Optimization of Sequential Enzymatic Hydrolysis in Porcine Blood and the Influence on Peptide Profile and Bioactivity of Prepared Hydrolysates
Abstract
1. Introduction
2. Results and Discussion
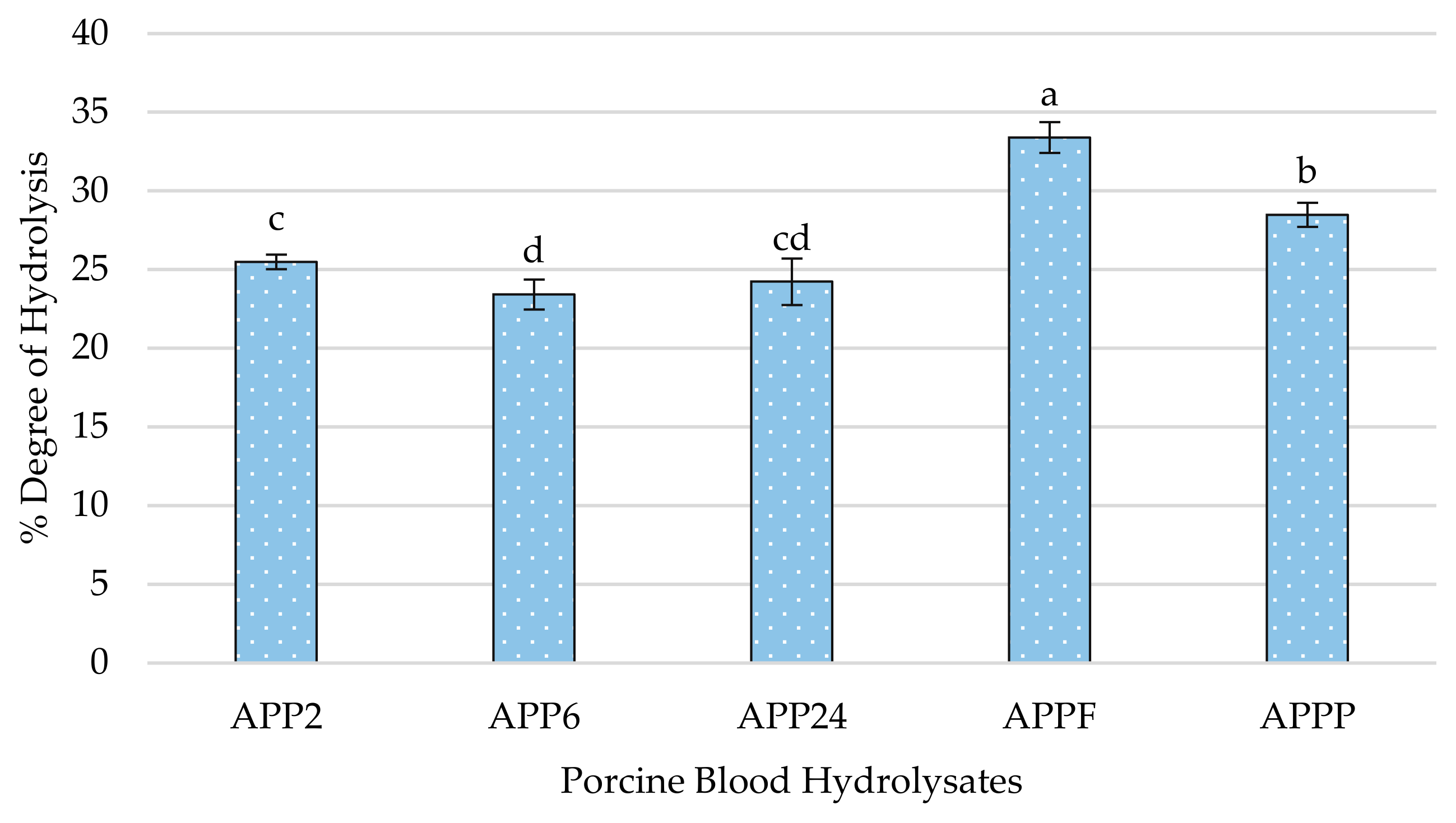
2.1. Degree of Hydrolysis and Molecular Size Profile of Generated Peptides of Porcine Blood Hydrolysates
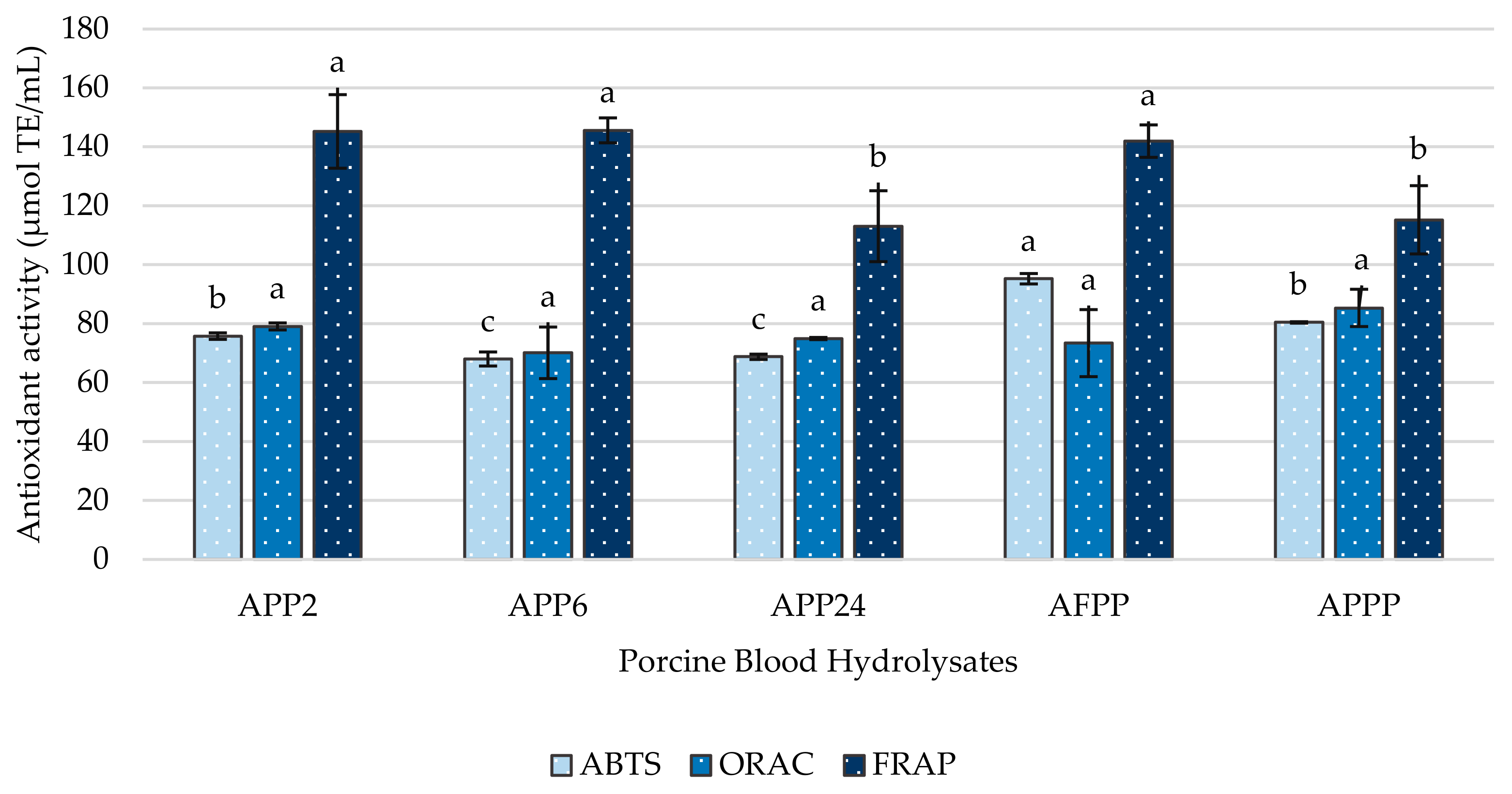
2.2. Antioxidant Capacity of Porcine Blood Hydrolysates
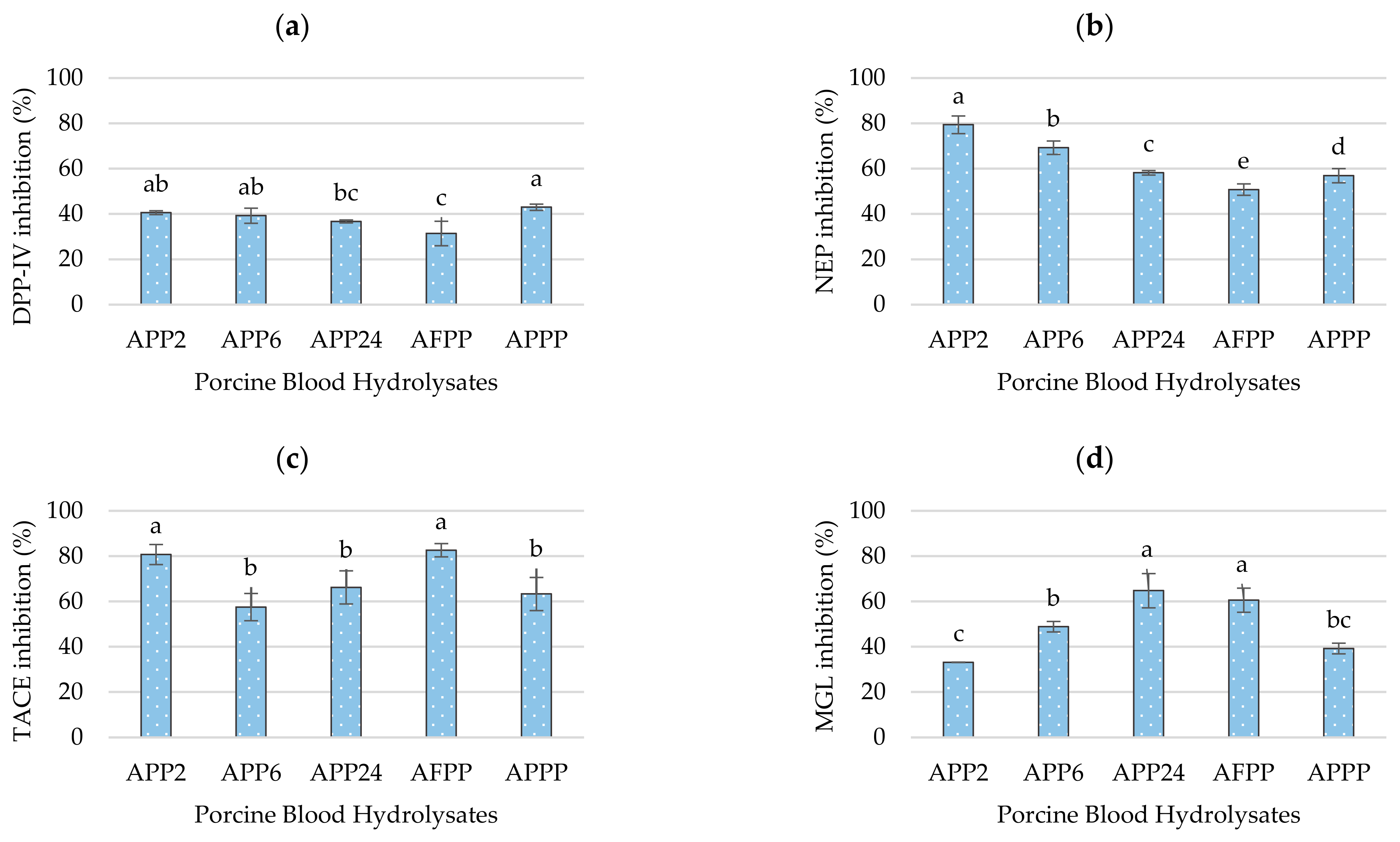
2.3. Hypoglycaemic Potential of Porcine Blood Hydrolysates
2.4. Anti-Inflammatory Potential of Porcine Blood Hydrolysates
2.5. Free Amino Acid Profile of Porcine Blood Hydrolysates (FAAs)
2.6. Peptidomics and In Silico Analysis of Identified Peptides from Porcine Blood Hydrolysates
3. Materials and Methods
3.1. Chemicals and Reagents
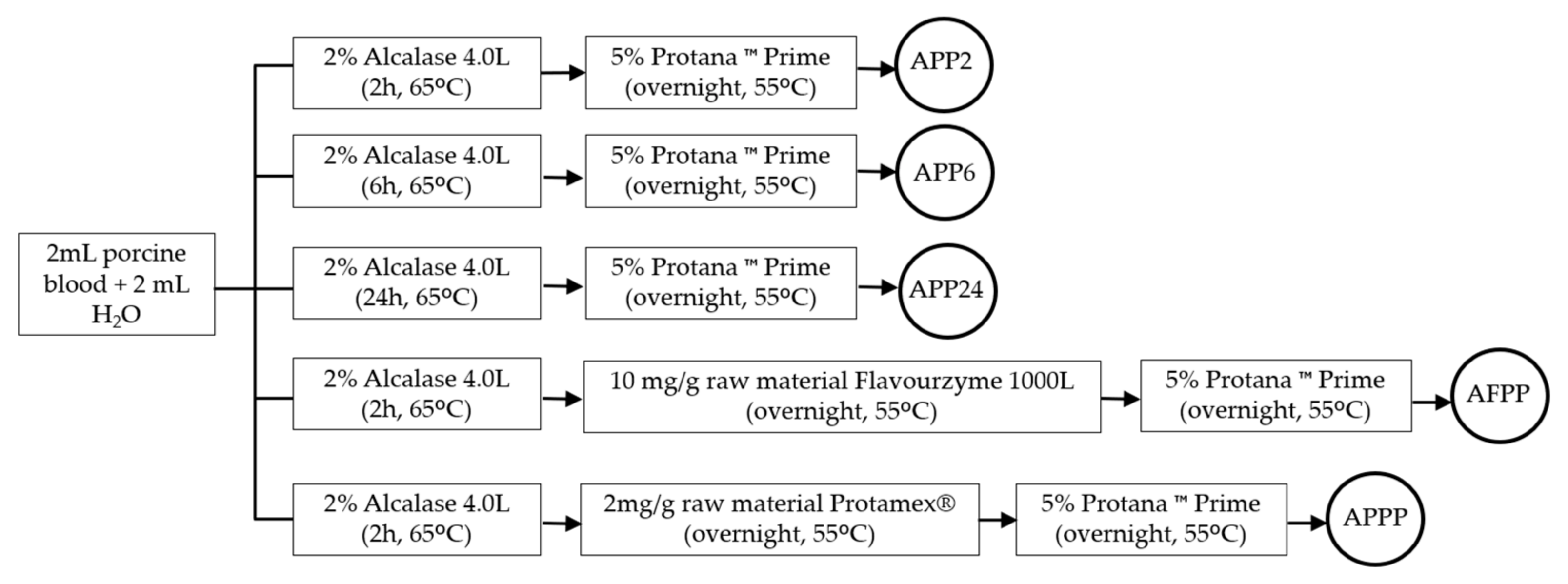
3.2. Preparation of Samples and Enzymatic Hydrolysis
3.3. Degree of Hydrolysis
- (i)
- “h” determination:
- (ii)
- DH calculation:
3.4. Ultrafiltration
3.5. Free Amino Acid Composition
3.6. Biological Activities
3.6.1. Antioxidant Activities
ABTS Radical Scavenging Capacity
Oxygen Radical Absorbance Capacity Assay (ORAC)
Ferric-Reducing Antioxidant Power (FRAP)
3.6.2. Dipeptidyl Peptidase IV (DPP-IV) Inhibitory Activity
3.6.3. Neprilysin (NEP) Inhibitory Activity
3.6.4. Tumor Necrosis α-Converting Enzyme (TACE) Inhibitory Activity
3.6.5. Monoacylglycerol Lipase (MGL) Inhibitory Activity
3.7. Peptide Characterization
3.7.1. Peptide Identification by Tandem Mass Spectrometry
3.7.2. Protein Analysis and Identification
3.7.3. In Silico Analysis
Software Tools
Simulated Gastrointestinal Digestion
3.8. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Del Hoyo, P.; Rendueles, M.; Díaz, M. Effect of Processing on Functional Properties of Animal Blood Plasma. Meat Sci. 2008, 78, 522–528. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority. Animal By-Products. Available online: https://www.efsa.europa.eu/en/topics/animal-by-products (accessed on 10 February 2025).
- Eurostat. Production of Animal Products. European Commission. Available online: https://ec.europa.eu/eurostat/databrowser/view/APRO_MT_PANN__custom_394311/default/table (accessed on 10 February 2025).
- Bah, C.S.F.; Bekhit, A.E.-D.A.; Carne, A.; McConnell, M.A. Slaughterhouse Blood: An Emerging Source of Bioactive Compounds. Compr. Rev. Food Sci. Food Saf. 2013, 12, 314–331. [Google Scholar] [CrossRef]
- Bashiri, B.; Cropotova, J.; Kvangarsnes, K.; Gavrilova, O.; Vilu, R. Environmental and Economic Life Cycle Assessment of Enzymatic Hydrolysis-Based Fish Protein and Oil Extraction. Resources 2024, 13, 61. [Google Scholar] [CrossRef]
- Daliri, E.B.-M.; Oh, D.H.; Lee, B.H. Bioactive Peptides. Foods 2017, 6, 32. [Google Scholar] [CrossRef]
- Karami, Z.; Akbari-Adergani, B. Bioactive Food Derived Peptides: A Review on Correlation between Structure of Bioactive Peptides and Their Functional Properties. J. Food Sci. Technol. 2019, 56, 535–547. [Google Scholar] [CrossRef]
- Borrajo, P.; Pateiro, M.; Barba, F.J.; Mora, L.; Franco, D.; Toldrá, F.; Lorenzo, J.M. Antioxidant and Antimicrobial Activity of Peptides Extracted from Meat By-Products: A Review. Food Anal. Methods 2019, 12, 2401–2415. [Google Scholar] [CrossRef]
- Gallego, M.; Aristoy, M.-C.; Toldrá, F. Dipeptidyl Peptidase IV Inhibitory Peptides Generated in Spanish Dry-Cured Ham. Meat Sci. 2014, 96, 757–761. [Google Scholar] [CrossRef]
