Abstract
Staphylococcus aureus (S. aureus) colonizes the nasal cavities of both healthy individuals and patients with chronic rhinosinusitis (CRS) with (CRSwNP) and without (CRSsNP) nasal polyps. Treatment-resistant S. aureus biofilms and intracellular persistence are common in CRS patients, requiring the expression of specific virulence factor genes to transition into these forms. We hypothesized that S. aureus isolates from non-diseased controls, CRSsNP patients, and CRSwNP patients would exhibit distinct virulence factor patterns contributing to persistence and intracellular survival in CRS patients. Nasal swabs from seventy-seven individuals yielded S. aureus cultures in eight non-diseased controls, eight CRSsNP patients, and five CRSwNP patients. Whole-genome sequencing analyzed stress, antimicrobial resistance, and virulence genes, including plasmids and prophages. Four virulence factor gene patterns emerged: a core set (hlgA, icaC, hlgB, hlgC, hld, and aur) present in all isolates, and accessory sets, including the enterotoxin gene cluster (seo, sem, seu, sei, and sen) and a partial/complete invasive virulence factor set (splE, splA, splB, lukE, and lukD) (p = 0.001). CRSwNP isolates exhibited incomplete carriage of the core set, with frequent loss of scn, icaC, and hlgA (p < 0.05). These findings suggest that S. aureus has clusters of virulence factors that may act in concert to support the survival and persistence of the bacteria, resulting in enhanced pathogenicity. This may manifest clinically with resistant disease and refractoriness to antibiotics.
1. Introduction
Staphylococcus aureus (S. aureus), a Gram-positive commensal bacterium, can be associated with a wide spectrum of pathology, ranging from asymptomatic colonization of the nares to being the leading cause of nosocomial bacteremia with an associated mortality of 15–60% [1]. Due to the diverse phenotypic behavior of S. aureus, it has been difficult to characterize its involvement in diseases such as chronic rhinosinusitis (CRS). S. aureus colonizes the nasal cavity in 64% of patients with nasal polyps (CRSwNP) compared with 33% of those without polyps (CRSsNP) and 20% of those without CRS [2,3]. A higher proportion of patients with CRSwNP demonstrate IgE towards S. aureus enterotoxins in their serum than those without CRS (22.6–32.5% vs. 6.7–14.3% of controls) [4]. Culture of S. aureus pre- and post-operatively in patients with CRS is a poor prognostic indicator for disease recurrence and recalcitrance [5]. However, the factors responsible for the enhanced pathogenicity of S. aureus strains prevalent in difficult-to-treat CRS disease remain poorly understood.
S. aureus can persist in the nasal cavity of both asymptomatic carriers and CRS patients, evading host immune responses and the effects of antimicrobial therapies [6]. In CRS, S. aureus appears to frequently adopt invasive strategies, including localizing intracellularly within host cells and forming extracellular biofilms, both of which are facilitated by the expression of specific virulence factors [7,8,9,10]. These virulence factors can be broadly categorized into three main groups: adherence factors, pore-forming toxins, and superantigens [11]. Adherence factors, or adhesins, enable S. aureus to bind to components of the host extracellular matrix. Key adhesins include staphylococcal protein A, fibronectin binding proteins A and B, and clumping factors A and B. These proteins play crucial roles in the initial attachment to host tissues and production of biofilms [12,13]. Pore-forming toxins include α toxin, leukocidins, β-hemolysin, and phenol-soluble modulins. These toxins disrupt host cell membranes and host cell components, leading to cell lysis and immune evasion. α toxin exhibits broad lytic activity against epithelial and endothelial cells, T cells, monocytes, macrophages, and platelets, as well as contributes to phagosome escape [14]. Leukocidins lyse neutrophils and erythrocytes, while β-hemolysin has cytotoxic effects on keratinocytes, polymorphonuclear leukocytes, monocytes, and T lymphocytes and is also implicated in biofilm development and phagosomal escape [11,15,16]. Phenol-soluble modulins are cytotoxic to both red and white blood cells and contribute to biofilm structuring and detachment [17,18]. Superantigens are potent T-cell mitogens that bind the variable β-chain of the T-cell receptor, triggering widespread T-cell activation, B-cell proliferation, increased local IgE production, and a robust type 2 inflammatory cytokine response. Staphylococcus enterotoxin B, a well characterized superantigen, has been shown to recruit mast cells to the epithelial and subepithelial layers of CRS patients’ nasal explant tissues and promote mast cell S. aureus uptake and degranulation [11,19,20].
Before the widespread adoption of whole-genome sequencing, S. aureus virulence and strain outbreaks were often inferred using accessory gene regulator (agr) locus typing and staphylococcal protein A gene (spa) typing, both of which provide insights into the virulence potential [21,22,23]. The agr locus plays a key role in regulating the expression of numerous S. aureus virulence factors. Loss-of-function or frameshift mutations within this region have been associated with the emergence of small colony variants (SCVs), which exhibit reduced virulence due to diminished expression of these factors [24]. Recent evidence has shown an increased number of mobile genetic elements (MGEs), including prophages and plasmids, in CRS isolates, providing additional virulence and antimicrobial resistance genes that may well support S. aureus survival and enhance pathogenicity [25,26].
Given the different selection pressures and distinct environments between the non-diseased, CRSsNP, and CRSwNP nasal mucosa, we hypothesized that differences in stress, antimicrobial resistance, and virulence genes exist among S. aureus isolates cultured from each condition, contributing to enhanced pathogenicity of the bacteria and manifesting clinically as disease recalcitrance and treatment resistance. To test this, we conducted a prospective investigation of S. aureus isolates cultured from control, CRSsNP, and CRSwNP patients, analyzing the genomic profile and the associated MGEs for stress, virulence, and antimicrobial resistance genes.
2. Results
2.1. Patient Demographics
Nasal swabs were obtained from 77 patients enrolled in the study, including 30 control subjects, 20 patients with CRSsNP, and 27 patients with CRSwNP. S. aureus was cultured in eight control subjects, eight CRSsNP patients, and five CRSwNP patients. A significantly higher proportion of asthmatic individuals was observed within the CRSwNP group (55%) compared with the control (13.3%) and CRSsNP (20%) groups (p = 0.001). Additionally, oral steroid use within the month preceding sample collection was significantly higher in the CRSwNP group (22.2%) compared to controls (6.67%) and CRSsNP patients (0%) (p = 0.03). Smoking prevalence was greater in the control group (13.3%), with no current smokers identified in either the CRSsNP or CRSwNP groups (p = 0.04). Other demographic variables did not differ significantly between the groups (Table 1).

Table 1.
Demographic profile of the study population.
