Identification and Characterization of WOX Gene Family in Flax (Linum usitatissimum L.) and Its Role Under Abiotic Stress
Abstract
1. Introduction
2. Results
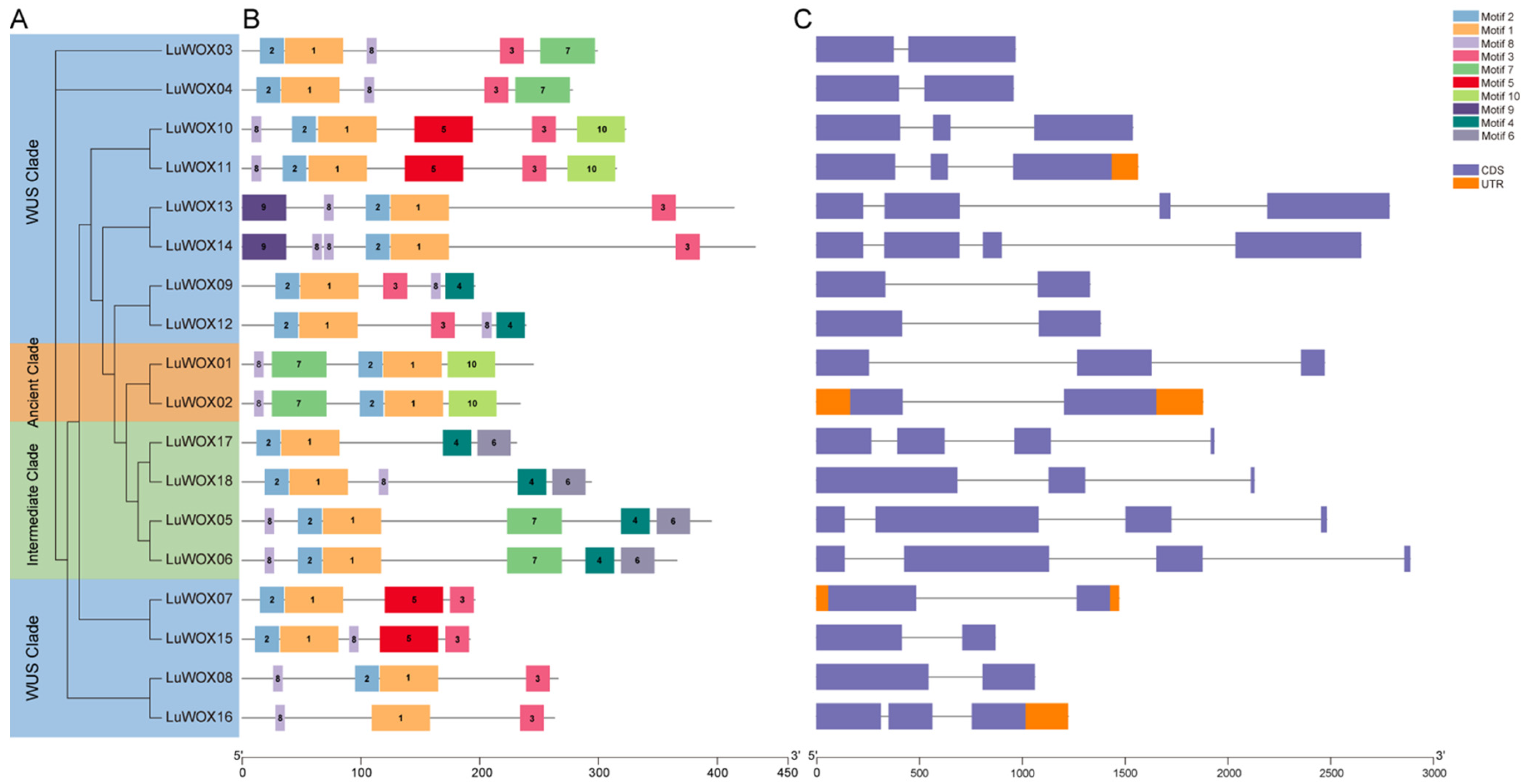
2.1. Phylogenetic Characterization and Identification of WOX Family Genes in Flax
2.2. Analysis of Gene Structure and Conservative Motif of LuWOX
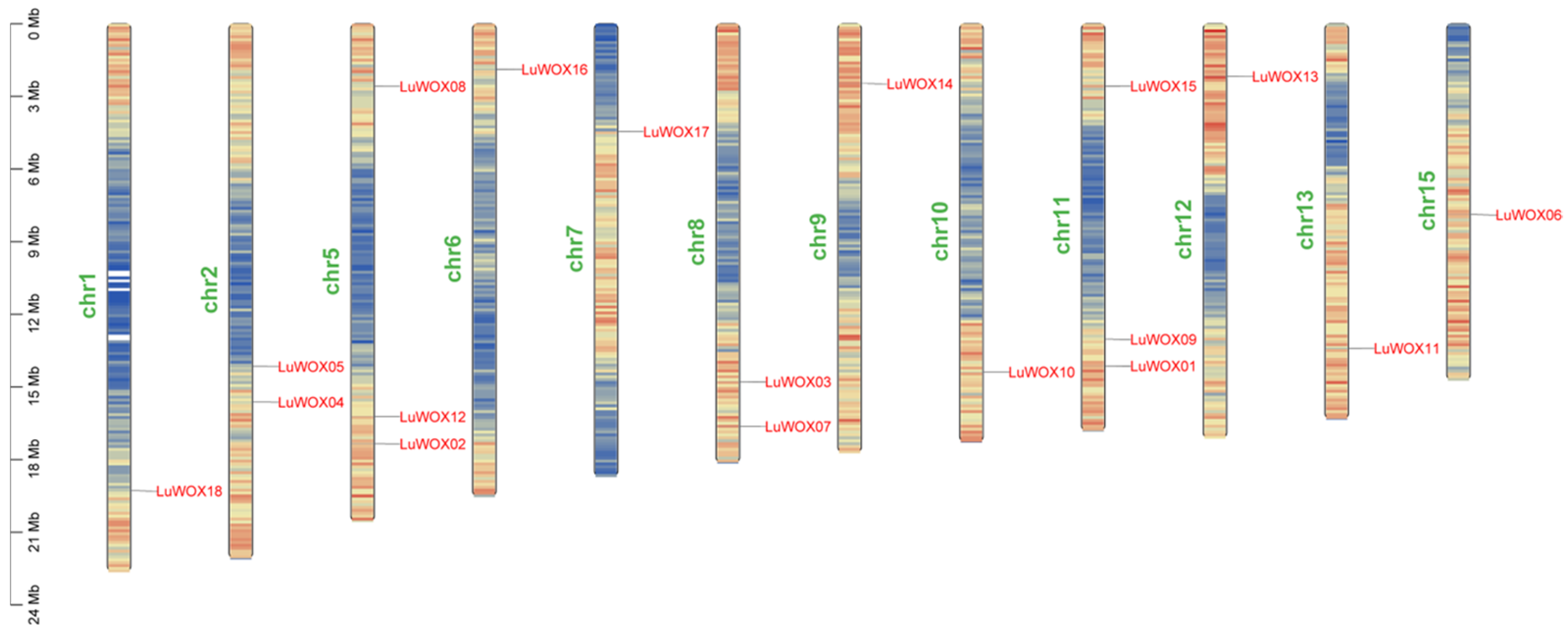
2.3. Chromosome Localization and Gene Reproduction Events of LuWOX Gene Family
2.4. Analysis of Cis-Acting Elements
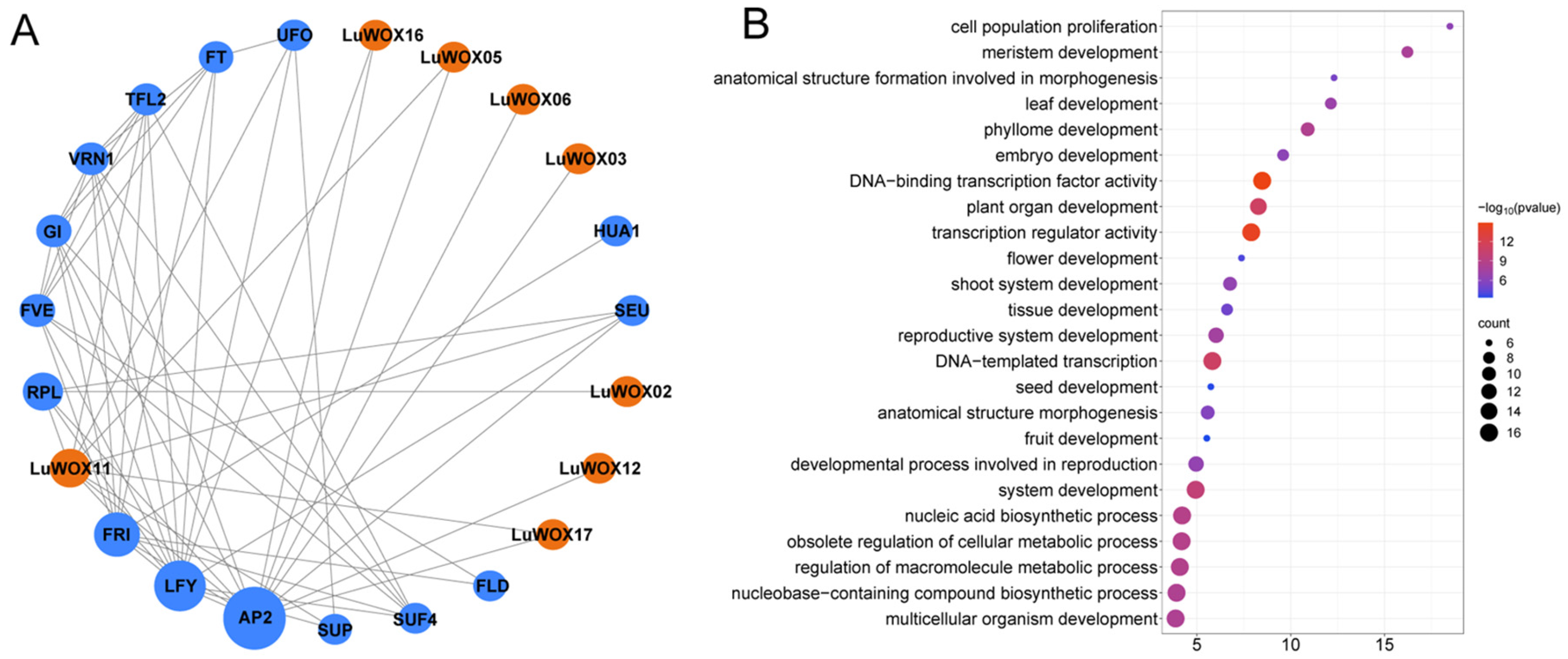
2.5. LuWOX Interactome Network and Gene Ontology Enrichment Profiling
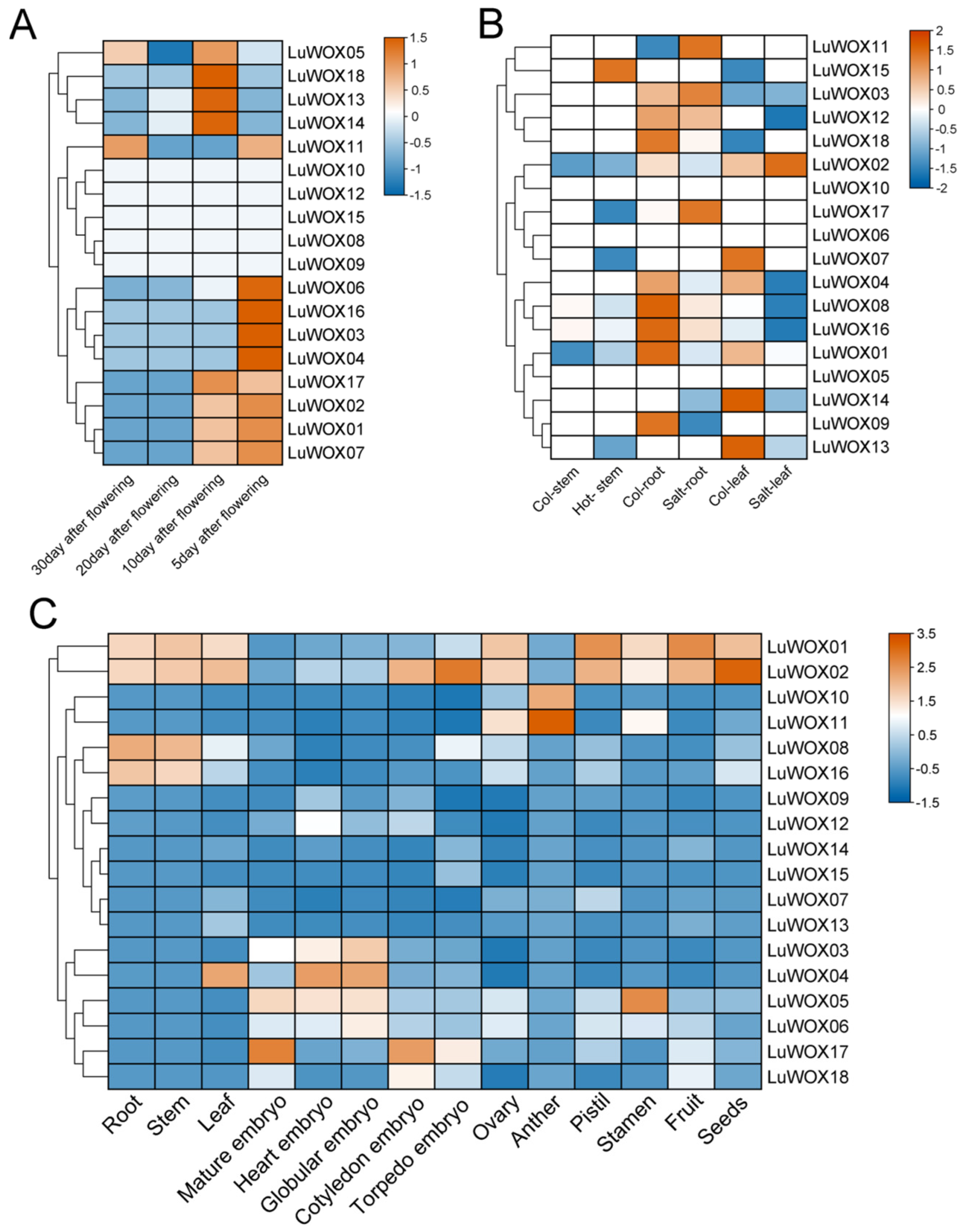
2.6. Analysis of LuWOX Gene Expression Pattern Based on RNA-Seq Data
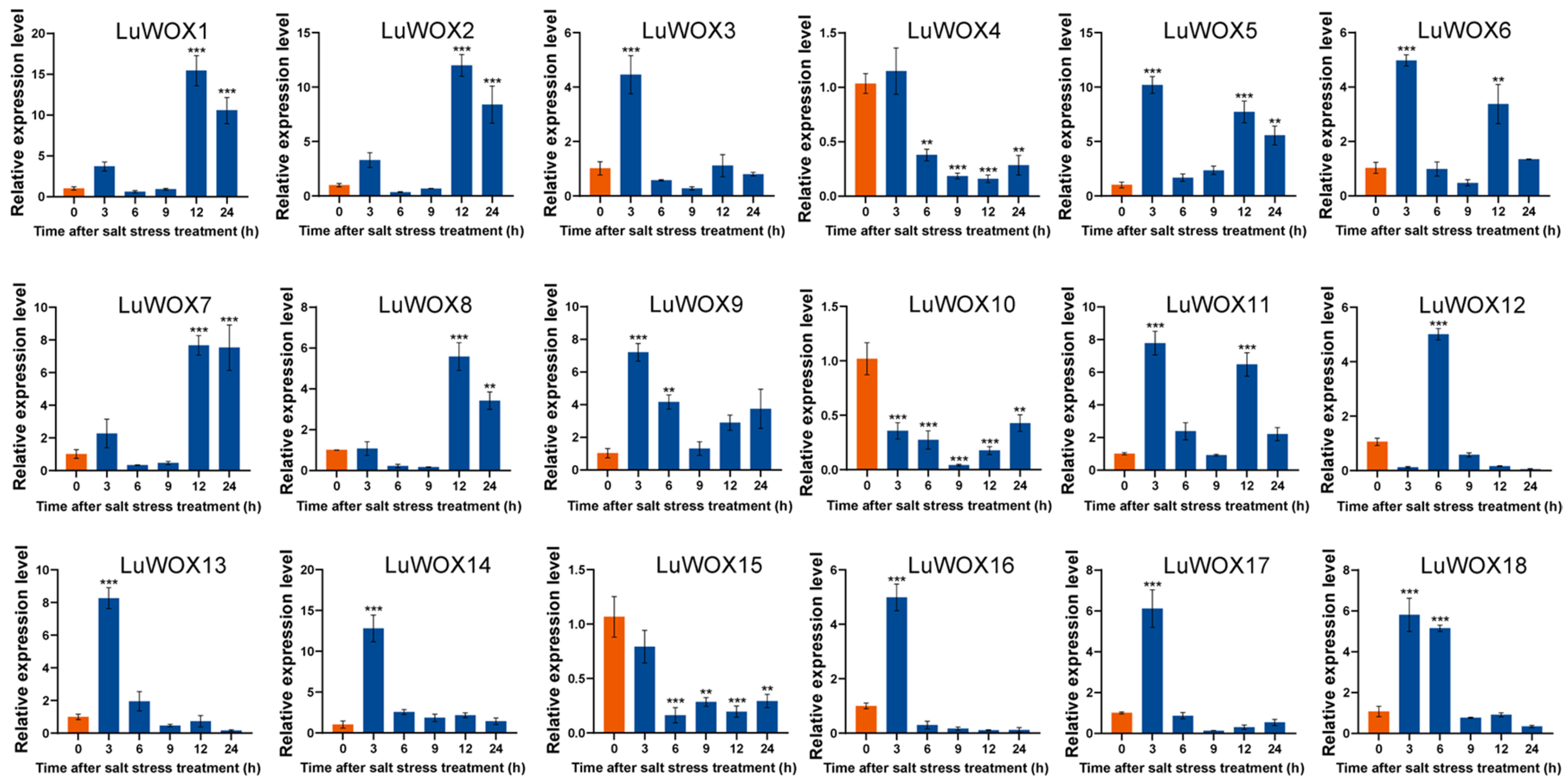
2.7. Expression Level of LuWOX Gene in Flax Under Abiotic Stress
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Identification of WOX Gene in Flax
4.3. Phylogeny, Chromosome Location, Conserved Domain, and Conserved Motif of LuWOX Gene
4.4. Genome-Wide Replication and Collinear Analysis of LuWOX Gene
4.5. Cis-Acting Element Analysis
4.6. Construction of Protein Interaction Network and GO Enrichment Analysis
4.7. Analysis of Expression Pattern of LuWOX Gene Family
4.8. RNA Extraction and Fluorescence Quantitative PCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- van der Graaff, E.; Laux, T.; Rensing, S.A. The WUS homeobox-containing (WOX) protein family. Genome Biol. 2009, 10, 248. [Google Scholar] [CrossRef] [PubMed]
- Lian, G.; Ding, Z.; Wang, Q.; Zhang, D.; Xu, J. Origins and evolution of WUSCHEL-related homeobox protein family in plant kingdom. Sci. World J. 2014, 2014, 534140. [Google Scholar] [CrossRef] [PubMed]
- Laux, T.; Mayer, K.F.X.; Berger, J.; Jürgens, G. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 1996, 122, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Tan, M.; Wang, X.; Jia, L.; Wang, M.; Huang, A.; You, L.; Li, C.; Zhang, Y.; Zhao, Y.; et al. WUS-RELATED HOMEOBOX 14 boosts de novo plant shoot regeneration. Plant Physiol. 2023, 192, 748–752. [Google Scholar] [CrossRef]
- Dolzblasz, A.; Nardmann, J.; Clerici, E.; Causier, B.; van der Graaff, E.; Chen, J.; Davies, B.; Werr, W.; Laux, T. Stem Cell Regulation by Arabidopsis WOX Genes. Mol. Plant 2016, 9, 1028–1039. [Google Scholar] [CrossRef]
