.; Safiullin, R.; Tikhomirova, V.; Kryukova, O.; Nechaeva, N.; Bulaeva, N.; Golukhova, E.; Ryzhikov, I.; Kost, O.; Afanasev, K.; et al. Human Angiotensin I-Converting Enzyme Produced by Different Cells: Classification of the SERS Spectra with Linear Discriminant Analysis. Biomedicines 2022, 10, 1389. [Google Scholar] [CrossRef]
- Durmanov, N.N.; Guliev, R.R.; Eremenko, A.V.; Boginskaya, I.A.; Ryzhikov, I.A.; Trifonova, E.A.; Putlyaev, E.V.; Mukhin, A.N.; Kalnov, S.L.; Balandina, M.V.; et al. Non-Labeled Selective Virus Detection with Novel SERS-Active Porous Silver Nanofilms Fabricated by Electron Beam Physical Vapor Deposition. Sens. Actuators B Chem. 2018, 257, 37–47. [Google Scholar] [CrossRef]
- Kurochkin, I.N.; Eremenko, A.V.; Evtushenko, E.G.; Nechaeva, N.L.; Durmanov, N.N.; Guliev, R.R.; Ryzhikov, I.A.; Boginskaya, I.A.; Sarychev, A.K.; Ivanov, A.V.; et al. SERS for Bacteria, Viruses, and Protein Biosensing. In Macro, Micro, and Nano-Biosensors; Springer International Publishing: Cham, Switzerland, 2021; pp. 75–94. [Google Scholar]
- Vasko, P.D.; Blackwell, J.; Koenig, J.L. Infrared and Raman Spectroscopy of Carbohydrates. Carbohydr. Res. 1971, 19, 297–310. [Google Scholar] [CrossRef]
- Mathlouthi, M.; Seuvre, A.M.; Koenig, J.L. F.T.-I.R. and Laser-Raman Spectra of d-Ribose and 2-Deoxy-D-Erythro-Pentose (“2-Deoxy-d-Ribose”). Carbohydr. Res. 1983, 122, 31–47. [Google Scholar] [CrossRef]
- Wells, H.A.; Atalla, R.H. An Investigation of the Vibrational Spectra of Glucose, Galactose and Mannose. J. Mol. Struct. 1990, 224, 385–424. [Google Scholar] [CrossRef]
- Wiercigroch, E.; Szafraniec, E.; Czamara, K.; Pacia, M.Z.; Majzner, K.; Kochan, K.; Kaczor, A.; Baranska, M.; Malek, K. Raman and Infrared Spectroscopy of Carbohydrates: A Review. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2017, 185, 317–335. [Google Scholar] [CrossRef] [PubMed]
- Bell, A.F.; Hecht, L.; Barron, L.D. Disaccharide Solution Stereochemistry from Vibrational Raman Optical Activity. J. Am. Chem. Soc. 1994, 116, 5155–5161. [Google Scholar] [CrossRef]
- Gałat, A. Study of the Raman Scattering and Infrared Absorption Spectra of Branched Polysaccharides. Acta Biochim. Pol. 1980, 27, 135–142. [Google Scholar] [PubMed]
- Dudek, M.; Zajac, G.; Szafraniec, E.; Wiercigroch, E.; Tott, S.; Malek, K.; Kaczor, A.; Baranska, M. Raman Optical Activity and Raman Spectroscopy of Carbohydrates in Solution. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2019, 206, 597–612. [Google Scholar] [CrossRef] [PubMed]
- Dumouilla, V.; Dussap, C.G. Online Analysis of D-Glucose and D-Mannose Aqueous Mixtures Using Raman Spectroscopy: An in Silico and Experimental Approach. Bioengineered 2021, 12, 4420–4431. [Google Scholar] [CrossRef] [PubMed]
- Kačuráková, M.; Mathlouthi, M. FTIR and Laser-Raman Spectra of Oligosaccharides in Water: Characterization of the Glycosidic Bond. Carbohydr. Res. 1996, 284, 145–157. [Google Scholar] [CrossRef]