A subgroup analysis was performed on subjects from whom S. aureus was cultured. A significantly higher Modified Lund–Mackay Score (MLMS) was observed for patients with S. aureus colonization (mean score 12.0) compared to those without S. aureus (mean score 7.5, p = 0.04). No other statistically significant differences were identified (Table 2).

Table 2.
Demographic profile based on a sub-analysis of the S. aureus culture status.
2.2. Bacterial Genome Sequencing
Illumina short-read, paired-end sequencing was performed on twenty-one S. aureus isolates, including eight from control subjects, eight from CRSsNP patients, and five from CRSwNP patients. Paired-end reads were assembled and analyzed using the Bactopia bacterial alignment and analysis pipeline [27]. The assembled genome sizes spanned between 2.67 and 3.61 Mbp.
2.3. Staphylococcal Protein A and agr Loci and Frameshift Assessment
An analysis of the highly variable region of spa identified three duplicated spa types: t008, t015, and t571. Two of these were shared between the control and CRSwNP groups, while the third was found in both the control and CRSsNP groups.
The agr locus was predominantly type I and found in 67% of the isolates. Type II was observed in 19% and type III in 14% of isolates (n = 21). One isolate from the CRSwNP group displayed a frameshift insertion mutation in the hld gene (encoding the delta hemolysin toxin) while another showed an absent hld gene (Table 3).

Table 3.
spa and agr types and assessment of the agr operon.
2.4. Stress, Antimicrobial Resistance, and Virulence Genes
Minimal differences were observed between the patient groups when comparing stress genes. The lmrS gene was present in 90% of isolates, while cadD was identified in 52%, with no statistically significant differences in genes detected using a Chi-square test (Figure 1a). Antimicrobial resistance genes exhibited greater variability, with tet(38) found in 95% of isolates and mepA in 90%, both conferring tetracycline resistance (n = 21). Penicillin resistance genes (blaI, blaZ, blaPC1, and blaR1) were identified in 75% of control isolates (n = 8), 50% of CRSsNP isolates (n = 8), and 100% of CRSwNP isolates (n = 5). The macrolide, lincosamide, and streptogramin resistance genes erm(T) and erm(A) were identified in 25% of control isolates (n = 8), 37% of CRSsNP isolates (n = 8), and 20% of CRSwNP isolates (n = 5). The presence of the fosfomycin resistance gene fosB approached significance (p = 0.08), with 62.5% of control isolates harboring this gene (n = 8) compared to 12.5% of CRSsNP isolates (n = 8) and 20% of CRSwNP isolates (n = 5) (Figure 1b).

Figure 1.
Stress and antimicrobial resistance gene carriage in control, CRSsNP, and CRSwNP S. aureus isolates. Each heatmap demonstrates the presence (blue) or absence (white) of (a) stress and (b) antimicrobial resistance genes from each isolate.
Four dominant virulence gene clusters were identified. All isolates contained the majority of a core set of virulence genes, including hlgA, icaC, hlgB, hlgC, hld, and aur. This core set was identified alone or supplemented by accessory virulence factor genes such as the enterotoxin gene cluster (seo, sem, seu, sei, and sen) or a full or partial cluster of invasive virulence genes (splE, splA, splB, lukE, and lukD). Among the isolates from the control group, 50% contained the invasive virulence genes and 37.5% carried the enterotoxin gene cluster (n = 8). Within the CRSsNP group, 25% of isolates showed a partial presence of the invasive virulence gene cluster, while 12.5% demonstrated an enterotoxin gene cluster (n = 8). In the CRSwNP group, 60% showed a complete enterotoxin gene cluster and 40% showed the invasive gene cluster (n = 5). The hld gene was absent in the CRSwNP2 isolate and had previously exhibited a frameshift mutation in CRSwNP3.
The Chi-square analysis revealed a statistically significant absence of hlgA in 40% of CRSwNP isolates (p = 0.03), icaC was absent in 40% of CRSwNP isolates (p = 0.03), and scn was absent in 60% of CRSwNP isolates and one control isolate (p = 0.02) (Figure 2).
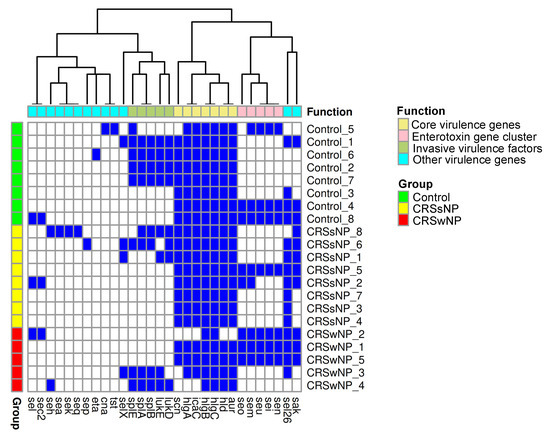
Figure 2.
Virulence gene carriage in control, CRSsNP, and CRSwNP S. aureus isolates. The heatmap demonstrates the presence (blue) or absence (white) of virulence genes in each isolate.
The cluster analysis method t-SNE confirmed the presence of the four distinct virulence gene cluster patterns (Figure 3). An initial PERMANOVA test for the significance of clusters based on the control, CRSsNP, and CRSwNP groups showed no significant associations (p = 0.26). However, when testing the significance of our four distinct virulence gene cluster patterns, PERMANOVA revealed a significant result (p = 0.001).
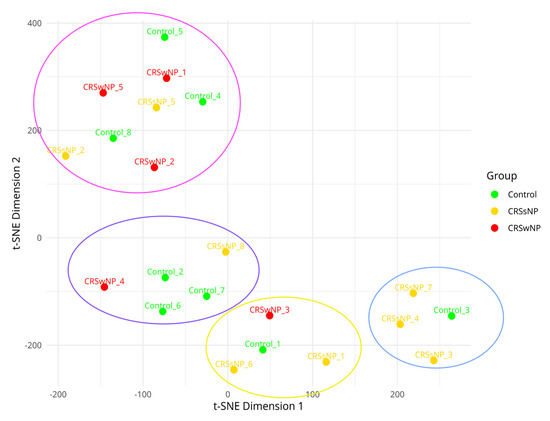
Figure 3.
T-distributed stochastic neighbor embedding of virulence factor presence and absence matrix demonstrating the clustering of virulence factor patterns. Isolates are labelled using their titles. The blue bubble represents the core virulence factors alone, and the pink bubble represents the core and enterotoxin gene cluster. The yellow bubble represents a partial invasive virulence gene cluster, and the purple bubble represents the core and invasive virulence gene clusters.
2.5. Mobile Genetic Element Identification
The mean number of plasmids in each group was calculated, demonstrating an average of 1.4 plasmids per isolate in the control group, 1.7 in the CRSsNP group, and 2.4 in the CRSwNP group; however, this difference was not statistically significant using one-way ANOVA (p = 0.15). Fourteen plasmid clusters were identified based on Blast high scoring pairs (HSPs), with no statistically significant differences between the plasmid cluster distribution and disease group (Figure 4a).