- Bravo, F.I.; Calvo, E.; López-Villalba, R.A.; Torres-Fuentes, C.; Muguerza, B.; García-Ruiz, A.; Morales, D. Valorization of Chicken Slaughterhouse Byproducts to Obtain Antihypertensive Peptides. Nutrients 2023, 15, 457. [Google Scholar] [CrossRef]
- Guha, S.; Majumder, K. Structural-features of food-derived bioactive peptides with anti-inflammatory activity: A brief review. J. Food Biochem. 2019, 43, e12531. [Google Scholar] [CrossRef]
- Hwang, J.; Son, W.-Y.; Jeong, E.J.; Kim, K.-S.; Shin, E.-C.; Song, D.-H.; Lee, K.-W.; Kim, H.-W. Comparative Exploration of Antioxidant Properties of Alcalase- and Trypsin-Hydrolyzed Porcine By-Products and Their Classification for Industrial Use. Appl. Sci. 2025, 15, 47. [Google Scholar] [CrossRef]
- Chen, Q.; Nie, X.; Huang, W.; Wang, C.; Lai, R.; Lu, Q.; He, Q.; Yu, X. Unlocking the Potential of Chicken Liver Byproducts: Identification of Antioxidant Peptides through In Silico Approaches and Anti-Aging Effects of a Selected Peptide in Caenorhabditis elegans. Int. J. Biol. Macromol. 2024, 272 Pt 1, 132833. [Google Scholar] [CrossRef] [PubMed]
- López-Pedrouso, M.; Valcarcel, J.; Lorenzo, J.M.; Toldrá, M.; Bou, R.; Franco, D.; Vázquez, J.A. Valorisation of Pork By-Products to Obtain Antioxidant and Antihypertensive Peptides. Food Chem. 2023, 423, 136351. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhu, L.; Wu, G.; Liu, T.; Qi, X.; Zhang, H. Food-Derived Dipeptidyl Peptidase IV Inhibitory Peptides: Production, Identification, Structure-Activity Relationship, and Their Potential Role in Glycemic Regulation. Crit. Rev. Food Sci. Nutr. 2022, 64, 2053–2075. [Google Scholar] [CrossRef] [PubMed]
- Romero-Garay, M.G.; Montalvo-González, E.; Hernández-González, C.; Soto-Domínguez, A.; Becerra-Verdín, E.M.; García-Magaña, M.L. Bioactivity of Peptides Obtained from Poultry By-Products: A Review. Food Chem. X 2022, 13, 100181. [Google Scholar] [CrossRef]
- Ghalamara, S.; Brazinha, C.; Silva, S.; Pintado, M. Exploring Fish Processing By-Products as an Alternative Source of Bioactive Peptides: A Review on Extraction and Food Applications. Curr. Food Sci. Tech. Rep. 2024, 2, 377–391. [Google Scholar] [CrossRef]
- Keskin Ulug, S.; Jahandideh, F.; Wu, J. Novel Technologies for the Production of Bioactive Peptides. Trends Food Sci. Technol. 2021, 108, 27–39. [Google Scholar] [CrossRef]
- Rao, P.S.; Bajaj, R.; Mann, B. Impact of Sequential Enzymatic Hydrolysis on Antioxidant Activity and Peptide Profile of Casein Hydrolysate. J. Food Sci. Technol. 2020, 57, 4562–4575. [Google Scholar] [CrossRef]
- Toldrá, M.; Lynch, S.A.; Couture, R.; Álvarez, C. Blood Proteins as Functional Ingredients. In Sustainable Meat Production and Processing; Elsevier: Amsterdam, The Netherlands, 2019; pp. 85–100. [Google Scholar] [CrossRef]
- Riedel, S.L.; Brigham, C. Polymers and Adsorbents from Agricultural Waste. In Byproducts from Agriculture and Fisheries: Adding Value for Food, Feed, Pharma and Fuels; Simpson, B.K., Aryee, A.N., Toldrá, F., Eds.; Wiley: Chichester, UK, 2019; pp. 523–544. [Google Scholar]
- Puigdellivol, J.; Comellas Berenger, C.; Pérez Fernández, M.Á.; Cowalinsky Millán, J.M.; Carreras Vidal, C.; Gil Gil, I.; Martínez Pagán, J.; Ruiz Nieto, B.; Jiménez Gómez, F.; Comas Figuerola, F.X.; et al. Effectiveness of a Dietary Supplement Containing Hydrolyzed Collagen, Chondroitin Sulfate, and Glucosamine in Pain Reduction and Functional Capacity in Osteoarthritis Patients. J. Diet. Suppl. 2019, 16, 379–389. [Google Scholar] [CrossRef]