- Tahir, M.; Majeed, M.I.; Nawaz, H.; Ali, S.; Rashid, N.; Kashif, M.; Ashfaq, I.; Ahmad, W.; Ghauri, K.; Sattar, F.; et al. Raman Spectroscopy for the Analysis of Different Exo-Polysaccharides Produced by Bacteria. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 237, 118408. [Google Scholar] [CrossRef]
- Boginskaya, I.; Nechaeva, N.; Tikhomirova, V.; Kryukova, O.; Evdokimov, V.; Bulaeva, N.; Golukhova, E.; Ryzhikov, I.; Kost, O.; Afanasev, K.; et al. Human Angiotensin I-Converting Enzyme Study by Surface-Enhanced Raman Spectroscopy. J. Raman Spectrosc. 2021, 52, 1529–1539. [Google Scholar] [CrossRef]
- Zhang, T.; Li, Y.; Lv, X.; Jiang, S.; Jiang, S.; Sun, Z.; Zhang, M.; Li, Y. Ultra-Sensitive and Unlabeled SERS Nanosheets for Specific Identification of Glucose in Body Fluids. Adv. Funct. Mater. 2024, 34, 2315668. [Google Scholar] [CrossRef]
- Sun, X. Glucose Detection through Surface-Enhanced Raman Spectroscopy: A Review. Anal. Chim. Acta 2022, 1206, 339226. [Google Scholar] [CrossRef]
- Zhu, J.; Du, H.; Zhang, Q.; Zhao, J.; Weng, G.; Li, J.; Zhao, J. SERS Detection of Glucose Using Graphene-Oxide-Wrapped Gold Nanobones with Silver Coating. J. Mater. Chem. C 2019, 7, 3322–3334. [Google Scholar] [CrossRef]
- Perez-Mayen, L.; Oliva, J.; Salas, P.; De la Rosa, E. Nanomolar Detection of Glucose Using SERS Substrates Fabricated with Albumin Coated Gold Nanoparticles. Nanoscale 2016, 8, 11862–11869. [Google Scholar] [CrossRef]
- Schorr, H.C.; Schultz, Z.D. Chemical Conjugation to Differentiate Monosaccharides by Raman and Surface Enhanced Raman Spectroscopy. Analyst 2023, 148, 2035–2044. [Google Scholar] [CrossRef]
- Xu, J.; Jian, X.; Guo, J.; Zhao, J.; Tang, J.; Zhao, Y.; Xu, J.; Gao, Z.; Song, Y.-Y. Selective SERS Identification and Quantification of Glucose Enantiomers on Homochiral MOFs Based Enzyme-Free Nanoreactors. Chem. Eng. J. 2023, 459, 141650. [Google Scholar] [CrossRef]
- Mrozek, M.F.; Zhang, D.; Ben-Amotz, D. Oligosaccharide Identification and Mixture Quantification Using Raman Spectroscopy and Chemometric Analysis. Carbohydr. Res. 2004, 339, 141–145. [Google Scholar] [CrossRef]
- Jin, L.; Wang, S.; Cheng, Y. A Raman Spectroscopy Analysis Method for Rapidly Determining Saccharides and Its Application to Monitoring the Extraction Process of Wenxin Granule Manufacturing Procedure. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 241, 118603. [Google Scholar] [CrossRef]
- Baburin, A.S.; Ivanov, A.I.; Ryzhikov, I.A.; Trofimov, I.V.; Gabidullin, A.R.; Moskalev, D.O.; Panfilov, Y.V.; Rodionov, I.A. Crystalline Structure Dependence on Optical Properties of Silver Thin Film over Time. In Proceedings of the 2017 Progress in Electromagnetics Research Symposium—Spring (PIERS), St Petersburg, Russia, 22–25 May 2017; pp. 1497–1502. [Google Scholar]
- ISO 21920-2:2021; Geometrical Product Specifications (GPS)—Surface Texture: Profile Part 2: Terms, Definitions and Surface Texture Parameters. ISO: Vernier, Switzerland, 2021; p. 78.
