Retinoblastoma: Molecular Evaluation of Tumor Samples, Aqueous Humor, and Peripheral Blood Using a Next-Generation Sequence Panel
Abstract
1. Introduction
2. Results
2.1. Clinical and Tumor Characteristics
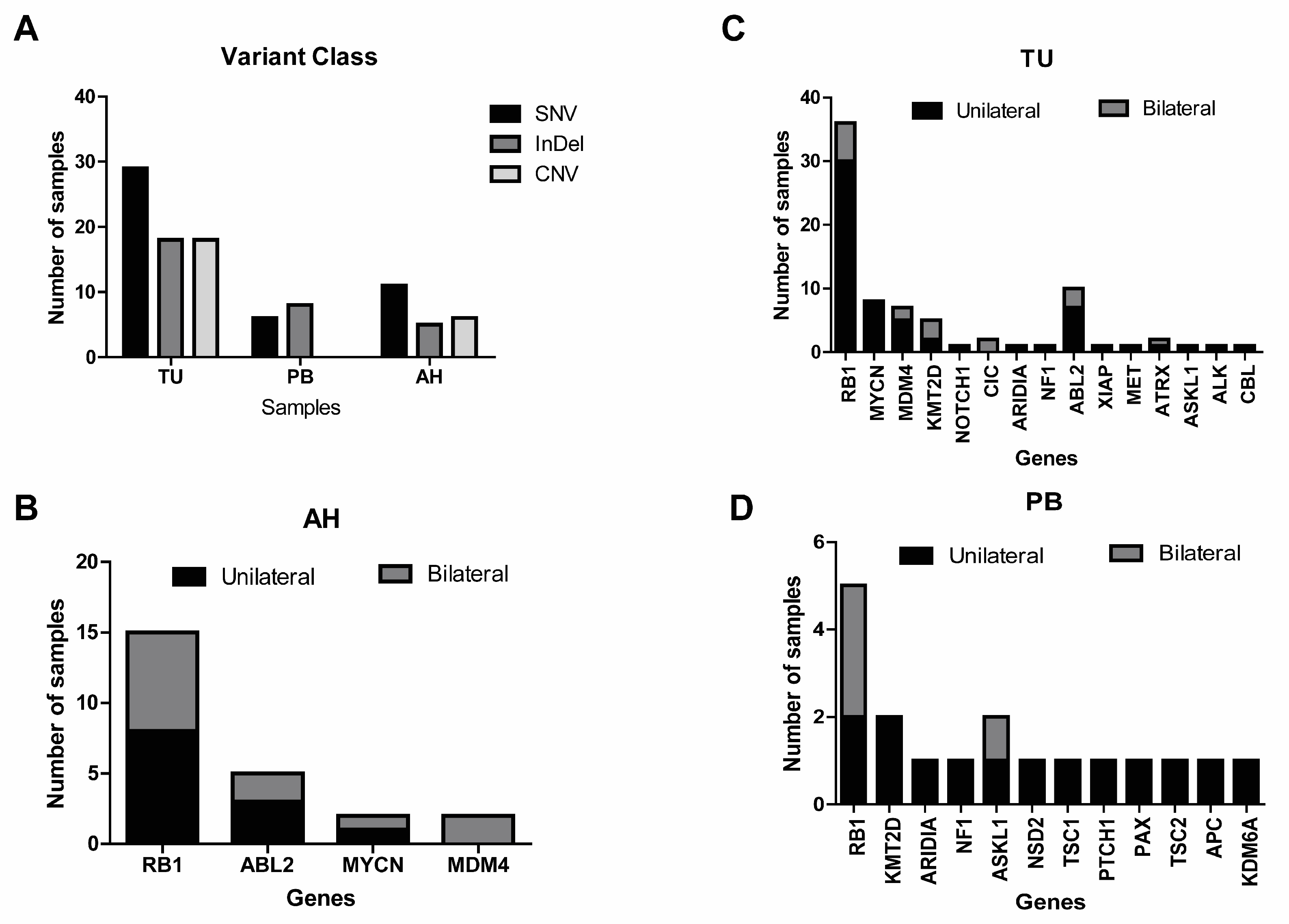
2.2. Identification of Genetic Alteration by Panel in Liquid Biopsy
2.3. Identification of Genetic Alterations in Tumor and Peripheral Blood
2.4. Genetic Analysis from Tumors, Aqueous Humor, and Peripheral Blood
3. Discussion
4. Methods
4.1. Patients and Samples
4.2. DNA and RNA Extraction from Tumor and Peripheral Blood
4.3. Cell-Free DNA/RNA Isolation from Aqueous Humor
4.4. cDNA Synthesis
4.5. Library Preparation and Next-Generation Sequencing (NGS) Run with the OCCRA® Panel
4.6. Oncomine Childhood Cancer Research Assay (OCCRA©) Sequencing Panel Analysis
4.7. Data and Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dimaras, H.; Corson, T.; Cobrinik, D.; White, A.; Zhao, J.; Munier, F.L.; Abramson, D.H.; Shields, C.L.; Chantada, G.L.; Njuguna, F.; et al. Retinoblastoma. Nat. Rev. Dis. Primers 2015, 1, 15021. [Google Scholar] [CrossRef] [PubMed]
- Yun, J.; Li, Y.; Xu, C.T.; Pan, B.R. Epidemiology and Rb1 gene of retinoblastoma. Int. J. Ophthalmol. 2011, 4, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Karcioglu, Z.A.; Gordon, R.A.; Karcioglu, G.L. Tumor seeding in ocular fine needle aspiration biopsy. Ophthalmology 1985, 92, 1763–1767. [Google Scholar] [CrossRef] [PubMed]
- Karcioglu, Z.A. Fine needle aspiration biopsy (FNAB) for retinoblastoma. Retina 2002, 22, 707–710. [Google Scholar] [CrossRef] [PubMed]
- Eide, N.; Walaas, L. Fine-needle aspiration biopsy and other biopsies in suspected intraocular malignant disease: A review. Acta Ophthalmol. 2009, 87, 588–601. [Google Scholar] [CrossRef]
- Eriksson, O.; Hagmar, B.; Ryd, W. Effects of fine-needle aspiration and other biopsy procedures on tumor dissemination in mice. Cancer 1984, 54, 73–78. [Google Scholar] [CrossRef]
- Joosse, S.A.; Pantel, K. Circulating DNA and Liquid Biopsies in the Management of Patients with Cancer. Cancer Res. 2022, 82, 2213–2215. [Google Scholar] [CrossRef]
- Munier, F.L.; Gaillard, M.-C.; Balmer, A.; Soliman, S.; Podilsky, G.; Moulin, A.P.; Beck-Popovic, M. Intravitreal chemotherapy for vitreous disease in retinoblastoma revisited: From prohibition to conditional indications. Br. J. Ophthalmol. 2012, 96, 1078–1083. [Google Scholar] [CrossRef]
- Munier, F.L.; Soliman, S.; Moulin, A.P.; Gaillard, M.-C.; Balmer, A.; Beck-Popovic, M. Profiling safety of intravitreal injections for retinoblastoma using an anti-reflux procedure and sterilisation of the needle track. Br. J. Ophthalmol. 2012, 96, 1084–1087. [Google Scholar] [CrossRef] [PubMed]