- Nielsen, P.M.; Petersen, D.; Dambmann, C. Improved Method for Determining Food Protein Degree of Hydrolysis. J. Food Sci. 2001, 66, 642–646. [Google Scholar] [CrossRef]
- Chiang, J.H.; Loveday, S.M.; Hardacre, A.K.; Parker, M.E. Effects of Enzymatic Hydrolysis Treatments on the Physicochemical Properties of Beef Bone Extract Using Endo- and Exoproteases. Int. J. Food Sci. Technol. 2019, 54, 111–120. [Google Scholar] [CrossRef]
- Carrera-Alvarado, G.; Toldrá, F.; Mora, L. Bile Acid-Binding Capacity of Peptide Extracts Obtained from Chicken Blood Hydrolysates Using HPLC. LWT Food Sci. Technol. 2023, 173, 114381. [Google Scholar] [CrossRef]
- Bao, Z.; Zhao, Y.; Wang, X.; Chi, Y. Effects of Degree of Hydrolysis (DH) on the Functional Properties of Egg Yolk Hydrolysate with Alcalase. J. Food Sci. Technol. 2017, 54, 669–678. [Google Scholar] [CrossRef] [PubMed]
- Guérard, F.; Dufossé, L.; De La Broise, D.; Binet, A. Enzymatic Hydrolysis of Proteins from Yellowfin Tuna (Thunnus albacares) Wastes Using Alcalase. J. Mol. Catal. B Enzym. 2001, 11, 1051–1059. [Google Scholar] [CrossRef]
- Zheng, Y.; Yang, F.; Yuan, X.; Ji, Y.; Li, H.; Li, H.; Yu, J.; Zulewska, J. Enzymatic Hydrolysis of Whey Proteins by the Combination of Alcalase and Neutrase: Kinetic Model and Hydrolysis Control. Int. Dairy J. 2024, 151, 105867. [Google Scholar] [CrossRef]
- Chen, J.; Yan, Y.; Zhang, L.; Zheng, J.; Guo, J.; Li, R.; Zeng, J. Purification of Novel Antioxidant Peptides from Myofibrillar Protein Hydrolysate of Chicken Breast and Their Antioxidant Potential in Chemical and H₂O₂-Stressed Cell Systems. Food Funct. 2021, 12, 4897–4908. [Google Scholar] [CrossRef]
- Cheung, I.W.Y.; Cheung, L.K.Y.; Tan, N.Y.; Li-Chan, E.C.Y. The Role of Molecular Size in Antioxidant Activity of Peptide Fractions from Pacific Hake (Merluccius productus) Hydrolysates. Food Chem. 2012, 134, 1297–1306. [Google Scholar] [CrossRef]
- Damgaard, T.; Lametsch, R.; Otte, J. Antioxidant Capacity of Hydrolyzed Animal By-Products and Relation to Amino Acid Composition and Peptide Size Distribution. J. Food Sci. Technol. 2015, 52, 6511–6519. [Google Scholar] [CrossRef]
- Di Bernardini, R.; Mullen, A.M.; Bolton, D.; Kerry, J.; O’Neill, E.; Hayes, M. Assessment of the Angiotensin-I-Converting Enzyme (ACE-I) Inhibitory and Antioxidant Activities of Hydrolysates of Bovine Brisket Sarcoplasmic Proteins Produced by Papain and Characterisation of Associated Bioactive Peptidic Fractions. Meat Sci. 2012, 90, 226–235. [Google Scholar] [CrossRef]
- Hernández-Ledesma, B.; Dávalos, A.; Bartolomé, B.; Amigo, L. Preparation of Antioxidant Enzymatic Hydrolysates from α-Lactalbumin and β-Lactoglobulin. J. Agric. Food Chem. 2005, 53, 588–593. [Google Scholar] [CrossRef]
- Ozturk-Kerimoglu, B.; Heres, A.; Mora, L.; Toldrá, F. Antioxidant Peptides Generated from Chicken Feet Protein Hydrolysates. J. Sci. Food Agric. 2023, 103, 7207–7217. [Google Scholar] [CrossRef]
- Singh, A.-K.; Yadav, D.; Sharma, N.; Jin, J.-O. Dipeptidyl Peptidase (DPP)-IV Inhibitors with Antioxidant Potential Isolated from Natural Sources: A Novel Approach for the Management of Diabetes. Pharmaceuticals 2021, 14, 586. [Google Scholar] [CrossRef] [PubMed]
- Ambhore, J.P.; Laddha, P.R.; Nandedkar, A.; Ajmire, P.V.; Chumbhale, D.S.; Navghare, A.B.; Kuchake, V.G.; Chaudhari, P.J. Medicinal Chemistry of Non-Peptidomimetic Dipeptidyl Peptidase IV (DPP IV) Inhibitors for Treatment of Type-2 Diabetes Mellitus: Insights on Recent Development. J. Mol. Struct. 2023, 1284, 135249. [Google Scholar] [CrossRef]