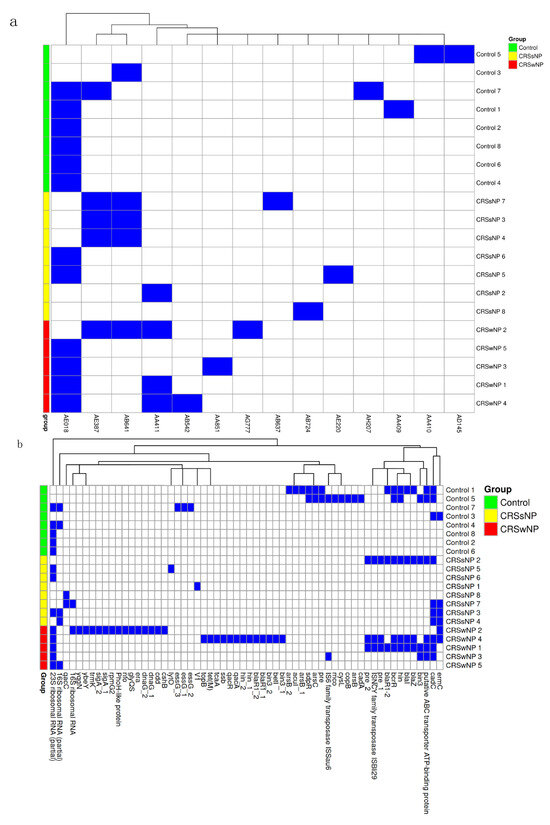
Figure 4.
Plasmid identification and genes. (a) Heatmap of Blast high-scoring pairs from MOB-suite. The presence of HSP is indicated by the blue rectangle and its absence is indicated by the white rectangle. (b) Heatmap of sequenced genes from each identified plasmid listed by group (blue = present and white = absent).
We further examined the individual genes encoded by plasmids. On average, plasmids contributed 15.87 genes to control isolates, 10.53 to CRSsNP isolates, and 30.6 to CRSwNP isolates, with no significant differences between groups (one-way ANOVA, p = 0.27). Approximately 59–67% of the identified genes were either hypothetical or not characterized. Notably, several antimicrobial resistance genes were detected on plasmids, including ermC, blaZ, blaI, blaR1, and tet(M). Erythromycin resistance genes were more frequently located on plasmids than in the core genome, appearing in one control, three CRSsNP, and two CRSwNP isolates. The penicillin resistance gene blaZ was found on plasmids from one control, one CRSsNP, and two CRSwNP isolates, none of which exhibited these genes in the core genome. Plasmids appeared to contribute few stress and virulence genes, with no clear concentration in any group (Figure 4b).
The mean number of prophages in each group was calculated, demonstrating 2.38 prophages per CRSsNP isolate, compared to 1.86 in control isolates and 1.60 in CRSwNP isolates; this difference was not significant using one-way ANOVA (p = 0.35). We also assessed the number of prophage genes per isolate, finding an average of 44.0 prophage genes in control isolates, 34.5 in the CRSsNP isolates, and 34.4 in the CRSwNP isolates. A large proportion of identified genes were hypothetical or not characterized (70–68%). Subsequently, we mapped known prophage genes to assess their contributions to virulence. The resulting heatmap showed a sparse distribution with no significant differences between the groups (Figure 5).
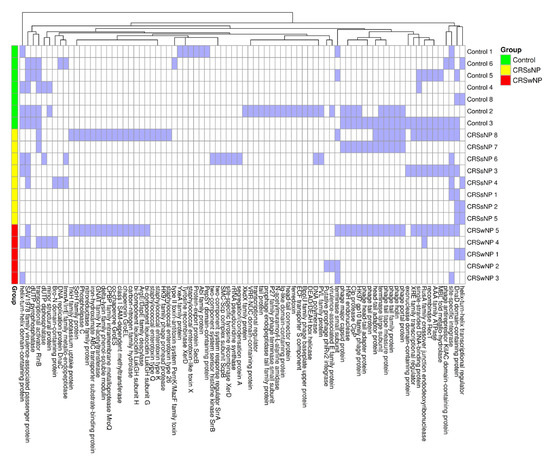
Figure 5.
Heatmap of PhiSpy with the identified phage genes listed by group (blue = present and white = absent).
3. Discussion
Our findings provide a detailed assessment of the S. aureus genome and MGEs in isolates from the middle meatuses of CRS patients with comparisons to disease-free control subjects and may have important implications for our understanding of virulence and antimicrobial resistance.
The demographic characteristics of our cohort align with those of previous studies, showing a significantly higher prevalence of asthma in the CRSwNP group compared with the CRSsNP and control groups [28,29]. Inhalant allergy prevalence was also elevated in the CRSwNP group (29.6%) compared to the CRSsNP (15.0%) and control (26.6%) groups, with higher rates across all groups relative to other studies, likely due to recruitment from surgical lists and rhinology clinics [28]. The MLMS was significantly higher in the CRSwNP group (13.5) compared to the CRSsNP (7.9) and control (3.6) groups, as expected based on the European Position Paper on Rhinosinusitis and Nasal Polyps 2020 diagnostic criteria, and were similar to previously reported values [30,31]. Smoking incidence was significantly higher in our control group, possibly due to primary prevention counselling in the CRS and asthma groups [29,32].
S. aureus colonization in the CRSwNP group was lower than expected, at 18.5%, although some studies have observed similarly low rates [3,33]. This discrepancy may be attributed to higher antimicrobial use in the CRSwNP group (23.5%). The subgroup analysis revealed a significant association between S. aureus colonization and a higher MLMS, consistent with other reports [34].
The S. aureus agr locus regulates the production of RNAII, RNAIII, and downstream virulence factors, functioning as a quorum-sensing system. Numerous studies classify the agr locus into groups I-IV based on polymorphisms in the agrB, agrC, and agrD genes, which influence virulence factor expression and are related to specific disease manifestations [21]. Most isolates belonged to group I, which is typically associated with commensal carriage, urinary infections, and bacteremia, with no significant differences detected between the groups [35,36]. The hld gene forms part of the RNAIII transcript and agr operon, and its absence or frameshift mutation abolishes the transcription of the accessory gene regulator [agrA] and downstream virulence factors [37]. The hld gene was absent or showed a frameshift mutation in 40% of CRSwNP isolates but was present in all other groups. Mutations in the agr locus and hld gene have been shown to create senescent bacteria known as small colony variants (SCVs), which show superior intracellular translocation in epithelial cells [38,39,40]. Furthermore, hld mutations have been shown to create larger biofilms in comparison to those with a functional hld gene [41]. Stress gene patterns across all groups displayed no significant differences. The penicillin resistance gene blaZ was present in 75% of control isolates, 25% of CRSsNP isolates, and 100% of CRSwNP isolates when reviewing the core and plasmid genomes. These findings contrast with those of Jervis Bardy et al., as they detected a persistent rate of blaZ carriage of 57% in both the CRSsNP and CRSwNP groups, suggesting differences in selection pressures between the studies [42]. The clustering analysis suggested the presence of four recurring virulence factor patterns, potentially reflecting distinct modes of pathogenicity across the different groups. These included a core set of virulence factors alone, a core set with an enterotoxin gene cluster, or a core set with either a partial or complete invasive virulence factor cluster. Notably, no isolates contained both the enterotoxin and invasive virulence factor clusters simultaneously.