- Lowe, K.; Wu, E.; Wang, N.; Hoerster, G.; Hastings, C.; Cho, M.-J.; Scelonge, C.; Lenderts, B.; Chamberlin, M.; Cushatt, J.; et al. Morphogenic Regulators Baby Boom and Wuschel Improve Monocot Transformation. Plant Cell 2016, 28, 1998–2015. [Google Scholar] [CrossRef]
- Nakata, M.; Okada, K. The three-domain model. Plant Signal. Behav. 2012, 7, 1423–1427. [Google Scholar] [CrossRef]
- Niu, H.; Liu, X.; Tong, C.; Wang, H.; Li, S.; Lu, L.; Pan, Y.; Zhang, X.; Weng, Y.; Li, Z. The WUSCHEL-related homeobox1 gene of cucumber regulates reproductive organ development. J. Exp. Bot. 2018, 69, 5373–5387. [Google Scholar] [CrossRef]
- Yang, R.; Wu, Z.; Bai, C.; Sun, Z.; Wang, M.; Huo, Y.; Zhang, H.; Wang, Y.; Zhou, H.; Dai, S.; et al. Overexpression of PvWOX3a in switchgrass promotes stem development and increases plant height. Hortic. Res. 2021, 8, 252. [Google Scholar] [CrossRef]
- Palovaara, J.; Hakman, I. Conifer WOX-related homeodomain transcription factors, developmental consideration and expression dynamic of WOX2 during Picea abies somatic embryogenesis. Plant Mol. Biol. 2008, 66, 533–549. [Google Scholar] [CrossRef]
- Ji, J.; Strable, J.; Shimizu, R.; Koenig, D.; Sinha, N.; Scanlon, M.J. WOX4 promotes procambial development. Plant Physiol. 2010, 152, 1346–1356. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, A.K.; Luijten, M.; Miyashima, S.; Lenhard, M.; Hashimoto, T.; Nakajima, K.; Scheres, B.; Heidstra, R.; Laux, T. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 2007, 446, 811–814. [Google Scholar] [CrossRef] [PubMed]
- Kong, D.; Hao, Y.; Cui, H. The WUSCHEL Related Homeobox Protein WOX7 Regulates the Sugar Response of Lateral Root Development in Arabidopsis thaliana. Mol. Plant 2016, 9, 261–270. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.-Q.; Li, Z.; Wen, S.-S.; Wang, J.-N.; Zhao, S.-T.; Lu, M.-Z. WUSCHEL-related homeobox gene PagWOX11/12a responds to drought stress by enhancing root elongation and biomass growth in poplar. J. Exp. Bot. 2020, 71, 1503–1513. [Google Scholar] [CrossRef]
- Minh-Thu, P.-T.; Kim, J.S.; Chae, S.; Jun, K.M.; Lee, G.-S.; Kim, D.-E.; Cheong, J.-J.; Song, S.I.; Nahm, B.H.; Kim, Y.-K. A WUSCHEL Homeobox Transcription Factor, OsWOX13, Enhances Drought Tolerance and Triggers Early Flowering in Rice. Mol. Cells 2018, 41, 781–798. [Google Scholar] [CrossRef]