- Nongonierma, A.B.; FitzGerald, R.J. Features of Dipeptidyl Peptidase IV (DPP-IV) Inhibitory Peptides from Dietary Proteins. J. Food Biochem. 2019, 43, e12451. [Google Scholar] [CrossRef] [PubMed]
- Carrera-Alvarado, G.; Toldrá, F.; Mora, L. Potential of Dry-Cured Ham Bones as a Sustainable Source to Obtain Antioxidant and DPP-IV Inhibitory Extracts. Antioxidants 2023, 12, 1151. [Google Scholar] [CrossRef]
- He, L.; Wang, X.; Wang, Y.; Luo, J.; Zhao, Y.; Han, G.; Han, L.; Yu, Q. Production and Identification of Dipeptidyl Peptidase IV (DPP-IV) Inhibitory Peptides from Discarded Cowhide Collagen. Food Chem. 2023, 405, 134793. [Google Scholar] [CrossRef]
- Zhu, L.; Li, Z.; Ma, M.; Huang, X.; Guyonnet, V.; Xiong, H. Exploration of Novel DPP-IV Inhibitory Peptides from Discarded Eggshell Membrane: An Integrated In Silico and In Vitro Study. Food Biosci. 2024, 59, 104036. [Google Scholar] [CrossRef]
- Xu, Q.; Zheng, L.; Huang, M.; Zhao, M. Exploring structural features of potent dipeptidyl peptidase IV (DPP-IV) inhibitory peptides derived from tilapia (Oreochromis niloticus) skin gelatin. Food Chem. 2022, 397, 133821. [Google Scholar] [CrossRef]
- Jin, R.; Shang, J.; Teng, X.; Zhang, L.; Liao, M.; Kang, J.; Meng, R.; Wang, D.; Ren, H.; Liu, N. Characterization of DPP-IV Inhibitory Peptides Using an in Vitro Cell Culture Model of the Intestine. J. Agric. Food Chem. 2021, 69, 2711–2718. [Google Scholar] [CrossRef]
- Bozkurt, B.; Nair, A.P.; Misra, A.; Scott, C.Z.; Mahar, J.H.; Fedson, S. Neprilysin Inhibitors in Heart Failure: The Science, Mechanism of Action, Clinical Studies, and Unanswered Questions. JACC Basic Transl. Sci. 2023, 8, 88–105. [Google Scholar] [CrossRef]
- Bayes-Genis, A.; Barallat, J.; Richards, A.M. A Test in Context: Neprilysin: Function, Inhibition, and Biomarker. J. Am. Coll. Cardiol. 2016, 68, 639–653. [Google Scholar] [CrossRef]
- Esser, N.; Zraika, S. Neprilysin Inhibition: A New Therapeutic Option for Type 2 Diabetes? Diabetologia 2019, 62, 1113–1122. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Mariscal, C.; Carrera-Alvarado, G.; Mora, L.; Toldrá, F. Neprilysin (NEP) and Angiotensin Converting Enzyme-I (ACE-I) Inhibitory Dipeptides from Chicken Carcass Hydrolysates. LWT Food Sci. Technol. 2025, 221, 117591. [Google Scholar] [CrossRef]
- Heres, A.; Gallego, M.; Mora, L.; Toldrá, F. Identification and Quantitation of Bioactive and Taste-Related Dipeptides in Low-Salt Dry-Cured Ham. Int. J. Mol. Sci. 2022, 23, 2507. [Google Scholar] [CrossRef] [PubMed]
- Gallego, M.; Mora, L.; Toldrá, F. Effect of Ultrasound and Enzymatic Pre-Treatments on the Profile of Bioactive Peptides of Beef Liver Hydrolysates. Food Res. Int. 2024, 197, 115240. [Google Scholar] [CrossRef]
- Cuffaro, D.; Scilabra, S.D.; Spanò, D.P.; Calligaris, M.; Nuti, E.; Rossello, A. A Disintegrin and Metalloproteinases (ADAMs) and Tumor Necrosis Factor-Alpha-Converting Enzyme (TACE). In Metalloenzymes; Elsevier: Amsterdam, The Netherlands, 2024; Chapter 3.5. [Google Scholar] [CrossRef]
- Chemaly, M.; McGilligan, V.; Gibson, M.; Clauss, M.; Watterson, S.; Alexander, H.D.; Bjourson, A.J.; Peace, A. Role of Tumour Necrosis Factor Alpha Converting Enzyme (TACE/ADAM17) and Associated Proteins in Coronary Artery Disease and Cardiac Events. Arch. Cardiovasc. Dis. 2017, 110, 700–711. [Google Scholar] [CrossRef]