The core set, including hlgA, hlgB, hlgC, icaC, scn, hld and aur, was present in nearly all isolates, though some CRSwNP isolates exhibited a partial loss of the core genes. The bi-component gamma-hemolysins (hlgB and hlgC) were well conserved across all groups. However, the hlgA gene was significantly absent in 40% of CRSwNP isolates [43]. The immune modulator aureolysin (aur), which cleaves the C3 complement protein, was present in all groups but absent in a single CRSwNP isolate [44]. The intercellular adhesion gene icaC, part of the intracellular adhesion (ica) locus responsible for biofilm formation, was unexpectedly absent in 40% of CRSwNP isolates, though it was present in the other groups. The icaC gene product mediates the transfer of polysaccharide intercellular adhesion molecules to the cell membrane, facilitating the development of longer poly-N-acetylglucosamine oligomers and larger biofilms [45,46]. The staphylococcal complement inhibitor, typically found on the immune evasion cluster (scn) of MGEs, was absent in 60% of CRSwNP isolates. The scn gene product binds and inactivates the complement component C3b, which is a potent activator of B cells. Consequently, scn gene absence could contribute to the increased IgE response against S. aureus seen in CRSwNP patients’ nasal tissues [3,25].
In addition to core virulence factors, an accessory cluster of invasive virulence factors was identified, consisting of lukE, lukD, splE, splA, and splB. This gene cluster was fully present in 50% of control isolates and 40% of CRSwNP isolates, and partially present (i.e., missing one or more genes) in 37.5% of CRSsNP isolates. The lukE/D gene product leukocidin E/D induces calcium channel activation in neutrophils, leading to cell death and compromised local immunity [43,47]. The splA gene product cleaves the mucin 16 glycoprotein, which forms a defensive mucosal barrier, while the splB gene product inhibits complement activation by cleaving several complement components (C3–C9), blocking opsonophagocytosis and the terminal complement cascade [48]. The target of splE remains unclear.
The enterotoxin gene cluster of superantigens (sei, sem, sen, seo, and seu) formed a further accessory group of virulence factors. They were present in 37.5% of control isolates, 60% of CRSwNP isolates, and 12.5% of CRSsNP isolates, with a further CRSsNP isolate showing partial presence. These enterotoxins are associated with long-term colonization in the nasal airway, cystic fibrosis lungs, and atopic dermatitis wounds [49]. They stimulate T-cell proliferation but appear to have unexpectedly low levels of immunoglobulins raised towards them in human serum when compared with other superantigens [49]. The precise function of these enterotoxins is yet to be fully elucidated.
Plasmid carriage sequentially increased from control to CRSsNP and CRSwNP isolates. Others have demonstrated an increasing plasmid copy number over time in S. aureus cultured from CRS patients, increasing their ability to produce antimicrobial-resistant biofilms [26]. We therefore studied the mobile genes each plasmid provided, identifying tetracycline, macrolide, and penicillin resistance genes. There appeared to be no differences in known plasmid-encoded genes between groups; however, a significant number of genes were classified as hypothetical or of unknown function. As a result, the functional significance of any observed differences might be difficult to interpret due to the presence of these genes with uncharacterized functions to date. We identified a lower carriage of prophages in the CRSwNP group than in the control and CRSsNP groups. Prophage encoded genes relevant to S. aureus virulence were sparsely detected, but there were no antimicrobial resistance genes identified on prophages, in keeping with others’ results [25]. The detection of these genes was infrequent and did not significantly contribute to the overall pattern of virulence factors previously noted.
This study has some limitations. Patient demographic data, including medication history, were recorded by questionnaire. Therefore, a one-month timeframe was used to record antibiotic and steroid use in order to reduce recall bias. It is possible that longer-term exposures could have influenced S. aureus carriage, antimicrobial resistance and virulence factor patterns, which warrant further study. While our findings highlight potential differences in antimicrobial resistance and virulence gene patterns between patients with CRS phenotypes and controls, the small sample size limits the strength of conclusions that can be drawn. This limitation was partly a result of our sampling technique, which prioritized the recovery of viable S. aureus isolates for future downstream experimental analyses, including transcriptomic and proteomic validation. Sequencing large numbers of genes often reveals significant heterogeneity, complicating statistical significance testing. More subtle differences may have been overlooked, which might have been identified in larger cohorts. Reflecting on our results, taking multiple swabs of each patient’s middle meatus may have identified multiple S. aureus isolates with different virulence factor patterns that could work synergistically. Furthermore, culture of the sinonasal tissue may have boosted the S. aureus culture yield; however, it would have been unethical to harvest tissue from control participants due to the inherent risks associated with the procedure. Nevertheless, our findings complement existing sequencing data and contribute to a more detailed understanding of mechanisms that enhance S. aureus pathogenicity in CRS patients.
4. Materials and Methods
4.1. Subjects
Patients undergoing a nasal endoscopic examination in rhinology clinics and those undergoing endoscopic sinus surgery procedures by the senior authors (H.A.S.J., P.G.H., and R.J.S.) at University Hospital Southampton between 22 April 2021 and 9 August 2022 were invited to participate. Subjects that met the European Position Paper on Rhinosinusitis and Nasal Polyps 2020 [30] criteria for the diagnosis of CRS were stratified into CRSsNP and CRSwNP, and those with other diagnoses, including allergic rhinitis, nasal masses, and anatomical nasal obstruction, were placed in the control group. The exclusion criteria included patients under 18 years of age; those with cystic fibrosis, primary ciliary dyskinesia, or immune deficiency syndromes; those with an inability to provide informed consent; and those with blood-borne viruses, including hepatitis B, hepatitis C, and human immunodeficiency virus. Demographic data, including age, sex, atopic status, antibiotic and steroid use in the past month, medical history, history of asthma, and smoking habits, were collected by questionnaire. Nasal swabs were taken either intraoperatively or under topical anesthetic from the middle meatal region in the clinic. Ethical approval was obtained for the study via the National Health Service (UK), London—Hampstead Research Ethics Committee (REC: 20/PR/0183). All participants provided written informed consent.