- Shvalya, V.; Filipič, G.; Zavašnik, J.; Abdulhalim, I.; Cvelbar, U. Surface-Enhanced Raman Spectroscopy for Chemical and Biological Sensing Using Nanoplasmonics: The Relevance of Interparticle Spacing and Surface Morphology. Appl. Phys. Rev. 2020, 7, 031307. [Google Scholar] [CrossRef]
- Grésillon, S.; Aigouy, L.; Boccara, A.C.; Rivoal, J.C.; Quelin, X.; Desmarest, C.; Gadenne, P.; Shubin, V.A.; Sarychev, A.K.; Shalaev, V.M. Experimental Observation of Localized Optical Excitations in Random Metal-Dielectric Films. Phys. Rev. Lett. 1999, 82, 4520–4523. [Google Scholar] [CrossRef]
- Lee, C.; Robertson, C.S.; Nguyen, A.H.; Kahraman, M.; Wachsmann-Hogiu, S. Thickness of a Metallic Film, in Addition to Its Roughness, Plays a Significant Role in SERS Activity. Sci. Rep. 2015, 5, 11644. [Google Scholar] [CrossRef]
- Krylov, V.B.; Nifantiev, N.E. Synthetic Oligosaccharides Mimicking Fungal Cell Wall Polysaccharides. Curr. Top. Microbiol. Immunol. 2019, 425, 1–16. [Google Scholar] [CrossRef]
- Yashunsky, D.V.; Tsvetkov, Y.E.; Grachev, A.A.; Chizhov, A.O.; Nifantiev, N.E. Synthesis of 3-Aminopropyl Glycosides of Linear β-(1→3)-D-Glucooligosaccharides. Carbohydr. Res. 2016, 419, 8–17. [Google Scholar] [CrossRef]
- Yashunsky, D.V.; Karelin, A.A.; Tsvetkov, Y.E.; Nifantiev, N.E. Synthesis of 3-Aminopropyl β-(1 → 6)-D-Glucotetraoside and Its Biotinylated Derivative. Carbohydr. Res. 2018, 455, 18–22. [Google Scholar] [CrossRef]
- Krylov, V.B.; Gerbst, A.G.; Argunov, D.A.; Dmitrenok, A.S.; Shashkov, A.S.; Kaczynski, Z.; Huebner, J.; Holst, O.; Nifantiev, N.E. Definitive Structural Assessment of Enterococcal Diheteroglycan. Chem. Eur. J. 2015, 21, 1749–1754. [Google Scholar] [CrossRef]
- Argunov, D.A.; Trostianetskaia, A.S.; Krylov, V.B.; Kurbatova, E.A.; Nifantiev, N.E. Convergent Synthesis of Oligosaccharides Structurally Related to Galactan I and Galactan II of Klebsiella Pneumoniae and Their Use in Screening of Antibody Specificity. Eur. J. Org. Chem. 2019, 2019, 4226–4232. [Google Scholar] [CrossRef]
- Krylov, V.B.; Argunov, D.A.; Solovev, A.S.; Petruk, M.I.; Gerbst, A.G.; Dmitrenok, A.S.; Shashkov, A.S.; Latgé, J.-P.; Nifantiev, N.E. Synthesis of Oligosaccharides Related to Galactomannans from Aspergillus fumigatus and Their NMR Spectral Data. Org. Biomol. Chem. 2018, 16, 1188–1199. [Google Scholar] [CrossRef]
- Argunov, D.A.; Aladysheva, U.S.; Krylov, V.B.; Nifantiev, N.E. Acid-Catalyzed Transformation of Pyranosides into Furanosides as a Tool for Preparation of Furanoside Synthetic Blocks. Org. Lett. 2024, 26, 8090–8094. [Google Scholar] [CrossRef]
- Maghsoodloo, S.; Huang, C.-Y. Comparing the Overlapping of Two Independent Confidence Intervals with a Single Confidence Interval for Two Normal Population Parameters. J. Stat. Plan. Inference 2010, 140, 3295–3305. [Google Scholar] [CrossRef]