- Murumkar, P.R.; Ghuge, R.B.; Chauhan, M.; Barot, R.R.; Sorathiya, S.; Choudhary, K.M.; Joshi, K.D.; Yadav, M.R. Recent Developments and Strategies for the Discovery of TACE Inhibitors. Expert Opin. Drug Discov. 2020, 15, 779–801. [Google Scholar] [CrossRef]
- Udechukwu, M.C.; Tsopmo, A.; Mawhinney, H.; He, R.; Kienesberger, P.C.; Udenigwe, C.C. Inhibition of ADAM17/TACE Activity by Zinc-Chelating Rye Secalin-Derived Tripeptides and Analogues. RSC Adv. 2017, 7, 26361–26369. [Google Scholar] [CrossRef]
- Zanfirescu, A.; Ungurianu, A.; Mihai, D.P.; Radulescu, D.; Nitulescu, G.M. Targeting Monoacylglycerol Lipase in Pursuit of Therapies for Neurological and Neurodegenerative Diseases. Molecules 2021, 26, 5668. [Google Scholar] [CrossRef]
- Omran, Z. New Disulfiram Derivatives as MAGL-Selective Inhibitors. Molecules 2021, 26, 3296. [Google Scholar] [CrossRef]
- Hayes, M.; Naik, A.; Mora, L.; Iñarra, B.; Ibarruri, J.; Bald, C.; Cariou, T.; Reid, D.; Gallagher, M.; Dragøy, R.; et al. Generation, Characterisation and Identification of Bioactive Peptides from Mesopelagic Fish Protein Hydrolysates Using in Silico and in Vitro Approaches. Mar. Drugs 2024, 22, 297. [Google Scholar] [CrossRef]
- López-Pedrouso, M.; Borrajo, P.; Pateiro, M.; Lorenzo, J.M.; Franco, D. Antioxidant Activity and Peptidomic Analysis of Porcine Liver Hydrolysates Using Alcalase, Bromelain, Flavourzyme and Papain Enzymes. Food Res. Int. 2020, 137, 109389. [Google Scholar] [CrossRef] [PubMed]
- Domínguez, H.; Iñarra, B.; Labidi, J.; Mendiola, D.; Bald, C. Comparison of Amino Acid Release Between Enzymatic Hydrolysis and Acid Autolysis of Rainbow Trout Viscera. Heliyon 2024, 10, e27030. [Google Scholar] [CrossRef] [PubMed]
- Power, O.; Nongonierma, A.B.; Jakeman, P.; FitzGerald, R.J. Food Protein Hydrolysates as a Source of Dipeptidyl Peptidase IV Inhibitory Peptides for the Management of Type 2 Diabetes. Proc. Nutr. Soc. 2014, 73, 34–46. [Google Scholar] [CrossRef] [PubMed]
- Hatanaka, T.; Inoue, Y.; Arima, J.; Kumagai, Y.; Usuki, H.; Kawakami, K.; Kimura, M.; Mukaihara, T. Production of Dipeptidyl Peptidase IV Inhibitory Peptides from Defatted Rice Bran. Food Chem. 2012, 134, 797–802. [Google Scholar] [CrossRef]
- AOAC. Protein (Crude) in animal feed. Dumas method. In Official Methods of Analysis; Oxford University Press: Oxford, UK, 1969; no. 968.06. [Google Scholar]
- Flores, M.; Aristoy, M.C.; Spanier, A.M.; Toldrá, F. Non-Volatile Components Effects on Quality of “Serrano” Dry-Cured Ham as Related to Processing Time. J. Food Sci. 1997, 62, 1235–1239. [Google Scholar] [CrossRef]
- Gallego, M.; Mora, L.; Hayes, M.; Reig, M.; Toldrá, F. Effect of Cooking and in Vitro Digestion on the Antioxidant Activity of Dry-Cured Ham By-Products. Food Res. Int. 2017, 97, 296–306. [Google Scholar] [CrossRef]
- Huang, S.-J.; Tsai, S.-Y.; Mau, J.-L. Antioxidant Properties of Methanolic Extracts from Agrocybe cylindracea. LWT Food Sci. Technol. 2006, 39, 378–386. [Google Scholar] [CrossRef]

| FAAs | Concentration (mg/mL of Hydrolysate) | ||||
|---|---|---|---|---|---|
| APP2 | APP6 | APP24 | AFPP | APPP | |
| Aspartic Acid (Asp) | 2.272 ± 0.001 c | 2.048 ± 0.007 d | 2.020 ± 0.000 d | 2.555 ± 0.036 a | 2.399 ± 0.007 b |
| Glutamic Acid (Glu) | 1.928 ± 0.001 b | 1.700 ± 0.005 c | 1.729 ± 0.001 c | 2.257 ± 0.033 a | 1.711 ± 0.025 c |
| Hydroxyproline (Hyp) | 0.051 ± 0.000 c | 0.053 ± 0.000 c | 0.065 ± 0.022 c | 0.088 ± 0.011 b | 0.212 ± 0.001 a |