- Denis, E.; Kbiri, N.; Mary, V.; Claisse, G.; e Silva, N.C.; Kreis, M.; Deveaux, Y. WOX14 promotes bioactive gibberellin synthesis and vascular cell differentiation in Arabidopsis. Plant J. 2017, 90, 560–572. [Google Scholar] [CrossRef]
- Park, S.O.; Zheng, Z.; Oppenheimer, D.G.; Hauser, B.A. The PRETTY FEW SEEDS2 gene encodes an Arabidopsis homeodomain protein that regulates ovule development. Development 2005, 132, 841–849. [Google Scholar] [CrossRef]
- Park, S.O.; Hwang, S.; Hauser, B.A. The phenotype of Arabidopsis ovule mutants mimics the morphology of primitive seed plants. R. Soc. Flagship Biol. Res. J. 2004, 271, 311–316. [Google Scholar] [CrossRef]
- Gu, R.; Song, X.; Liu, X.; Yan, L.; Zhou, Z.; Zhang, X. Genome-wide analysis of CsWOX transcription factor gene family in cucumber (Cucumis sativus L.). Sci. Rep 2020, 10, 6216. [Google Scholar] [CrossRef]
- Zhu, J.; Shi, H.; Lee, B.H.; Damsz, B.; Cheng, S.; Stirm, V.; Zhu, J.K.; Hasegawa, P.M.; Bressan, R.A. An Arabidopsis homeodomain transcription factor gene, HOS9, mediates cold tolerance through a CBF-independent pathway. Proc. Natl. Acad. Sci. USA 2004, 101, 9873–9878. [Google Scholar] [CrossRef]
- Hao, Q.; Zhang, L.; Yang, Y.; Shan, Z.; Zhou, X.-A. Genome-Wide Analysis of the WOX Gene Family and Function Exploration of GmWOX18 in Soybean. Plants 2019, 8, 215. [Google Scholar] [CrossRef] [PubMed]
- Sajjad, M.; Wei, X.; Liu, L.; Li, F.; Ge, X. Transcriptome Analysis Revealed GhWOX4 Intercedes Myriad Regulatory Pathways to Modulate Drought Tolerance and Vascular Growth in Cotton. Int. J. Mol. Sci. 2021, 22, 898. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wen, S.; Wang, R.; Wang, C.; Gao, B.; Lu, M. PagWOX11/12a activates PagCYP736A12 gene that facilitates salt tolerance in poplar. Plant Biotechnol. J. 2021, 19, 2249–2260. [Google Scholar] [CrossRef] [PubMed]
- Guarneri, N.; Willig, J.; Willemsen, V.; Goverse, A.; Sterken, M.G.; Nibbering, P.; Torres, J.L.L.; Smant, G. WOX11-mediated cell size control in Arabidopsis attenuates growth and fecundity of endoparasitic cyst nematodes. Plant J. 2024, 120, 540–551. [Google Scholar] [CrossRef]
