Genome-Wide Identification of the BTB Domain-Containing Protein Gene Family in Pepper (Capsicum annuum L.)
Abstract
1. Introduction
2. Results
2.1. Identification of BTB Genes in Pepper
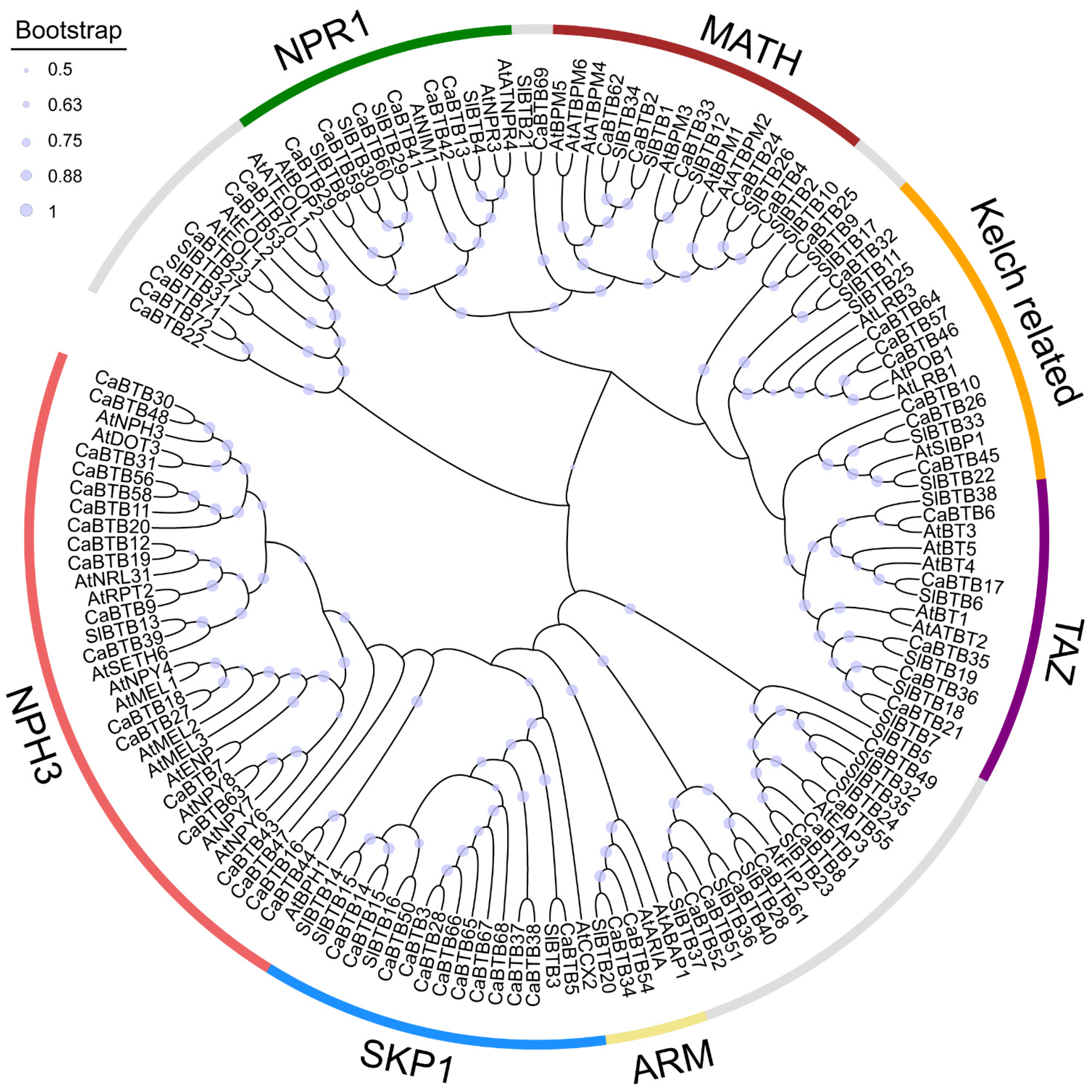
2.2. Phylogenetic Analysis of BTB Family
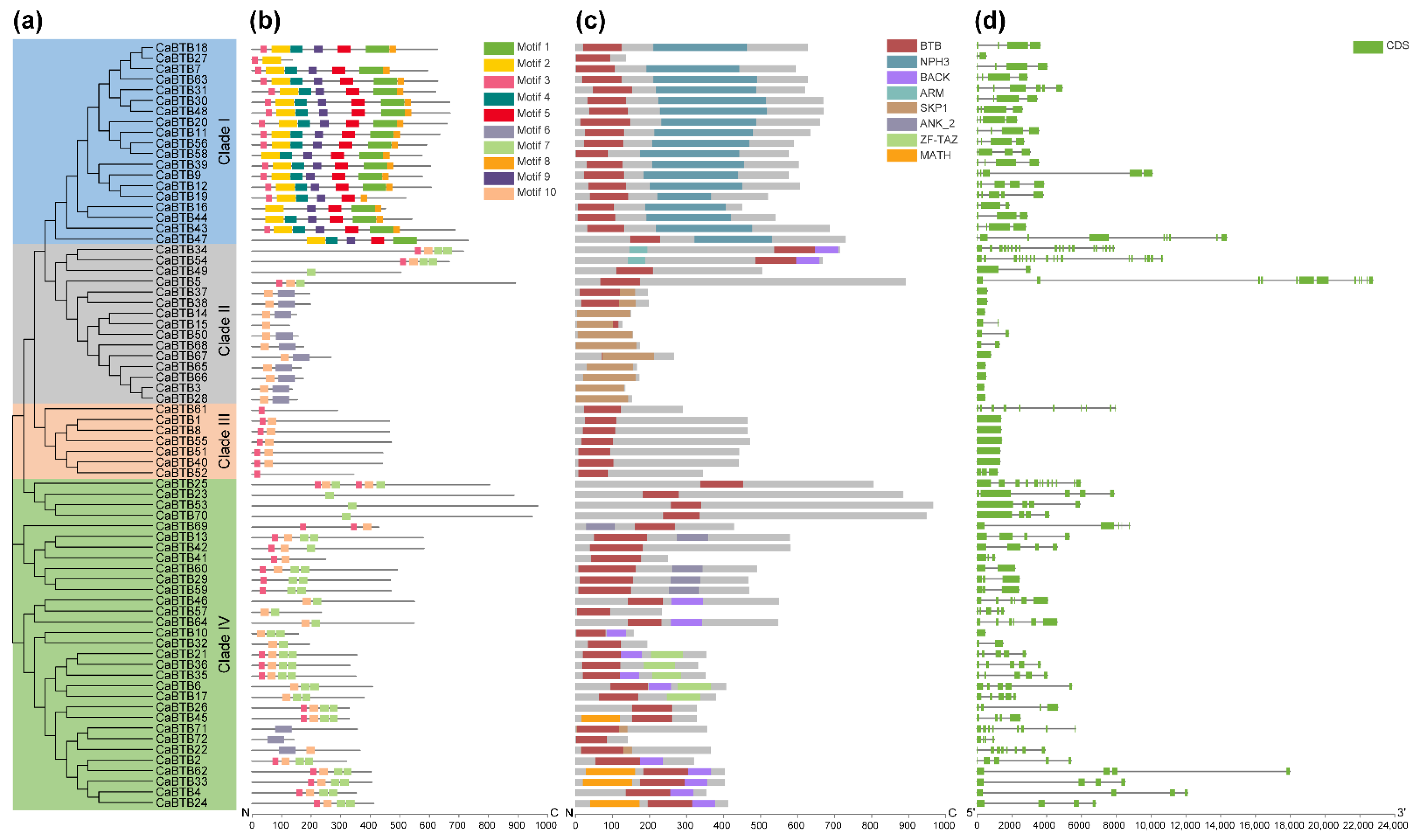