| Serine (Ser) | 2.725 ± 0.001 c | 2.185 ± 0.008 d | 1.922 ± 0.001 e | 3.457 ± 0.048 a | 2.893 ± 0.002 b |
| Asparagine (Asn) | 2.405 ± 0.004 c | 2.095 ± 0.010 d | 1.992 ± 0.000 e | 3.406 ± 0.048 a | 2.785 ± 0.003 b |
| Glycine (Gly) | 1.479 ± 0.002 c | 1.127 ± 0.008 e | 1.289 ± 0.001 d | 1.713 ± 0.023 a | 1.521 ± 0.003 b |
| Glutamine (Gln) | 0.545 ± 0.001 e | 0.925 ± 0.003 d | 1.164 ± 0.000 c | 1.794 ± 0.027 a | 1.465 ± 0.001 b |
| β-Alanine (βAla) | 0.047 ± 0.000 d | 0.050 ± 0.002 c | 0.046 ± 0.000 c | 0.072 ± 0.001 a | 0.081 ± 0.000 b |
| Taurine (Tau) | 0.028 ± 0.001 c | 0.026 ± 0.000 d | 0.028 ± 0.000 c | 0.031 ± 0.001 b | 0.035 ± 0.000 a |
| Histidine (His) | 3.805 ± 0.058 c | 3.214 ± 0.008 e | 3.506 ± 0.002 d | 5.410 ± 0.010 a | 4.629 ± 0.029 b |
| γ-Aminobutyric Acid (γaba) | 0.069 ± 0.012 a | 0.004 ± 0.001 b | 0.025 ± 0.002 b | 0.064 ± 0.011 a | 0.048 ± 0.022 a |
| Threonine (Thr) | 1.790 ± 0.015 c | 1.507 ± 0.012 d | 1.542 ± 0.005 d | 3.243 ± 0.047 a | 2.136 ± 0.057 b |
| Alanine (Ala) | 4.319 ± 0.009 c | 3.543 ± 0.16 e | 3.991 ± 0.001 d | 6.110 ± 0.087 a | 5.241 ± 0.051 b |
| Arginine (Arg) | 1.734 ± 0.004 c | 1.674 ± 0.010 d | 1.780 ± 0.003 c | 3.452 ± 0.045 a | 2.381 ± 0.054 b |
| Proline (Pro) | 0.561 ± 0.002 d | 0.577 ± 0.006 c | 0.585 ± 0.004 c | 1.020 ± 0.014 a | 0.680 ± 0.003 a |
| Tirosine (Tyr) | 2.127 ± 0.002 a | 1.451 ± 0.008 d | 1.518 ± 0.002 c | 1.528 ± 0.025 c | 1.923 ± 0.031 b |
| Valine (Val) | 4.918 ± 0.007 c | 4.348 ± 0.007 d | 4.418 ± 0.010 d | 7.996 ± 0.137 a | 5.959 ± 0.015 b |
| Metionine (Met) | 0.769 ± 0.001 c | 0.646 ± 0.005 e | 0.705 ± 0.000 d | 0.900 ± 0.013 a | 0.824 ± 0.001 b |
| Isoleucine (Ile) | 0.429 ± 0.001 c | 0.374 ± 0.006 d | 0.278 ± 0.000 e | 0.920 ± 0.014 a | 0.538 ± 0.000 b |
| Leucine (Leu) | 8.693 ± 0.005 c | 7.332 ± 0.018 d | 7.033 ± 0.000 e | 12.165 ± 0.176 a | 9.035 ± 0.004 b |
| Phenylalanine (Phe) | 3.800 ± 0.002 c | 3.299 ± 0.009 d | 3.191 ± 0.001 e | 5.701 ± 0.087 a | 4.241 ± 0.002 b |
| Tryptophan (Trp) | 1.283 ± 0.002 b | 1.006 ± 0.018 d | 0.910 ± 0.001 e | 1.508 ± 0.024 a | 1.155 ± 0.001 c |
| Ornitine (Orn) | 0.111 ± 0.000 a | 0.046 ± 0.009 b | 0.045 ± 0.000 b | 0.045 ± 0.000 b | 0.051 ± 0.000 b |
| Lysine (Lys) | 6.403 ± 0.002 c | 5.677 ± 0.018 e | 6.057 ± 0.001 d | 8.851 ± 0.130 a | 7.686 ± 0.001 b |
| Total | 52.291 ± 0.110 c | 44.908 ± 0.089 e | 45.170 ± 1.151 d | 74.286 ± 1.113 a | 59.628 ± 0.205 b |
| Sample | Peptide Sequence | PRR a | Allergenicity | FRS Score b | Chel. Score b | Hydrophobicity | Toxicity | Net Charge | CPPpred |
|---|---|---|---|---|---|---|---|---|---|
| APP2 | DRPFPDF | 0.9478 | non-allergen | 0.4197 | 0.2804 | −0.3 | non-toxic | −1 | 0.0751 |
| QPFPIRP | 0.9270 | allergen | 0.4315 | 0.2672 | −0.19 | non-toxic | 1 | 0.1219 | |
| DGPLPRP | 0.8566 | non-allergen | 0.4772 | 0.2914 | −0.29 | non-toxic | 0 | 0.2438 | |
| LFPNPPPPPQP | 0.8564 | non-allergen | 0.6587 | 0.3414 | −0.06 | non-toxic | 0 | 0.1297 | |
| ADPVNFPKL | 0.8482 | non-allergen | 0.4004 | 0.2633 | −0.07 | non-toxic | 0 | 0.1646 | |
| ASQPDVDGFLVGGASLKPEF | 0.8193 | non-allergen | 0.4769 | 0.2594 | −0.01 | non-toxic | −2 | 0.1123 | |
| GPGGPGTWKPGRPEPG | 0.802 | allergen | 0.5683 | 0.1886 | −0.17 | non-toxic | 1 | 0.2960 | |
| SMGKGYYLKGKIGKVPVRF | 0.7891 | allergen | 0.4137 | 0.1610 | −0.14 | non-toxic | 5 | 0.2669 | |
| DPQSPWDR | 0.7839 | allergen | 0.4375 | 0.2600 | −0.49 | non-toxic | −1 | 0.1224 | |