- Huis, R.; Hawkins, S.; Neutelings, G. Selection of reference genes for quantitative gene expression normalization in flax (Linum usitatissimum L.). BMC Plant Biol. 2010, 10, 71. [Google Scholar] [CrossRef]
- Chytilová, M.; Mudroňová, D.; Nemcová, R.; Gancarčíková, S.; Buleca, V.; Koščová, J.; Tkáčiková, Ľ. Anti-inflammatory and immunoregulatory effects of flax-seed oil and Lactobacillus plantarum—Biocenol LP96 in gnotobiotic pigs challenged with enterotoxigenic Escherichia coli. Res. Vet. Sci. 2013, 95, 103–109. [Google Scholar] [CrossRef]
- Santos, H.O.; Price, J.C.; Bueno, A.A. Beyond Fish Oil Supplementation: The Effects of Alternative Plant Sources of Omega-3 Polyunsaturated Fatty Acids upon Lipid Indexes and Cardiometabolic Biomarkers-An Overview. Nutrients 2020, 12, 3159. [Google Scholar] [CrossRef]
- Brandt, R.; Cabedo, M.; Xie, Y.; Wenkel, S. Homeodomain leucine-zipper proteins and their role in synchronizing growth and development with the environment. J. Integr. Plant Biol. 2014, 56, 518–526. [Google Scholar] [CrossRef]
- Costanzo, E.; Trehin, C.; Vandenbussche, M. The role of WOX genes in flower development. Ann. Bot. 2014, 114, 1545–1553. [Google Scholar] [CrossRef]
- Wang, M.-M.; Liu, M.-M.; Ran, F.; Guo, P.-C.; Ke, Y.-Z.; Wu, Y.-W.; Wen, J.; Li, P.-F.; Li, J.-N.; Du, H. Global Analysis of WOX Transcription Factor Gene Family in Brassica napus Reveals Their Stress-and Hormone-Responsive Patterns. Int. J. Mol. Sci. 2018, 19, 3470. [Google Scholar] [CrossRef]
- Rathour, M.; Sharma, A.; Kaur, A.; Upadhyay, S.K. Genome-wide characterization and expression and co-expression analysis suggested diverse functions of WOX genes in bread wheat. Heliyon 2020, 6, e5762. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zong, J.; Liu, J.; Yin, J.; Zhang, D. Genome-wide analysis of WOX gene family in rice, sorghum, maize, Arabidopsis and poplar. J. Integr. Plant Biol. 2010, 52, 1016–1026. [Google Scholar] [CrossRef] [PubMed]
- Zheng, R.; Peng, Y.; Chen, J.; Zhu, X.; Xie, K.; Ahmad, S.; Zhao, K.; Peng, D.; Liu, Z.-J.; Zhou, Y. The Genome-Level Survey of the WOX Gene Family in Melastoma dodecandrum Lour. Int. J. Mol. Sci. 2023, 24, 17349. [Google Scholar] [CrossRef] [PubMed]