- Berry, J.L.; Xu, L.; Murphree, A.L.; Krishnan, S.; Stachelek, K.; Zolfaghari, E.; McGovern, K.; Lee, T.C.; Carlsson, A.; Kuhn, P.; et al. Potential of Aqueous Humor as a Surrogate Tumor Biopsy for Retinoblastoma. JAMA Ophthalmol. 2017, 135, 1221–1230. [Google Scholar] [CrossRef] [PubMed]
- Gerrish, A.; Stone, E.; Clokie, S.; Ainsworth, J.R.; Jenkinson, H.; McCalla, M.; Hitchcott, C.; Colmenero, I.; Allen, S.; Parulekar, M.; et al. Non-invasive diagnosis of retinoblastoma using cell-free DNA from aqueous humour. Br. J. Ophthalmol. 2019, 103, 721–724. [Google Scholar] [CrossRef]
- Berry, J.L.; Xu, L.; Polski, A.; Jubran, R.; Kuhn, P.; Kim, J.W.; Hicks, J. Aqueous Humor Is Superior to Blood as a Liquid Biopsy for Retinoblastoma. Ophthalmology 2020, 127, 552–554. [Google Scholar] [CrossRef]
- Im, D.H.; Pike, S.; Reid, M.W.; Peng, C.-C.; Sirivolu, S.; Grossniklaus, H.E.; Hubbard, G.B.; Skalet, A.H.; Bellsmith, K.N.; Shields, C.L.; et al. A Multicenter Analysis of Nucleic Acid Quantification Using Aqueous Humor Liquid Biopsy in Retinoblastoma: Implications for Clinical Testing. Ophthalmol. Sci. 2023, 3, 100289. [Google Scholar] [CrossRef]
- Ghose, N.; Kaliki, S. Liquid biopsy in Retinoblastoma: A review. Semin. Ophthalmol. 2022, 37, 813–819. [Google Scholar] [CrossRef] [PubMed]
- Raval, V.; Racher, H.; Wrenn, J.; Singh, A.D. Aqueous humor as a surrogate biomarker for retinoblastoma tumor tissue. J. Am. Assoc. Pediatr. Ophthalmol. Strabismus 2022, 26, 137.e1–137.e5. [Google Scholar] [CrossRef]
- Berry, J.L.; Xu, L.; Kooi, I.; Murphree, A.L.; Prabakar, R.K.; Reid, M.; Stachelek, K.; Le, B.H.A.; Welter, L.; Reiser, B.J.; et al. Genomic cfDNA Analysis of Aqueous Humor in Retinoblastoma Predicts Eye Salvage: The Surrogate Tumor Biopsy for Retinoblastoma. Mol. Cancer Res. 2018, 16, 1701–1712. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Kim, M.E.; Polski, A.; Prabakar, R.K.; Shen, L.; Peng, C.-C.; Reid, M.W.; Chévez-Barrios, P.; Kim, J.W.; Shah, R.; et al. Establishing the Clinical Utility of ctDNA Analysis for Diagnosis, Prognosis, and Treatment Monitoring of Retinoblastoma: The Aqueous Humor Liquid Biopsy. Cancers 2021, 13, 1282. [Google Scholar] [CrossRef] [PubMed]
- Kletke, S.N.; Soliman, S.; Racher, H.; Mallipatna, A.; Shaikh, F.; Mireskandari, K.; Gallie, B.L. A typical anterior retinoblastoma: Diagnosis by aqueous humor cell-free DNA analysis. Ophthalmic Genet. 2022, 43, 862–865. [Google Scholar] [CrossRef] [PubMed]