| GPGSVPGTGSPGGLKPG | 0.7815 | non-allergen | 0.4537 | 0.2112 | 0.01 | non-toxic | 1 | 0.1529 | |
| DPENFRL | 0.7746 | allergen | 0.3757 | 0.2620 | −0.38 | non-toxic | −1 | 0.1295 | |
| YQEPVLGPVRGPFPIIV | 0.7744 | allergen | 0.5392 | 0.1770 | 0.07 | non-toxic | 0 | 0.1219 | |
| AAGPPISEGKYF | 0.7593 | non-allergen | 0.4157 | 0.2176 | 0 | non-toxic | 0 | 0.0736 | |
| GPGSVPGTGSPGGLKP | 0.7557 | non-allergen | 0.4383 | 0.2098 | 0 | non-toxic | 1 | 0.1546 | |
| VDGPLPRP | 0.7519 | non-allergen | 0.4872 | 0.2716 | −0.18 | non-toxic | 0 | 0.2618 | |
| HPDDFNP | 0.7510 | allergen | 0.3928 | 0.2717 | −0.29 | non-toxic | −1.5 | 0.0494 | |
| APP6 | GYFVFGPTGCNLEGFF | 0.9328 | non-allergen | 0.4997 | 0.2137 | 0.17 | non-toxic | −1 | 0.0618 |
| DPQSPWDR | 0.7839 | allergen | 0.4375 | 0.26000 | −0.49 | non-toxic | −1 | 0.1224 | |
| YQEPVLGPVRGPFPIIV | 0.7744 | allergen | 0.5392 | 0.1770 | 0.07 | non-toxic | 0 | 0.1219 | |
| APP24 | AAEGACQQCSCRPMWCLTCMGKWFASRQDPQRPDTWL | 0.9733 | non-allergen | - | - | −0.2 | toxic | 1 | 0.4553 |
| QPFPIRP | 0.9270 | allergen | 0.4315 | 0.2672 | −0.19 | non-toxic | 1 | 0.1219 | |
| DPQSPWDR | 0.7839 | allergen | 0.4375 | 0.2600 | −0.49 | non-toxic | −1 | 0.1224 | |
| YQEPVLGPVRGPFPIIV | 0.7744 | allergen | 0.5392 | 0.1770 | 0.07 | non-toxic | 0 | 0.1219 | |
| GPGSVPGTGSPGGLKP | 0.7557 | non-allergen | 0.4383 | 0.2098 | 0 | non-toxic | 1 | 0.1546 | |
| HPDDFNP | 0.7510 | allergen | 0.3928 | 0.2717 | −0.29 | non-toxic | −1.5 | 0.0494 | |
| AFPP | GGGFGGGGGIGGGGGGGGGGGG | 0.8075 | non-allergen | 0.4887 | 0.1860 | 0.21 | non-toxic | 0 | 0.1023 |
| DPQSPWDR | 0.7839 | allergen | 0.4375 | 0.2600 | −0.49 | non-toxic | −1 | 0.1224 | |
| YQEPVLGPVRGPFPIIV | 0.7744 | allergen | 0.5392 | 0.1770 | 0.07 | non-toxic | 0 | 0.1219 | |
| DDPQSPWDR | 0.7303 | non-allergen | 0.3971 | 0.2742 | −0.52 | non-toxic | −2 | 0.1006 | |
| APPP | DPQSPWDR | 0.7839 | allergen | 0.4375 | 0.2600 | −0.49 | non-toxic | −1 | 0.1224 |
| DPENFRL | 0.7746 | allergen | 0.3757 | 0.2620 | −0.38 | non-toxic | −1 | 0.1295 | |
| YQEPVLGPVRGPFPIIV | 0.7744 | allergen | 0.5392 | 0.1770 | 0.07 | non-toxic | 0 | 0.1219 | |
| HPDDFNP | 0.7510 | allergen | 0.3928 | 0.2717 | −0.29 | non-toxic | −1.5 | 0.0494 |
| Sample | Peptide Sequence | PRR | GID-Enzymes Action | Active Fragment | Bioactivity of the Fragment | CPP Fragment |
|---|---|---|---|---|---|---|
| APP2 | DRPFPDF | 0.9478 | DR–PF–PDF | DR | ACE inhibitor | 0.4317 |
| DPP IV inhibitor | ||||||
| PF | ACE inhibitor | 0.0466 | ||||
| DPP IV inhibitor | ||||||
| DPP-III inhibitor | ||||||
| QPFPIRP | 0.927 | QPF–PIR–P | - | - | - | |
| DGPLPRP | 0.8566 | DGPL–PR–P | PR | ACE inhibitor | 0.8096 | |
| DPP-III inhibitor | ||||||
| LFPNPPPPPQP | 0.8564 | L–F–PN–PPPPPQP | PN | DPP IV inhibitor | 0.1469 | |
| ADPVNFPKL | 0.8482 | ADPVN–F–PK–L | PK | Antioxidative | 0.5177 | |
| DPP IV inhibitor | ||||||
| ASQPDVDGFLVGGASLKPEF | 0.8193 | ASQPDVDGF–L–VGGASL–K–PEF | - | - | - | |
| GPGGPGTWKPGRPEPG | 0.8020 | GPGGPGTW–K–PGR–PEPG | - | - | - | |
| SMGKGYYLKGKIGKVPVR | 0.7891 | SM–GK–GY–Y–L–K–GK–IGK–VPVR–F | GK | ACE inhibitor | 0.5298 | |
| GY | ACE inhibitor | 0.0443 | ||||
| Inhibitor of tripeptidyl peptidase II | ||||||
| DPP IV inhibitor | ||||||
| SM | DPP-III inhibitor | 0.0482 | ||||
| DPQSPWDR | 0.7839 | DPQSPW–DR- | DR | ACE inhibitor | 0.4317 | |
| DPP IV inhibitor | ||||||
| GPGSVPGTGSPGGLKPG | 0.7815 | GPGSVPGTGSPGGL–K–PG | - | - | - | |