- Riccucci, E.; Vanni, C.; Vangelisti, A.; Fambrini, M.; Giordani, T.; Cavallini, A.; Mascagni, F.; Pugliesi, C. Genome-Wide Analysis of WOX Multigene Family in Sunflower (Helianthus annuus L.). Int. J. Mol. Sci. 2023, 24, 3352. [Google Scholar] [CrossRef]
- Wang, Z.; Cai, Q.; Xia, H.; Han, B.; Li, M.; Wang, Y.; Jiao, C.; Wang, D.; Zhu, J.; Yuan, W.; et al. Genome-Wide Identification and Comparative Analysis of WOX Genes in Four Euphorbiaceae Species and Their Expression Patterns in Jatropha curcas. Front. Genet. 2022, 13, 878554. [Google Scholar] [CrossRef]
- Gambino, G.; Minuto, M.; Boccacci, P.; Perrone, I.; Vallania, R.; Gribaudo, I. Characterization of expression dynamics of WOX homeodomain transcription factors during somatic embryogenesis in Vitis vinifera. J. Exp. Bot. 2011, 62, 1089–1101. [Google Scholar] [CrossRef]
- Yin, S.; Zhao, L.; Liu, J.; Sun, Y.; Li, B.; Wang, L.; Ren, Z.; Chen, C. Pan-genome Analysis of WOX Gene Family and Function Exploration of CsWOX9 in Cucumber. Int. J. Mol. Sci. 2023, 24, 17568. [Google Scholar] [CrossRef]
- Xu, G.; Guo, C.; Shan, H.; Kong, H. Divergence of duplicate genes in exon-intron structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1187–1192. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef]
- Jiao, Y.; Li, J.; Tang, H.; Paterson, A.H. Integrated syntenic and phylogenomic analyses reveal an ancient genome duplication in monocots. Plant Cell 2014, 26, 2792–2802. [Google Scholar] [CrossRef]
- Clark, J.W.; Donoghue, P. Whole-Genome Duplication and Plant Macroevolution. Trends Plant Sci. 2018, 23, 933–945. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Garcia, C.M.; Finer, J.J. Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 2014, 217, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Zhang, X. Genome-wide dynamic network analysis reveals the potential genes for MeJA-induced growth-to-defense transition. BMC Plant Biol. 2021, 21, 450. [Google Scholar] [CrossRef] [PubMed]
- Emenecker, R.J.; Strader, L.C. Auxin-Abscisic Acid Interactions in Plant Growth and Development. Biomolecules 2020, 10, 281. [Google Scholar] [CrossRef]
- Nobeli, I.; Favia, A.D.; Thornton, J.M. Protein promiscuity and its implications for biotechnology. Nat. Biotechnol. 2009, 27, 157–167. [Google Scholar] [CrossRef]