- Linn Murphree, A. Intraocular retinoblastoma: The case for a new group classification. Ophthalmol. Clin. N. Am. 2005, 18, 41–53, viii. [Google Scholar] [CrossRef] [PubMed]
- Hiemenz, M.C.; Ostrow, D.G.; Busse, T.M.; Buckley, J.; Maglinte, D.T.; Bootwalla, M.; Done, J.; Ji, J.; Raca, G.; Ryutov, A.; et al. A Comprehensive Next-Generation Sequencing Panel for Pediatric Malignancies. J. Mol. Diagn. 2018, 20, 765–776. [Google Scholar] [CrossRef]
- Lohmann, D.R.; Gerick, M.; Brandt, B.; Oelschläger, U.; Lorenz, B.; Passarge, E.; Horsthemke, B. Constitutional RB1-gene mutations in patients with isolated unilateral retinoblastoma. Am. J. Hum. Genet. 1997, 61, 282–294. [Google Scholar] [CrossRef] [PubMed]
- Tomar, S.; Sethi, R.; Sundar, G.; Quah, T.C.; Quah, B.L.; Lai, P.S. Mutation spectrum of RB1 mutations in retinoblastoma cases from Singapore with implications for genetic management and counselling. PLoS ONE 2017, 12, e0178776. [Google Scholar] [CrossRef] [PubMed]
- Rushlow, D.; Piovesan, B.; Zhang, K.; Prigoda-Lee, N.L.; Marchong, M.N.; Clark, R.D.; Gallie, B.L. Detection of mosaic RB1 mutations in families with retinoblastoma. Hum. Mutat. 2009, 30, 842–851. [Google Scholar] [CrossRef] [PubMed]
- Gupta, H.; Malaichamy, S.; Mallipatna, A.; Murugan, S.; Jeyabalan, N.; Babu, V.S.; Ghosh, A.; Ghosh, A.; Santhosh, S.; Seshagiri, S.; et al. Retinoblastoma genetics screening and clinical management. BMC Med. Genom. 2021, 14, 188. [Google Scholar] [CrossRef]
- Kim, M.E.; Polski, A.; Xu, L.; Prabakar, R.K.; Peng, C.-C.; Reid, M.W.; Shah, R.; Kuhn, P.; Cobrinik, D.; Hicks, J.; et al. Comprehensive Somatic Copy Number Analysis Using Aqueous Humor Liquid Biopsy for Retinoblastoma. Cancers 2021, 13, 3340. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.E.; Xu, L.; Prabakar, R.K.; Shen, L.; Peng, C.-C.; Kuhn, P.; Gai, X.; Hicks, J.; Berry, J.L. Aqueous Humor as a Liquid Biopsy for Retinoblastoma: Clear Corneal Paracentesis and Genomic Analysis. J. Vis. Exp. 2021, 175, e62939. [Google Scholar] [CrossRef]
- Sradhanjali, S.; Rout, P.; Tripathy, D.; Kaliki, S.; Rath, S.; Modak, R.; Mittal, R.; Chowdary, T.K.; Reddy, M.M. The Oncogene MYCN Modulates Glycolytic and Invasive Genes to Enhance Cell Viability and Migration in Human Retinoblastoma. Cancers 2021, 13, 5248. [Google Scholar] [CrossRef]
- Oliveira Reis, A.; Ribamar de Carvalho, I.N.S.; Damasceno, P.B.S. Influence of MDM2 and MDM4 on development and survival in hereditary retinoblastoma. Pediatr. Blood Cancer 2012, 59, 39–43. [Google Scholar] [CrossRef]