- Baburin, A.S.; Ivanov, A.I.; Lotkov, E.S.; Sorokina, O.S.; Boginskaya, I.A.; Sergeev, E.V.; Buzaverov, K.A.; Konstantinova, T.G.; Moskalev, D.O.; Issabayeva, Z.; et al. Epitaxial Silver Films Morphology and Optical Properties Evolution over Two Years. Coatings 2020, 10, 911. [Google Scholar] [CrossRef]

| Bands, cm−1 | Band Assignment | Ref. | |||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | ||
| 136 | 136 | ||||||
| 203 | |||||||
| 212 | |||||||
| 226 | |||||||
| 306 | 302 | - | - | - | 302 | ||
| - | - | - | - | 311 | - | ||
| 437 | 437 | 437 | - | - | 437 | δ(CCO) | [65] |
| - | - | - | - | 469 | - | δ(CCO) | [64] |
| - | 506 | - | - | - | - | ||
| - | - | - | - | - | 524 | ||
| 565 | - | - | - | 560 | |||
| - | - | 574 | - | - | - | ||
| - | 615 | - | - | - | 619 | ||
| - | - | - | - | 637 | - | ||
| 678 | - | 682 | - | - | - | ||
| 727 | 732 | ||||||
| - | - | - | 763 | - | - | δ(COH) | [65] |
| - | 807 | 807 | - | 803 | |||
| 816 | - | - | |||||
| - | - | - | - | 877 | 873 | δ(COH) | [65] |
| - | - | - | 882 | - | - | ||
| 891 | - | 891 | - | - | |||
| 904 | - | - | - | - | |||
| 973 | 973 | 973 | 973 | - | 969 | ν(CO) | [65] |
| 1008 | |||||||
| 1060 | 1060 | 1064 | 1068 | 1072 | 1068 | ω(CH2) | [65] |
| 1124 | |||||||
| 1141 | - | 1136 | - | 1136 | - | δ(COH) | [65] |
| - | - | - | 1175 | - | 1175 | ||
| - | - | - | 1208 | - | |||
| - | - | 1225 | - | - | - | ||
| 1242 | - | - | - | - | - | ||
| - | - | - | 1276 | 1276 | 1280 | ||
| - | - | 1292 | - | - | - | ||
| 1321 | |||||||
| - | - | - | 1375 | 1375 | 1371 | ω(CH2) | [66] |
| 1384 | 1384 | 1384 | - | - | - | ω(CH2) | [66] |
| - | - | - | - | 1449 | - | δ(CH2) | [64] |
| 1466 | 1462 | 1462 | - | - | 1462 | δ(CH2) | [65] |
| - | - | - | 1470 | - | - | δ(CH2) | [61] |
| - | - | 1539 | - | 1539 | |||
| - | - | 1551 | - | - | - | ||
| - | - | - | 1603 | 1603 | 1599 | ||
| 1627 | 1627 | 1627 | - | - | - | ||
| - | - | - | - | 1663 | 1671 | ||
| - | - | - | -- | - | 1750 | ||
| 2871 | - | - | - | 2864 | - | ||
| - | - | - | - | - | 2898 | ν(CH) | [61] |
| 2942 | 2945 | 2945 | 2945 | 2945 | 2948 | ν(CH2) | [61] |
| Analyte | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|
| 1 | 140, 311, 468, 637, 1012, 1016, 1068, 1375, 1461, 1469, 1538, 2894, 2941 | 140, 311, 468, 637, 1012, 1016, 1110, 1174, 1461, 1469, 1538, 2894, 2941, 2944 | 140, 311, 468, 637, 1012, 1016, 1068, 1072, 1110, 1174, 1375, 1461, 1469, 1538, 2894, 2941, 2944 | 140, 311, 468, 637, 1012, 1016, 1068, 1072, 1110, 1174, 1375, 1461, 1469, 1538, 2894, 2941, 2944 | 1012, 1016, 1068, 1072, 1110, 1174, 1375, 1461, 1469, 1538, 2894, 2941, 2944 |