4.2. Specimen Testing and Genomic Sequencing
Bacterial swabs (M40 transystem, Copan, Brescia, Italy) were spread onto S. aureus 24 h brilliance agar plates (Oxoid, Basingstoke, UK). Blue coagulase-positive colonies were tested for catalase and DNase and subjected to MALDI-TOF to confirm their identity as S. aureus species. Positive S. aureus cultures were grown to logarithmic growth in Rosewell Park Memorial Institute Medium 1640 (Life Technologies, Paisley, UK) at 37 °C and frozen in the presence of 25% glycerol (VWR, Lutterworth, UK) at −80 °C. All 21 collected S. aureus strains were prepared in RNA Shield (Zymo Research, Freiburg im Breisgau, Germany) and underwent 30× Illumina short-read sequencing (MicrobesNG, Birmingham, UK).
4.3. Alignment and Assembly of the Genome
Twenty-one paired-end sequencing reads were assembled and annotated using Bactopia (v3.0.0). Quality control was performed using Bbtools (v38.96), Fastp (v0.23.2), FastQC (v0.11.9), and Lighter (v1.1.2), followed by genome assembly with Shovill (v1.1.0) and annotation with Prokka (v1.14.6) and Bakta (v1.8.2) [50,51,52,53].
4.4. Typing of the spa and agr Loci and Detection of Frameshifts
The Bactopia workflow staphtyper was used to analyze our assemblies [54]. This elucidated the staphylococcal protein A gene (spa) type and accessory gene regulator (agr) locus type. The agr locus was further investigated by aligning our sequences to the S. aureus Newman strain (GenBank accession: GCA_040702955.1) using snippy (v4.6.0), followed by manual inspection using Integrated Genome Viewer (v2.18.2) [55].
4.5. Stress, Virulence, and Antimicrobial Resistance Genes
Stress, virulence, and antimicrobial resistance genes were assessed using the Bactopia workflow AMRFinderPlus (v3.11.18) [56]. Detected genes were imported into the statistical package R (v4.3.1) [57]. Genes with greater than 90% sequence coverage and homology were transformed into presence/absence matrices. Isolates were then clustered within their respective groups using complete linkage clustering and heatmaps were generated using the pheatmap package (v1.0.12) [58].
The patterns of virulence factor genes were further analyzed using t-distributed stochastic neighbor embedding (t-SNE) in R using the Rtsne package (v0.17) and subsequently visualized using ggplot2 (v3.4.0) [59,60]. The significance of clusters was assessed by PERMANOVA using the vegan package (v2.6-10) [61].
4.6. Plasmid Detection
Plasmids were detected using the Bactopia workflow MOB-suite (v3.1.7), generating a Blast HSP table for each isolate along with a FASTA file of each plasmid’s sequence [62,63]. The FASTA files were interrogated using Prokka (v1.14.6) to generate a list of the genes present in each plasmid [52]. The resulting data had hypothetical genes removed and were transformed into a gene presence/absence matrix. This matrix was subsequently visualized using pheatmap [58].
4.7. Prophage Detection
Prophages were detected using PhiSpy (v4.2.21) executed with the –-output_choice 9 flag, providing the number of prophages and predicted phage-associated genes from the genome [64]. The resulting predicted prophage genes were used to create a presence/absence matrix, which was visualized using the pheatmap package [58].
4.8. Statistical Analysis
A statistical analysis of patient demographics was performed using SPSS (v29.0.2.0, IBM, Portsmouth, UK). Data normality was assessed through histogram plots and normality tests. Pearson’s Chi-square test and one-way ANOVA were used to compare categorical and numerical demographic data, respectively. Differences in genomic information were analyzed in R, with categorical data such as gene presence assessed using the Chi-square analysis and numerical data assessed using one-way ANOVA [57].
5. Conclusions
In summary, our findings support the presence of a core virulence factor set consisting of hlgA, hlgB, hlgC, hld, icaC, aur, and scn, which is detected in nearly all S. aureus isolates, but appears to be incompletely carried in some CRSwNP isolates, with the loss of scn, icaC, and hlgA. The loss of these genes may support bacterial survival by facilitating the formation of SCVs and biofilms and the promotion of IgE production through B-cell activation. This core virulence set is observed both independently and in conjunction with accessory virulence factor sets, including the enterotoxin or complete/partial invasive virulence gene clusters. These previously unrecognized virulence patterns likely represent distinct phenotypes that act synergistically in promoting bacterial colonization and persistence, and manifest clinically with disease recalcitrance and refractoriness to antibiotics.
Author Contributions
S.P.G., A.F.W. and R.J.S. contributed to the study conception and design. R.J.S., P.G.H. and H.A.S.J. were responsible for tissue sample and data collection. Sample preparation, data collection, and analysis were performed by S.P.G. and L.C.L. The first draft of the manuscript was written by S.P.G. and all authors advised on previous drafts. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the following: Royal College of Surgeons of England through the Dr Shapurji H Modi Memorial Research Fellowship, a pump priming grant provided by the Royal College of Surgeons Edinburgh (grant number SPPG/21/158), and a British Rhinological Society research grant.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and was approved by the Ethics Committee of the National Health Service (UK), London—Hampstead Research Ethics Committee (REC reference: 20/PR/0183, Approved 21 July 2020) for studies involving humans.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Paired-end reads of the reported S. aureus strains and supporting data have been deposited in the European Nucleotide Archive at EMBL-EBI under accession number PRJEB81780 “https://www.ebi.ac.uk/ena/browser/view/PRJEB81780 (accessed on 28 October 2024)”.