- Jones, J.K.; Zhang, H.; Lyne, A.M. ABL1 and ABL2 promote medulloblastoma leptomeningeal dissemination. Neuro-Oncol. Adv. 2023, 5, vdad095. [Google Scholar] [CrossRef]
- Cabral de Carvalho Corrêa, D.; Tesser-Gamba, F.; Dias Oliveira, I.; da Silva, N.S.; Capellano, A.M.; Alves, M.T.d.S.; Silva, F.A.B.; Dastoli, P.A.; Cavalheiro, S.; de Toledo, S.R.C. Molecular profiling of pediatric and adolescent ependymomas: Identification of genetic variants using a next-generation sequencing panel. J. Neurooncol. 2021, 155, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Guimarães, G.M.; Tesser-Gamba, F.; Petrilli, A.S.; Donato-Macedo, C.; Alves, M.; de Lima, F.; Garcia-Filho, R.; Oliveira, R.; Toledo, S. Molecular profiling of osteosarcoma in children and adolescents from different age groups using a next-generation sequencing panel. Cancer Genet. 2021, 258–259, 85–92. [Google Scholar] [CrossRef] [PubMed]

| Retinoblastoma | ||||
|---|---|---|---|---|
| Unilateral | Bilateral | Trilateral | All | |
| Number of patients (%) | 58 (77%) | 17 (22%) | 1 (1%) | 76 (100%) |
| Number of eyes | 58 | 34 | 2 | 94 |
| Mean age diagnosis (Months) | 32 | 20 | 15 | 29 |
| Gender | ||||
| Male | 32 (55%) | 10 (59%) | 0 | 43 (57%) |
| Female | 28 (48%) | 7 (41%) | 1 (100%) | 33 (43%) |
| Familial RB | 0 | 2 (11%) | 2 (3%) | |
| Tumor | ||||
| Intraocular | 48 (83%) | 11 (65%) | 1 (100%) | 60 (79%) |
| Extraocular | 10 (17%) | 6 (35%) | 0 | 16 (21%) |
| Primary treatment | ||||
| Enucleation | 53 (91%) | 8 (47%) | 0 | 61 (80%) |
| Therapy | 5 (9%) | 9 (53%) | 1 (100%) | 15 (20%) |
| Recurrence | 4 (7%) | 7 (41%) | 1 (100%) | 12 (16%) |
| Mortality | 5 (9%) | 3 (18%) | 0 | 8 (10%) |
| IIRC group | ||||
| A | 1 (2%) | 4 (12%) | 0 | 5 (5%) |
| B | 2 (3%) | 4 (12%) | 0 | 6 (6%) |
| C | 0 | 1 (3%) | 0 | 1 (1%) |
| D | 0 | 4 (12%) | 1 (50%) | 5 (5%) |
| E | 46 (79%) | 11 (32%) | 1 (50%) | 58 (62%) |
| Without group | 9 (15%) | 10 (29%) | 0 | 19 (21%) |
| Patient ID | Laterality | Initial Treatment | Gene | Chromosome | Coding | Aminoacid Change | Variant Classification | Gene Classification | Variant Class | Variant Effect | Clinical Significance |
|---|---|---|---|---|---|---|---|---|---|---|---|
| RB32 | UL | PE | RB1 | 13q14.2 | c.160G>T | p.Glu54Ter | Deletion | Lost | SNV | Nonsense | Pathogenic |
| 13q14.2 | c.1735C>T | p.Arg579Ter | Deletion | Lost | SNV | Nonsense | Pathogenic | ||||
| RB36 | UL | PE | RB1 | 13q14.2 | c.1072C>T | p.Arg358Ter | Deletion | Lost | SNV | Nonsense | Pathogenic |
| 13q14.2 | c.1341_1342insA | p.Leu448fs | Deletion | Lost | INDEL | Frameshift Insertion | Patogenic | ||||