| DPENFRL | 0.7746 | - | - | - | ||
| YQEPVLGPVRGPFPIIV | 0.7744 | Y–QEPVL–GPVR–GPF–PIIV | GPF | DPP IV inhibitor | 0.0477 | |
| AAGPPISEGKYF | 0.7593 | AAGPPISEGK–Y–F- | - | - | ||
| GPGSVPGTGSPGGLKP | 0.7557 | GPGSVPGTGSPGGL–K–P | - | - | ||
| VDGPLPRP | 0.7519 | VDGPL–PR–P | PR | ACE inhibitor | 0.8096 | |
| DPP-III inhibitor | ||||||
| HPDDFNP | 0.7510 | H–PDDF–N–P | - | - | - | |
| APP6 | GYFVFGPTGCNLEGFF | 0.9328 | GY–F–VF–GPTGCN–L–EGF–F | GY | ACE inhibitor | 0.0443 |
| DPP IV inhibitor | ||||||
| Inhibitor of tripeptidyl peptidase II | ||||||
| VF | ACE inhibitor | 0.0751 | ||||
| DPP IV inhibitor | ||||||
| Calpain 1 inhibitor | ||||||
| Inhibitor of tripeptidyl peptidase II | ||||||
| Inhibitor of neprilysin-2 | ||||||
| DPQSPWDR | 0.7839 | DPQSPW–DR | DR | ACE inhibitor | 0.4317 | |
| DPP IV inhibitor | ||||||
| YQEPVLGPVRGPFPIIV | 0.7744 | Y–QEPVL–GPVR–GPF–PIIV | GPF | DPP IV inhibitor | 0.0477 | |
| APP24 | AAEGACQQCSCRPMWCLTCMGKWFASRQDPQRPDTWL | 0.9733 | AAEGACQQCSCR–PM–W–CL–TCM–GK–W–F–ASR–QDPQR–PDTW–L- | GK | ACE inhibitor | 0.5298 |
| PM | ACE inhibitor | 0.1096 | ||||
| DPP IV inhibitor | ||||||
| QPFPIRP | 0.9270 | QPF–PIR–P | - | - | - | |
| DPQSPWDR | 0.7839 | DPQSPW–DR- | DR | ACE inhibitor | 0.4317 | |
| DPP IV inhibitor | ||||||
| YQEPVLGPVRGPFPIIV | 0.7744 | Y–QEPVL–GPVR–GPF–PIIV | GPF | DPP IV inhibitor | 0.0477 | |
| GPGSVPGTGSPGGLKP | 0.7557 | GPGSVPGTGSPGGL–K–P | - | - | ||
| HPDDFNP | 0.7510 | H–PDDF–N–P | - | - | - | |
| AFPP | GGGFGGGGGIGGGGGGGGGGGG | 0.8075 | GGGF–GGGGGIGGGGGGGGGGGG | - | - | - |
| DPQSPWDR | 0.7839 | DPQSPW–DR- | DR | ACE inhibitor | 0.4317 | |
| DPP IV inhibitor | ||||||
| YQEPVLGPVRGPFPIIV | 0.7744 | Y–QEPVL–GPVR–GPF–PIIV | GPF | DPP IV inhibitor | 0.0477 | |
| DDPQSPWDR | 0.7303 | DDPQSPW–DR- | DR | ACE inhibitor | 0.4317 | |
| DPP IV inhibitor | ||||||
| APPP | DPQSPWDR | 0.7839 | DPQSPW–DR | DR | ACE inhibitor | 0.4317 |
| DPP IV inhibitor | ||||||
| DPENFRL | 0.7746 | DPEN–F–R–L- | - | - | - | |
| YQEPVLGPVRGPFPIIV | 0.7744 | Y–QEPVL–GPVR–GPF–PIIV | GPF | DPP IV inhibitor | 0.0477 | |
| HPDDFNP | 0.7510 | H–PDDF–N–P | - | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moreno-Mariscal, C.; Moroni, F.; Pérez-Sánchez, J.; Mora, L.; Toldrá, F. Optimization of Sequential Enzymatic Hydrolysis in Porcine Blood and the Influence on Peptide Profile and Bioactivity of Prepared Hydrolysates. Int. J. Mol. Sci. 2025, 26, 3583. https://doi.org/10.3390/ijms26083583
Moreno-Mariscal C, Moroni F, Pérez-Sánchez J, Mora L, Toldrá F. Optimization of Sequential Enzymatic Hydrolysis in Porcine Blood and the Influence on Peptide Profile and Bioactivity of Prepared Hydrolysates. International Journal of Molecular Sciences. 2025; 26(8):3583. https://doi.org/10.3390/ijms26083583
Chicago/Turabian StyleMoreno-Mariscal, Cristina, Federico Moroni, Jaume Pérez-Sánchez, Leticia Mora, and Fidel Toldrá. 2025. "Optimization of Sequential Enzymatic Hydrolysis in Porcine Blood and the Influence on Peptide Profile and Bioactivity of Prepared Hydrolysates" International Journal of Molecular Sciences 26, no. 8: 3583. https://doi.org/10.3390/ijms26083583
APA StyleMoreno-Mariscal, C., Moroni, F., Pérez-Sánchez, J., Mora, L., & Toldrá, F. (2025). Optimization of Sequential Enzymatic Hydrolysis in Porcine Blood and the Influence on Peptide Profile and Bioactivity of Prepared Hydrolysates. International Journal of Molecular Sciences, 26(8), 3583. https://doi.org/10.3390/ijms26083583