Acknowledgments
We are indebted to all the study participants, who provided the swabs used to isolate CRS-specific strains of S. aureus. We are grateful for the assistance received from the staff of the Biomedical Imaging Unit at the Faculty of Medicine, in particular David Johnston. We would also like to thank David Cleary for his advice and assistance in the bioinformatics approach used in this study. This study was supported by the Southampton National Institute of Health Research (NIHR) Respiratory Biomedical Research Unit and the Southampton NIHR Wellcome Trust Clinical Research Facility.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Abbreviations
The following abbreviations are used in this manuscript:
| agr | Accessory gene regulator |
| ANOVA | Analysis of Variance |
| aur | Aureolysin gene |
| blaZ | Beta-lactamase gene |
| cadD | Cadmium resistance gene |
| CRS | Chronic rhinosinusitis |
| CRSsNP | Chronic rhinosinusitis without nasal polyps |
| CRSwNP | Chronic rhinosinusitis with nasal polyps |
| DNA | Deoxyribonucleic acid |
| erm(A), erm (T), erm (C) | Erythromycin resistance methylase genes |
| fosB | Fosfomycin resistance gene |
| hlgA, hlgB, hlgC | Gamma hemolysin genes |
| HSP | High scoring pairs |
| hld | Delta-hemolysin gene |
| icaC | Biofilm-associated intercellular adhesion gene |
| lmrS | Lincomycin resistance protein of S. aureus |
| lukE, lukD | Leukocidin genes |
| MALDI-TOF | Matrix-assisted laser desorption/ionization time of flight |
| MGE | Mobile genetic element |
| MLMS | Modified Lund–Mackay Score |
| PERMANOVA | Permutational multivariate analysis of variance |
| RNA | Ribonucleic acid |
| RNAII | Second transcript of the agr operon |
| RNAIII | Third transcript of the agr operon |
| S. aureus | Staphylococcus aureus |
| SCV | Small colony variant |
| sei, sem, sen, seo, seu | Enterotoxin gene cluster |
| spa | Staphylococcal protein A gene |
| splA, splB, splE | Serine protease genes |
| tet(38), tet(M) | Tetracycline resistance genes |
| t-SNE | t-distributed stochastic neighbor embedding |
References
- Cosgrove, S.E.; Sakoulas, G.; Perencevich, E.N.; Schwaber, M.J.; Karchmer, A.W.; Carmeli, Y. Comparison of mortality associated with methicillin-resistant and methicillin-susceptible Staphylococcus aureus bacteremia: A meta-analysis. Clin. Infect. Dis. 2003, 36, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Vickery, T.W.; Ramakrishnan, V.R.; Suh, J.D. The Role of Staphylococcus aureus in Patients with Chronic Sinusitis and Nasal Polyposis. Curr. Allergy Asthma Rep. 2019, 19, 21. [Google Scholar] [CrossRef] [PubMed]
- Van Zele, T.; Gevaert, P.; Watelet, J.B.; Claeys, G.; Holtappels, G.; Claeys, C.; van Cauwenberge, P.; Bachert, C. Staphylococcus aureus colonization and IgE antibody formation to enterotoxins is increased in nasal polyposis. J. Allergy Clin. Immunol. 2004, 114, 981–983. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.Y.; Miao, J.L.; Lu, H.Q.; Qi, Q.H.; Chen, X.I.; Xu, J.; Lin, Z.P.; Chen, Z.B.; Yin, M.; Cheng, L. Serum levels of specific IgE to Staphylococcus aureus enterotoxins in patients with chronic rhinosinusitis. Exp. Ther. Med. 2015, 9, 1523–1527. [Google Scholar] [CrossRef]
- Maniakas, A.; Asmar, M.H.; Renteria Flores, A.E.; Nayan, S.; Alromaih, S.; Mfuna Endam, L.; Desrosiers, M.Y. Staphylococcus aureus on Sinus Culture Is Associated With Recurrence of Chronic Rhinosinusitis After Endoscopic Sinus Surgery. Front. Cell Infect. Microbiol. 2018, 8, 150. [Google Scholar] [CrossRef]
- Watkins, K.E.; Unnikrishnan, M. Evasion of host defenses by intracellular Staphylococcus aureus. Adv. Appl. Microbiol. 2020, 112, 105–141. [Google Scholar]
- Tan, N.C.; Foreman, A.; Jardeleza, C.; Douglas, R.; Vreugde, S.; Wormald, P.J. Intracellular Staphylococcus aureus: The Trojan horse of recalcitrant chronic rhinosinusitis? Int. Forum Allergy Rhinol. 2013, 3, 261–266. [Google Scholar] [CrossRef]
- Singhal, D.; Foreman, A.; Jervis-Bardy, J.; Wormald, P.J. Staphylococcus aureus biofilms: Nemesis of endoscopic sinus surgery. Laryngoscope 2011, 121, 1578–1583. [Google Scholar] [CrossRef]
- Hayes, S.M.; Howlin, R.; Johnston, D.A.; Webb, J.S.; Clarke, S.C.; Stoodley, P.; Harries, P.G.; Wilson, S.J.; Pender, S.L.; Faust, S.N.; et al. Intracellular residency of Staphylococcus aureus within mast cells in nasal polyps: A novel observation. J. Allergy Clin. Immunol. 2015, 135, 1648–1651. [Google Scholar] [CrossRef]
- Foster, T.J.; Geoghegan, J.A.; Ganesh, V.K.; Hook, M. Adhesion, invasion and evasion: The many functions of the surface proteins of Staphylococcus aureus. Nat. Rev. Microbiol. 2014, 12, 49–62. [Google Scholar] [CrossRef]
- Shaghayegh, G.; Cooksley, C.; Ramezanpour, M.; Wormald, P.J.; Psaltis, A.J.; Vreugde, S. Chronic Rhinosinusitis, S. aureus Biofilm and Secreted Products, Inflammatory Responses, and Disease Severity. Biomedicines 2022, 10, 1362. [Google Scholar] [CrossRef] [PubMed]
- Moormeier, D.E.; Bayles, K.W. Staphylococcus aureus biofilm: A complex developmental organism. Mol. Microbiol. 2017, 104, 365–376. [Google Scholar] [CrossRef] [PubMed]
- Loffler, B.; Tuchscherr, L.; Niemann, S.; Peters, G. Staphylococcus aureus persistence in non-professional phagocytes. Int. J. Med. Microbiol. 2014, 304, 170–176. [Google Scholar] [CrossRef]
- Oliveira, D.; Borges, A.; Simoes, M. Staphylococcus aureus Toxins and Their Molecular Activity in Infectious Diseases. Toxins 2018, 10, 252. [Google Scholar] [CrossRef]
- Yoong, P.; Torres, V.J. The effects of Staphylococcus aureus leukotoxins on the host: Cell lysis and beyond. Curr. Opin. Microbiol. 2013, 16, 63–69. [Google Scholar] [CrossRef]