- Ma, Z.; Hu, L.; Jiang, W. Understanding AP2/ERF Transcription Factor Responses and Tolerance to Various Abiotic Stresses in Plants: A Comprehensive Review. Int. J. Mol. Sci. 2024, 25, 893. [Google Scholar] [CrossRef]
- Schoof, H.; Lenhard, M.; Haecker, A.; Mayer, K.F.; Jürgens, G.; Laux, T. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 2000, 100, 635–644. [Google Scholar] [CrossRef]
- Haecker, A.; Groß-Hardt, R.; Geiges, B.; Sarkar, A.; Breuninger, H.; Herrmann, M.; Laux, T. Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 2004, 131, 657–668. [Google Scholar] [CrossRef]
- Sun, R.; Zhang, X.; Ma, D.; Liu, C. Identification and Evolutionary Analysis of Cotton (Gossypium hirsutum) WOX Family Genes and Their Potential Function in Somatic Embryogenesis. Int. J. Mol. Sci. 2023, 24, 11077. [Google Scholar] [CrossRef]
- Wang, K.; Shi, L.; Liang, X.; Zhao, P.; Wang, W.; Liu, J.; Chang, Y.; Hiei, Y.; Yanagihara, C.; Du, L.; et al. The gene TaWOX5 overcomes genotype dependency in wheat genetic transformation. Nat. Plants 2022, 8, 110–117. [Google Scholar] [CrossRef]
- Tang, Y.; Li, H.; Guan, Y.; Li, S.; Xun, C.; Dong, Y.; Huo, R.; Guo, Y.; Bao, X.; Pei, E.; et al. Genome-Wide Identification of the Physic Nut WUSCHEL-Related Homeobox Gene Family and Functional Analysis of the Abiotic Stress Responsive Gene JcWOX5. Front. Genet. 2020, 11, 670. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Feng, Y.; Jiang, L.; Zhang, G.; Wu, T.; Zhang, X.; Xu, X.; Wang, Y.; Han, Z. Genome-wide identification of WOX family members in nine Rosaceae species and a functional analysis of MdWOX13-1 in drought resistance. Plant Sci. 2023, 328, 111564. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Qi, Y.; Wang, L.; Wang, L.; Yan, X.; Dang, Z.; Li, W.; Zhao, W.; Pei, X.; Li, X.; et al. Genomic Comparison and Population Diversity Analysis Provide Insights into the Domestication and Improvement of Flax. iScience 2020, 23, 100967. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Hernandez, M.; Berardini, T.; Chen, G.; Crist, D.; Doyle, A.; Huala, E.; Knee, E.; Lambrecht, M.; Miller, N.; Mueller, L.A.; et al. TAIR: A resource for integrated Arabidopsis data. Funct. Integr. Genom. 2002, 2, 239–253. [Google Scholar] [CrossRef]