| RB37 | UL | PE | RB1 | 13q14.2 | c.1333C>T | p.Arg445Ter | Deletion | Lost | SNV | Nonsense | Patogenic |
| RB40 | BL | IVC | RB1 | 13q14.2 | c.1333C>T | p.Arg445Ter | Deletion | Lost | SNV | Nonsense | Patogenic |
| RB43 | BL | IVC | RB1 | 13q14.2 | c.361C>T | p.Gln121Ter | Deletion | Lost | SNV | Nonsense | |
| RB46 | BL | IVC | RB1 | 13q14.2 | c.1072C>T | p.Arg358Ter | Deletion | Lost | SNV | Nonsense | Patogenic |
| RB47 OD | BL | PE | RB1 | 13q14.2 | c.933delT | p.Pro312fs | Deletion | Lost | INDEL | Frameshift Deletion | Patogenic |
| RB47 OE | BL | PE | RB1 | 13q14.2 | c.933delT | p.Pro312fs | Deletion | Lost | INDEL | Frameshift Deletion | Patogenic |
| RB48 | BL | PE | RB1 | 13q14.2 | c.2486delC | p.Ser829Ter | Deletion | Lost | INDEL | nonsense | Patogenic |
| RB53 | UL | PE | RB1 | 13q14.2 | c.958C>T | p.Arg320Ter | Deletion | Lost | SNV | Nonsense | Patogenic |
| RB54 | UL | PE | RB1 | 13q14.2 | c.1318G>T | p.Glu440Ter | Truncating mutation | Lost | SNV | Nonsense | Patogenic |
| RB64 | UL | PE | RB1 | 13q14.2 | c.1363C>T | p.Arg455Ter | Truncating mutation | Lost | SNV | Nonsense | Patogenic |
| RB65 | BL | PE | RB1 | 13q14.2 | c.1597G>T | p.Glu533Ter | Deletion | Lost | SNV | Nonsense | Patogenic |
| RB70 | UL | PE | RB1 | 13q14.2 | c.2478delT | p.Pro827GlnfsTer6 | Truncating mutation | Lost | INDEL | Frameshift Deletion | Patogenic |
| RB73 | UL | PE | RB1 | 13q14.2 | c.1654C>T | p.Arg552Ter | Truncating mutation | Lost | SNV | Nonsense | Patogenic |
| Coding | Aminoacid Change | Classificação Variante | Classificação Gene | Variant Class | Variant Effect | Clinical Significance | TU | PB |
|---|---|---|---|---|---|---|---|---|
| c.1654C>T | p.Arg552Ter | Deletion | Lost | SNV | Nonsense | Pathogenic | 1 | 0 |
| c.1333C>T | p.Arg455Ter | Deletion | Lost | SNV | Nonsense | Pathogenic | 4 | 0 |
| c.1363C>T | p.Arg455Ter | Deletion | Lost | SNV | Nonsense | Pathogenic | 3 | 0 |
| c.958C>T | p.Arg320Ter | Deletion | Lost | SNV | Nonsense | Pathogenic | 5 | 0 |
| c.1666C>T | p.Arg556Ter | Deletion | Lost | SNV | Nonsense | Pathogenic | 2 | 1 |
| c.1072C>T | p.Arg358Ter | Deletion | Lost | SNV | Nonsense | Pathogenic | 3 | 0 |
| c.1060C>T | p.Gln354Ter | Deletion | Lost | SNV | Nonsense | Pathogenic | 1 | 0 |
| c.844G>T | p.Glu282Ter | Deletion | Lost | SNV | Nonsense | Pathogenic | 1 | 0 |
| c.1735C>T | p.Arg579Ter | Deletion | Lost | SNV | Nonsense | Pathogenic | 1 | 0 |
| c.160G>T | p.Glu54Ter | Deletion | Lost | SNV | Nonsense | Pathogenic | 1 | 0 |