- Morinaga, N.; Kaihou, Y.; Noda, M. Purification, cloning and characterization of variant LukE-LukD with strong leukocidal activity of staphylococcal bi-component leukotoxin family. Microbiol. Immunol. 2003, 47, 81–90. [Google Scholar] [CrossRef]
- Peschel, A.; Otto, M. Phenol-soluble modulins and staphylococcal infection. Nat. Rev. Microbiol. 2013, 11, 667–673. [Google Scholar] [CrossRef]
- Otto, M. Phenol-soluble modulins. Int. J. Med. Microbiol. 2014, 304, 164–169. [Google Scholar] [CrossRef]
- Van Zele, T.; Gevaert, P.; Holtappels, G.; van Cauwenberge, P.; Bachert, C. Local immunoglobulin production in nasal polyposis is modulated by superantigens. Clin. Exp. Allergy 2007, 37, 1840–1847. [Google Scholar] [CrossRef]
- Hayes, S.M.; Biggs, T.C.; Goldie, S.P.; Harries, P.G.; Walls, A.F.; Allan, R.N.; Pender, S.L.F.; Salib, R.J. Staphylococcus aureus internalization in mast cells in nasal polyps: Characterization of interactions and potential mechanisms. J. Allergy Clin. Immunol. 2020, 145, 147–159. [Google Scholar] [CrossRef]
- Tan, L.; Huang, Y.; Shang, W.; Yang, Y.; Peng, H.; Hu, Z.; Wang, Y.; Rao, Y.; Hu, Q.; Rao, X.; et al. Accessory Gene Regulator (agr) Allelic Variants in Cognate Staphylococcus aureus Strain Display Similar Phenotypes. Front. Microbiol. 2022, 13, 700894. [Google Scholar] [CrossRef] [PubMed]
- Sangvik, M.; Olsen, R.S.; Olsen, K.; Simonsen, G.S.; Furberg, A.S.; Sollid, J.U. Age- and gender-associated Staphylococcus aureus spa types found among nasal carriers in a general population: The Tromso Staph and Skin Study. J. Clin. Microbiol. 2011, 49, 4213–4218. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, I.; Millon, B.; Meugnier, H.; Vandenesch, F.; Maurin, M.; Pavese, P.; Boisset, S. High prevalence of spa type t571 among methicillin-susceptible Staphylococcus aureus from bacteremic patients in a French University Hospital. PLoS ONE 2018, 13, e0204977. [Google Scholar] [CrossRef]
- Dunman, P.M.; Murphy, E.; Haney, S.; Palacios, D.; Tucker-Kellogg, G.; Wu, S.; Brown, E.L.; Zagursky, R.J.; Shlaes, D.; Projan, S.J. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J. Bacteriol. 2001, 183, 7341–7353. [Google Scholar] [CrossRef]
- Nepal, R.; Houtak, G.; Shaghayegh, G.; Bouras, G.; Shearwin, K.; Psaltis, A.J.; Wormald, P.J.; Vreugde, S. Prophages encoding human immune evasion cluster genes are enriched in Staphylococcus aureus isolated from chronic rhinosinusitis patients with nasal polyps. Microb. Genom. 2021, 7, 000726. [Google Scholar] [CrossRef]
- Houtak, G.; Bouras, G.; Nepal, R.; Shaghayegh, G.; Cooksley, C.; Psaltis, A.J.; Wormald, P.J.; Vreugde, S. The intra-host evolutionary landscape and pathoadaptation of persistent Staphylococcus aureus in chronic rhinosinusitis. Microb. Genom. 2023, 9, 001128. [Google Scholar] [CrossRef]
- Petit, R.A., III; Read, T.D. Bactopia: A Flexible Pipeline for Complete Analysis of Bacterial Genomes. mSystems 2020, 5, 10–1128. [Google Scholar] [CrossRef]
- Philpott, C.M.; Erskine, S.; Hopkins, C.; Kumar, N.; Anari, S.; Kara, N.; Sunkaraneni, S.; Ray, J.; Clark, A.; Wilson, A.; et al. Prevalence of asthma, aspirin sensitivity and allergy in chronic rhinosinusitis: Data from the UK National Chronic Rhinosinusitis Epidemiology Study. Respir. Res. 2018, 19, 129. [Google Scholar] [CrossRef]
- Jarvis, D.; Newson, R.; Lotvall, J.; Hastan, D.; Tomassen, P.; Keil, T.; Gjomarkaj, M.; Forsberg, B.; Gunnbjornsdottir, M.; Minov, J.; et al. Asthma in adults and its association with chronic rhinosinusitis: The GA2LEN survey in Europe. Allergy 2012, 67, 91–98. [Google Scholar] [CrossRef]
- Fokkens, W.J.; Lund, V.J.; Hopkins, C.; Hellings, P.W.; Kern, R.; Reitsma, S.; Toppila-Salmi, S.; Bernal-Sprekelsen, M.; Mullol, J.; Alobid, I.; et al. European Position Paper on Rhinosinusitis and Nasal Polyps 2020. Rhinology 2020, 58 (Suppl. S29), 1–464. [Google Scholar] [CrossRef]
- Hopkins, C.; Browne, J.P.; Slack, R.; Lund, V.; Brown, P. The Lund-Mackay staging system for chronic rhinosinusitis: How is it used and what does it predict? Otolaryngol. Head. Neck Surg. 2007, 137, 555–561. [Google Scholar] [CrossRef] [PubMed]
- Phillips, K.M.; Hoehle, L.; Bergmark, R.W.; Caradonna, D.S.; Gray, S.T.; Sedaghat, A.R. Reversal of Smoking Effects on Chronic Rhinosinusitis after Smoking Cessation. Otolaryngol. Head. Neck Surg. 2017, 157, 737–742. [Google Scholar] [CrossRef] [PubMed]
- Wagner Mackenzie, B.; Baker, J.; Douglas, R.G.; Taylor, M.W.; Biswas, K. Detection and quantification of Staphylococcus in chronic rhinosinusitis. Int. Forum Allergy Rhinol. 2019, 9, 1462–1469. [Google Scholar] [CrossRef] [PubMed]
- Foreman, A.; Wormald, P.J. Different biofilms, different disease? A clinical outcomes study. Laryngoscope 2010, 120, 1701–1706. [Google Scholar] [CrossRef]
- Shopsin, B.; Mathema, B.; Alcabes, P.; Said-Salim, B.; Lina, G.; Matsuka, A.; Martinez, J.; Kreiswirth, B.N. Prevalence of agr specificity groups among Staphylococcus aureus strains colonizing children and their guardians. J. Clin. Microbiol. 2003, 41, 456–459. [Google Scholar] [CrossRef]
- Jarraud, S.; Lyon, G.J.; Figueiredo, A.M.; Lina, G.; Vandenesch, F.; Etienne, J.; Muir, T.W.; Novick, R.P. Exfoliatin-producing strains define a fourth agr specificity group in Staphylococcus aureus. J. Bacteriol. 2000, 182, 6517–6522. [Google Scholar] [CrossRef]
- Janzon, L.; Arvidson, S. The role of the delta-lysin gene (hld) in the regulation of virulence genes by the accessory gene regulator (agr) in Staphylococcus aureus. EMBO J. 1990, 9, 1391–1399. [Google Scholar] [CrossRef]