- Li, Y.C.; Lu, Y.C. BLASTP-ACC: Parallel Architecture and Hardware Accelerator Design for BLAST-Based Protein Sequence Alignment. IEEE Trans. Biomed. Circuits Syst. 2019, 13, 1771–1782. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; López, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Khaja, R.; MacDonald, J.R.; Zhang, J.; Scherer, S.W. Methods for identifying and mapping recent segmental and gene duplications in eukaryotic genomes. Methods Mol. Biol. 2006, 338, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef]
- Carpenter, C.M.; Frank, D.N.; Williamson, K.; Arbet, J.; Wagner, B.D.; Kechris, K.; Kroehl, M.E. tidyMicro: A pipeline for microbiome data analysis and visualization using the tidyverse in R. BMC Bioinform. 2021, 22, 41. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 2019, 47, e47. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]





| Gene | Gene ID in Genome | Number of Amino Acids | Molecular Weight (KDa) | PI | Instability Index | Aliphatic Index | Grand Average of Hydropathicity (GRAVY) | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|
| LuWOX01 | L.us.o.m.scaffold170.48 | 245 | 26.88 | 6.75 | 39.55 | 70.08 | −0.629 | Nuclear |
| LuWOX02 | L.us.o.m.scaffold255.15 | 234 | 25.94 | 5.63 | 55.21 | 65.04 | −0.832 | Nuclear |
| LuWOX03 | L.us.o.m.scaffold218.7 | 299 | 33.44 | 8.69 | 54.07 | 63.95 | −0.638 | Nuclear |
| LuWOX04 | L.us.o.m.scaffold50.47 | 278 | 30.72 | 8.76 | 63.10 | 60.72 | −0.764 | Nuclear |
| LuWOX05 | L.us.o.m.scaffold33.69 | 395 | 43.91 | 7.38 | 56.58 | 59.65 | −0.677 | Nuclear |
| LuWOX06 | L.us.o.m.scaffold107.61 | 366 | 40.69 | 7.86 | 60.13 | 59.59 | −0.763 | Nuclear |
| LuWOX07 | L.us.o.m.scaffold155.99 | 196 | 22.01 | 9.64 | 74.39 | 63.72 | −0.793 | Nuclear |
| LuWOX08 | L.us.o.m.scaffold98.176 | 266 | 29.85 | 8.69 | 56.80 | 58.68 | −0.973 | Nuclear |
| LuWOX09 | L.us.o.m.scaffold8.54 | 196 | 22.58 | 8.42 | 51.73 | 56.28 | −0.825 | Nuclear |
| LuWOX10 | L.us.o.m.scaffold133.89 | 323 | 35.57 | 5.68 | 50.37 | 47.43 | −0.930 | Nuclear |
| LuWOX11 | L.us.o.m.scaffold83.172 | 315 | 34.71 | 5.56 | 51.80 | 47.40 | −0.909 | Nuclear |
| LuWOX12 | L.us.o.m.scaffold2.503 | 239 | 27.73 | 8.69 | 63.42 | 51.84 | −0.954 | Nuclear |
| LuWOX13 | L.us.o.m.scaffold72.52 | 414 | 45.71 | 7.10 | 59.57 | 61.55 | −0.698 | Nuclear |