| c.526C>T | p.Gln176Ter | Deletion | Lost | SNV | Nonsense | Pathogenic | 1 | 0 |
| c.361C>T | p.Gln121Ter | Truncating mutation | Lost | SNV | Nonsense | Pathogenic | 2 | 1 |
| c.1318G>T | p.Glu440Ter | Truncating mutation | Lost | SNV | Nonsense | Pathogenic | 1 | 0 |
| c.1597G>T | p.Glu533Ter | Truncating mutation | Lost | SNV | Nonsense | Pathogenic | 1 | 1 |
| c.1330C>T | p.Gln444Ter | Truncating mutation | Lost | SNV | Nonsense | Pathogenic | 1 | 1 |
| c.2308C>T | p.Gln770Ter | Truncating mutation | Lost | SNV | Nonsense | Pathogenic | 1 | 0 |
| c.2532_2541delGTTCCAGAAA | p.Phe845Ter | Deletion | Lost | InDel | Nonsense | Pathogenic | 1 | 0 |
| c.1942delT | p.Ser648fs | Deletion | Lost | InDel | Frameshift deletion | Pathogenic | 1 | 0 |
| c.795delA | p.Lys265fs | Deletion | Lost | InDel | Frameshift deletion | Pathogenic | 1 | 0 |
| c.733_739delCCCATTA | p.Pro245fs | Deletion | Lost | InDel | Frameshift deletion | Pathogenic | 1 | 0 |
| c.660_661delAT | p.Val222fs | Deletion | Lost | InDel | Frameshift deletion | Pathogenic | 1 | 0 |
| c.1827delT | p.Val610Ter | Deletion | Lost | InDel | Nonsense | Pathogenic | 1 | 0 |
| c.1341_1342insA | p.Leu448fs | Deletion | Lost | InDel | Frameshift insertion | Pathogenic | 1 | 0 |
| c.933delT | p.Pro312fs | Deletion | Lost | InDel | Frameshift deletion | Pathogenic | 1 | 1 |
| c.2486delC | p.Ser829Ter | Deletion | Lost | InDel | Nonsense | Pathogenic | 1 | 0 |
| c.613_614insGAAG | p.Val205fs | Deletion | Lost | InDel | Frameshift insertion | Pathogenic | 1 | 0 |
| c.2478delT | p.Pro827GlnfsTer6 | Truncating mutation | Lost | InDel | Frameshift deletion | Pathogenic | 1 | 0 |
| Patient ID | Gender | Laterality | Sample | Nucleotide Change | Amino acid Change | Variant Classification | Gene Classification | Variant Class | Consequences |
|---|---|---|---|---|---|---|---|---|---|
| RB 32 | M | UL | TU/AH | c.160G>T | p.Glu54Ter | Deletion | Lost | SNV | Nonsense |
| c.1735C>T | p.Arg579Ter | Deletion | Lost | SNV | Nonsense | ||||
| RB 36 | M | UL | TU/AH | c.1072C>T | p.Arg358Ter | Deletion | Lost | SNV | Nonsense |
| c.1341_1342insA | p.Leu448fs | Deletion | Lost | InDel | Frameshift insertion | ||||
| RB 37 | M | UL | TU/AH | c.1333C>T | p.Arg445Ter | Deletion | Lost | SNV | Nonsense |
| RB 43 | F | BL | TU/AH/PB | c.361C>T | p.Gln121Ter | Truncating mutation | Lost | SNV | Nonsense |
| RB 47 | M | BL | TU/AH/PB | c.933delT | p.Pro312fs | Deletion | Lost | InDel | Frameshift deletion |
| RB 48 | F | BL | TU/AH | c.2486delC | p.Ser829Ter | Deletion | Lost | InDel | Nonsense |
| RB 53 | F | UL | TU/AH | c.958C>T | p.Arg320Ter | Truncating mutation | Lost | SNV | Nonsense |