| 2 | 140, 1012, 1016, 1110, 1174, 1375, 1538, 2941, 2944, 1461 | 140, 311, 468, 637, 1012, 1016, 1068, 1072, 1110, 1174, 1375, 1461, 1538, 2894, 2941, 2944 | 140, 311, 468, 637, 1012, 1016, 1068, 1072, 1110, 1174, 1375, 1461, 1469, 1538, 2894, 2941, 2944 | 140, 311, 468, 637, 1012, 1068, 1110, 1174, 1375, 1461, 1469, 1538, 2894, 2941, 2944, 1016 | |
| 3 | 140, 311, 468, 637, 1012, 1016, 1068, 1072, 1110, 1174, 1375, 1461, 1538, 2894, 2941, 2944 | 140, 311, 468, 637, 1012, 1016, 1068, 1072, 1110, 1174, 1375, 1461, 1469, 1538, 2894, 2941, 2944 | 140, 311, 468, 637, 1110, 1375, 1461, 1469, 1538, 2894, 2944 | ||
| 4 | 311, 468, 637, 1012, 1016, 1068, 1110, 1174, 1375, 1461, 1469, 1538, 2894, 2941, 2944 | 140, 311, 468, 637, 1012, 1016, 1068, 1072, 1110, 1174, 1375, 1461, 1469, 1538, 2894, 2941, 2944 | |||
| 5 | 140, 311, 468, 637, 1012, 1016, 1068, 1072, 1110, 1174, 1375, 1461, 1469, 1538, 2894, 2941, 2944 |
| Precision | Recall | F1-Score | Quantity | |
|---|---|---|---|---|
| (1–3) | 0.97 | 0.97 | 0.97 | 33 |
| (4–6) | 0.97 | 0.97 | 0.97 | 31 |
| Accuracy | 0.97 | 64 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zvyagina, J.Y.; Safiullin, R.R.; Boginskaya, I.A.; Slipchenko, E.A.; Afanas‘ev, K.N.; Sedova, M.V.; Krylov, V.B.; Yashunsky, D.V.; Argunov, D.A.; Nifantiev, N.E.; et al. Selective Detection of Fungal and Bacterial Glycans with Galactofuranose (Galf) Residues by Surface-Enhanced Raman Scattering and Machine Learning Methods. Int. J. Mol. Sci. 2025, 26, 4218. https://doi.org/10.3390/ijms26094218
Zvyagina JY, Safiullin RR, Boginskaya IA, Slipchenko EA, Afanas‘ev KN, Sedova MV, Krylov VB, Yashunsky DV, Argunov DA, Nifantiev NE, et al. Selective Detection of Fungal and Bacterial Glycans with Galactofuranose (Galf) Residues by Surface-Enhanced Raman Scattering and Machine Learning Methods. International Journal of Molecular Sciences. 2025; 26(9):4218. https://doi.org/10.3390/ijms26094218
Chicago/Turabian StyleZvyagina, Julia Yu., Robert R. Safiullin, Irina A. Boginskaya, Ekaterina A. Slipchenko, Konstantin N. Afanas‘ev, Marina V. Sedova, Vadim B. Krylov, Dmitry V. Yashunsky, Dmitry A. Argunov, Nikolay E. Nifantiev, and et al. 2025. "Selective Detection of Fungal and Bacterial Glycans with Galactofuranose (Galf) Residues by Surface-Enhanced Raman Scattering and Machine Learning Methods" International Journal of Molecular Sciences 26, no. 9: 4218. https://doi.org/10.3390/ijms26094218
APA StyleZvyagina, J. Y., Safiullin, R. R., Boginskaya, I. A., Slipchenko, E. A., Afanas‘ev, K. N., Sedova, M. V., Krylov, V. B., Yashunsky, D. V., Argunov, D. A., Nifantiev, N. E., Ryzhikov, I. A., Merzlikin, A. M., & Lagarkov, A. N. (2025). Selective Detection of Fungal and Bacterial Glycans with Galactofuranose (Galf) Residues by Surface-Enhanced Raman Scattering and Machine Learning Methods. International Journal of Molecular Sciences, 26(9), 4218. https://doi.org/10.3390/ijms26094218