| LuWOX14 | L.us.o.m.scaffold103.108 | 432 | 47.91 | 7.44 | 52.36 | 62.13 | −0.756 | Nuclear |
| LuWOX15 | L.us.o.m.scaffold258.26 | 192 | 21.81 | 9.71 | 54.33 | 70.10 | −0.741 | Nuclear |
| LuWOX16 | L.us.o.m.scaffold212.30 | 263 | 29.86 | 9.37 | 64.60 | 63.80 | −0.983 | Nuclear |
| LuWOX17 | L.us.o.m.scaffold6.173 | 231 | 24.66 | 5.65 | 64.59 | 67.97 | −0.345 | Nuclear |
| LuWOX18 | L.us.o.m.scaffold141.67 | 294 | 31.13 | 5.96 | 61.38 | 66.73 | −0.366 | Nuclear |
| Duplicated Gene Pairs | Non-Synonymous (Ka) | Synonymous (Ks) | Ka/Ks |
|---|---|---|---|
| LuWOX01 & LuWOX02 | 0.07464 | 0.3158 | 0.2363 |
| LuWOX04 & LuWOX03 | 0.08564 | 0.2392 | 0.3579 |
| LuWOX06 & LuWOX05 | 0.04869 | 0.2712 | 0.1795 |
| LuWOX15 & LuWOX07 | 0.08562 | 0.1661 | 0.5151 |
| LuWOX08 & LuWOX16 | 0.04944 | 0.0838 | 0.5899 |
| LuWOX09 & LuWOX12 | 0.04245 | 0.2428 | 0.1748 |
| LuWOX10 & LuWOX11 | 0.02632 | 0.0976 | 0.2697 |
| LuWOX13 & LuWOX14 | 0.06725 | 0.1652 | 0.4069 |
| LuWOX17 & LuWOX18 | 0.02931 | 0.1340 | 0.2186 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, X.; Lu, J.; Wang, H.; Tang, L.; Li, S.; Zang, Z.; Wu, G.; Zhang, J. Identification and Characterization of WOX Gene Family in Flax (Linum usitatissimum L.) and Its Role Under Abiotic Stress. Int. J. Mol. Sci. 2025, 26, 3571. https://doi.org/10.3390/ijms26083571
Song X, Lu J, Wang H, Tang L, Li S, Zang Z, Wu G, Zhang J. Identification and Characterization of WOX Gene Family in Flax (Linum usitatissimum L.) and Its Role Under Abiotic Stress. International Journal of Molecular Sciences. 2025; 26(8):3571. https://doi.org/10.3390/ijms26083571
Chicago/Turabian StyleSong, Xixia, Jianyu Lu, Hang Wang, Lili Tang, Shuyao Li, Zhenyuan Zang, Guangwen Wu, and Jian Zhang. 2025. "Identification and Characterization of WOX Gene Family in Flax (Linum usitatissimum L.) and Its Role Under Abiotic Stress" International Journal of Molecular Sciences 26, no. 8: 3571. https://doi.org/10.3390/ijms26083571
APA StyleSong, X., Lu, J., Wang, H., Tang, L., Li, S., Zang, Z., Wu, G., & Zhang, J. (2025). Identification and Characterization of WOX Gene Family in Flax (Linum usitatissimum L.) and Its Role Under Abiotic Stress. International Journal of Molecular Sciences, 26(8), 3571. https://doi.org/10.3390/ijms26083571