- Peterson, M.L.; Schlievert, P.M. Glycerol monolaurate inhibits the effects of Gram-positive select agents on eukaryotic cells. Biochemistry 2006, 45, 2387–2397. [Google Scholar] [CrossRef]
- Haslinger-Loffler, B.; Kahl, B.C.; Grundmeier, M.; Strangfeld, K.; Wagner, B.; Fischer, U.; Cheung, A.L.; Peters, G.; Schulze-Osthoff, K.; Sinha, B. Multiple virulence factors are required for Staphylococcus aureus-induced apoptosis in endothelial cells. Cell Microbiol. 2005, 7, 1087–1097. [Google Scholar] [CrossRef]
- Sendi, P.; Proctor, R.A. Staphylococcus aureus as an intracellular pathogen: The role of small colony variants. Trends Microbiol. 2009, 17, 54–58. [Google Scholar] [CrossRef]
- Vuong, C.; Saenz, H.L.; Gotz, F.; Otto, M. Impact of the agr quorum-sensing system on adherence to polystyrene in Staphylococcus aureus. J. Infect. Dis. 2000, 182, 1688–1693. [Google Scholar] [CrossRef] [PubMed]
- Bardy, J.J.; Sarovich, D.S.; Price, E.P.; Steinig, E.; Tong, S.; Drilling, A.; Ou, J.; Vreugde, S.; Wormald, P.J.; Psaltis, A.J. Staphylococcus aureus from patients with chronic rhinosinusitis show minimal genetic association between polyp and non-polyp phenotypes. BMC Ear Nose Throat Disord. 2018, 18, 16. [Google Scholar] [CrossRef] [PubMed]
- Staali, L.; Colin, D.A. Bi-component HlgC/HlgB and HlgA/HlgB gamma-hemolysins from S. aureus: Modulation of Ca(2+) channels activity through a differential mechanism. Toxicon 2021, 201, 74–85. [Google Scholar] [CrossRef] [PubMed]
- Laarman, A.J.; Ruyken, M.; Malone, C.L.; van Strijp, J.A.; Horswill, A.R.; Rooijakkers, S.H. Staphylococcus aureus metalloprotease aureolysin cleaves complement C3 to mediate immune evasion. J. Immunol. 2011, 186, 6445–6453. [Google Scholar] [CrossRef]
- Atkin, K.E.; MacDonald, S.J.; Brentnall, A.S.; Potts, J.R.; Thomas, G.H. A different path: Revealing the function of staphylococcal proteins in biofilm formation. FEBS Lett. 2014, 588, 1869–1872. [Google Scholar] [CrossRef]
- Sedarat, Z.; Taylor-Robinson, A.W. Biofilm Formation by Pathogenic Bacteria: Applying a Staphylococcus aureus Model to Appraise Potential Targets for Therapeutic Intervention. Pathogens 2022, 11, 388. [Google Scholar] [CrossRef]
- Alonzo, F., III; Benson, M.A.; Chen, J.; Novick, R.P.; Shopsin, B.; Torres, V.J. Staphylococcus aureus leucocidin ED contributes to systemic infection by targeting neutrophils and promoting bacterial growth in vivo. Mol. Microbiol. 2012, 83, 423–435. [Google Scholar] [CrossRef]
- Dasari, P.; Nordengrun, M.; Vilhena, C.; Steil, L.; Abdurrahman, G.; Surmann, K.; Dhople, V.; Lahrberg, J.; Bachert, C.; Skerka, C.; et al. The Protease SplB of Staphylococcus aureus Targets Host Complement Components and Inhibits Complement-Mediated Bacterial Opsonophagocytosis. J. Bacteriol. 2022, 204, e0018421. [Google Scholar] [CrossRef]
- Fischer, A.J.; Kilgore, S.H.; Singh, S.B.; Allen, P.D.; Hansen, A.R.; Limoli, D.H.; Schlievert, P.M. High Prevalence of Staphylococcus aureus Enterotoxin Gene Cluster Superantigens in Cystic Fibrosis Clinical Isolates. Genes 2019, 10, 1036. [Google Scholar] [CrossRef]
- Song, L.; Florea, L.; Langmead, B. Lighter: Fast and memory-efficient sequencing error correction without counting. Genome Biol. 2014, 15, 509. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Schwengers, O.; Jelonek, L.; Dieckmann, M.A.; Beyvers, S.; Blom, J.; Goesmann, A. Bakta: Rapid and standardized annotation of bacterial genomes via alignment-free sequence identification. Microb. Genom. 2021, 7, 000685. [Google Scholar]
- Harmsen, D.; Claus, H.; Witte, W.; Rothganger, J.; Claus, H.; Turnwald, D.; Vogel, U. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 2003, 41, 5442–5448. [Google Scholar] [CrossRef]
- Thorvaldsdottir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef]
- Feldgarden, M.; Brover, V.; Haft, D.H.; Prasad, A.B.; Slotta, D.J.; Tolstoy, I.; Tyson, G.H.; Zhao, S.; Hsu, C.H.; McDermott, P.F.; et al. Validating the AMRFinder Tool and Resistance Gene Database by Using Antimicrobial Resistance Genotype-Phenotype Correlations in a Collection of Isolates. Antimicrob. Agents Chemother. 2019, 63, 10–1128. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation For Statistical Computing: Vienna, Austria, 2023. [Google Scholar]
- Kolde, R. Pheatmap: Pretty Heatmaps, R package version 1.0.12; Github: Tallinn, Estonia, 2018. [Google Scholar]
- Krijthe, J.H. T-Distributed Stochastic Neighbor Embedding Using a Barnes-Hut Implementation, R package version 0.17; Github Leiden University: Leiden, The Netherlands, 2015. [Google Scholar]
- Wickham, H. Ggplot2; Springer Science + Business Media, LLC: New York, NY, USA, 2016. [Google Scholar]
- Oksanen, J.S.G.; Blanchet, F.; Kindt, R.; Legendre, P.; Minchin, P.; O’Hara, R.S.P.; Stevens, M.; Szoecs, E.; Wagner, H.; Barbour, M.; et al. Vegan: Community Ecology Package, R package version 2.6-10; CRAN University of Oulu: Oulu, Finland, 2024. [Google Scholar]
- Robertson, J.; Nash, J.H.E. MOB-suite: Software tools for clustering, reconstruction and typing of plasmids from draft assemblies. Microb. Genom. 2018, 4, e000206. [Google Scholar] [CrossRef] [PubMed]
- Robertson, J.; Bessonov, K.; Schonfeld, J.; Nash, J.H.E. Universal whole-sequence-based plasmid typing and its utility to prediction of host range and epidemiological surveillance. Microb. Genom. 2020, 6, e000435. [Google Scholar] [CrossRef]
- Akhter, S.; Aziz, R.K.; Edwards, R.A. PhiSpy: A novel algorithm for finding prophages in bacterial genomes that combines similarity- and composition-based strategies. Nucleic Acids Res. 2012, 40, e126. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).