| RB 55 | F | UL | TU | c.361C>T | p.Gln121Ter | Truncating mutation | Lost | SNV | Nonsense |
| RB 56 | M | UL | TU | c.958C>T | p.Arg320Ter | Truncating mutation | Lost | SNV | Nonsense |
| RB 58 | F | UL | TU | c.958C>T | p.Arg320Ter | Truncating mutation | Lost | SNV | Nonsense |
| c.1072C>T | p.Arg358Ter | Deletion | Lost | SNV | Nonsense | ||||
| RB 59 | M | UL | TU | c.958C>T | p.Arg320Ter | Truncating mutation | Lost | SNV | Nonsense |
| RB 64 | F | UL | TU/PB | c.1363C>T | p.Arg455Ter | Truncating mutation | Lost | SNV | Nonsense |
| RB 65 | M | BL | TU/AH/PB | c.1597G>T | p.Glu533Ter | Truncating mutation | Lost | SNV | Nonsense |
| RB 68 | M | UL | TU/PB | c.1330C>T | p.Gln444Ter | Truncating mutation | Lost | SNV | Nonsense |
| RB 70 | F | UL | TU/AH | c.2478delT | p.Pro827GlnfsTer6 | Truncating mutation | Lost | InDel | Frameshift deletion |
| RB 71 | M | UL | TU | c.2308C>T | p.Gln770Ter | Truncating mutation | Lost | SNV | Nonsense |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mendes, T.B.; Oliveira, I.D.; Gamba, F.T.; Lima, F.T.; Morales, B.F.S.C.; Macedo, C.R.D.; Teixeira, L.F.; de Toledo, S.R.C. Retinoblastoma: Molecular Evaluation of Tumor Samples, Aqueous Humor, and Peripheral Blood Using a Next-Generation Sequence Panel. Int. J. Mol. Sci. 2025, 26, 3523. https://doi.org/10.3390/ijms26083523
Mendes TB, Oliveira ID, Gamba FT, Lima FT, Morales BFSC, Macedo CRD, Teixeira LF, de Toledo SRC. Retinoblastoma: Molecular Evaluation of Tumor Samples, Aqueous Humor, and Peripheral Blood Using a Next-Generation Sequence Panel. International Journal of Molecular Sciences. 2025; 26(8):3523. https://doi.org/10.3390/ijms26083523
Chicago/Turabian StyleMendes, Thais Biude, Indhira Dias Oliveira, Francine Tesser Gamba, Fernanda Teresa Lima, Bruna Fernanda Silva Cardoso Morales, Carla Renata Donato Macedo, Luiz Fernando Teixeira, and Silvia Regina Caminada de Toledo. 2025. "Retinoblastoma: Molecular Evaluation of Tumor Samples, Aqueous Humor, and Peripheral Blood Using a Next-Generation Sequence Panel" International Journal of Molecular Sciences 26, no. 8: 3523. https://doi.org/10.3390/ijms26083523
APA StyleMendes, T. B., Oliveira, I. D., Gamba, F. T., Lima, F. T., Morales, B. F. S. C., Macedo, C. R. D., Teixeira, L. F., & de Toledo, S. R. C. (2025). Retinoblastoma: Molecular Evaluation of Tumor Samples, Aqueous Humor, and Peripheral Blood Using a Next-Generation Sequence Panel. International Journal of Molecular Sciences, 26(8), 3523. https://doi.org/10.3390/ijms26083523