2.3. Structure Characterization and Motif Composition of CaBTB Genes
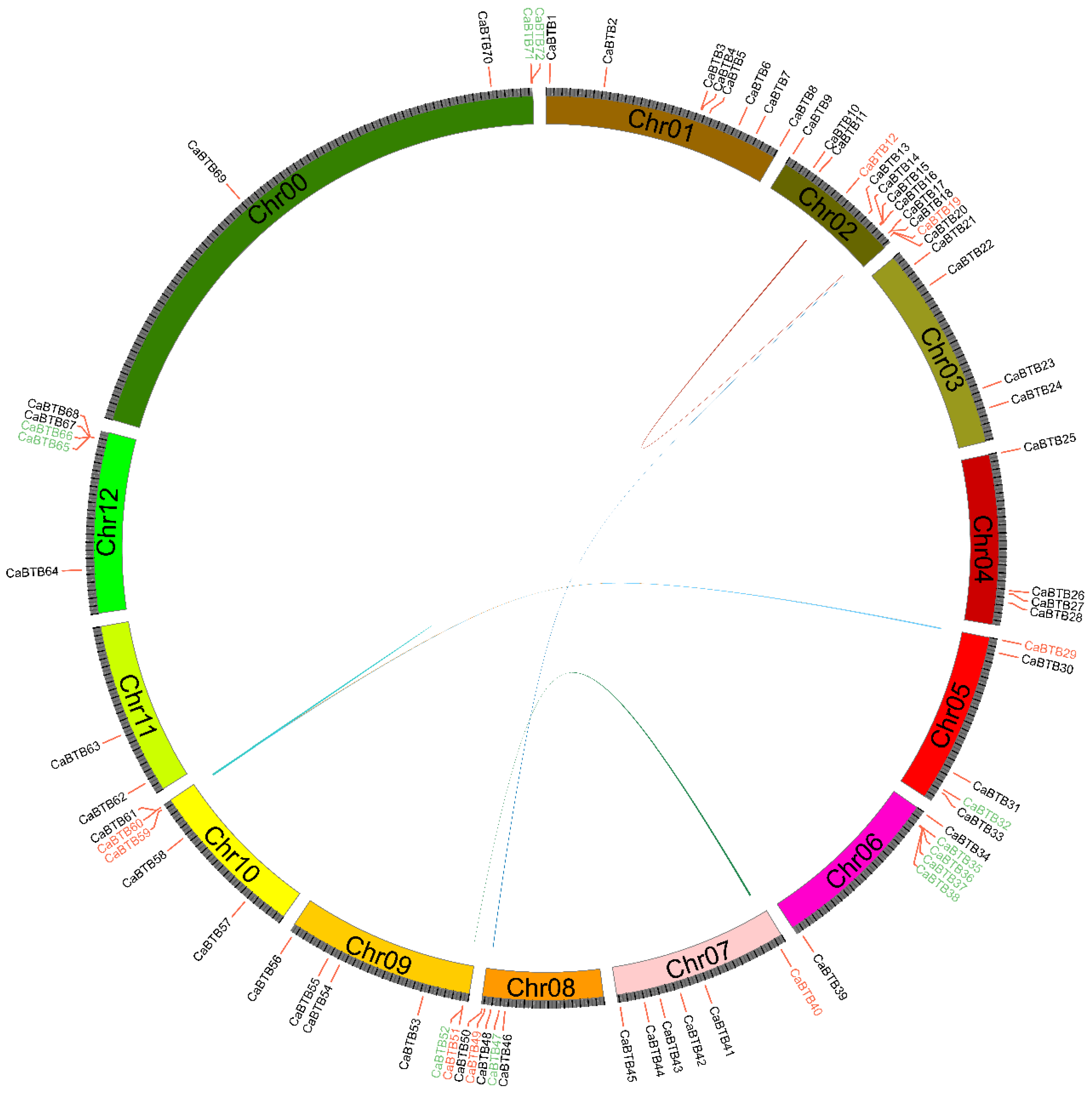
2.4. Chromosomal Localization and Duplication Analysis of CaBTB Genes
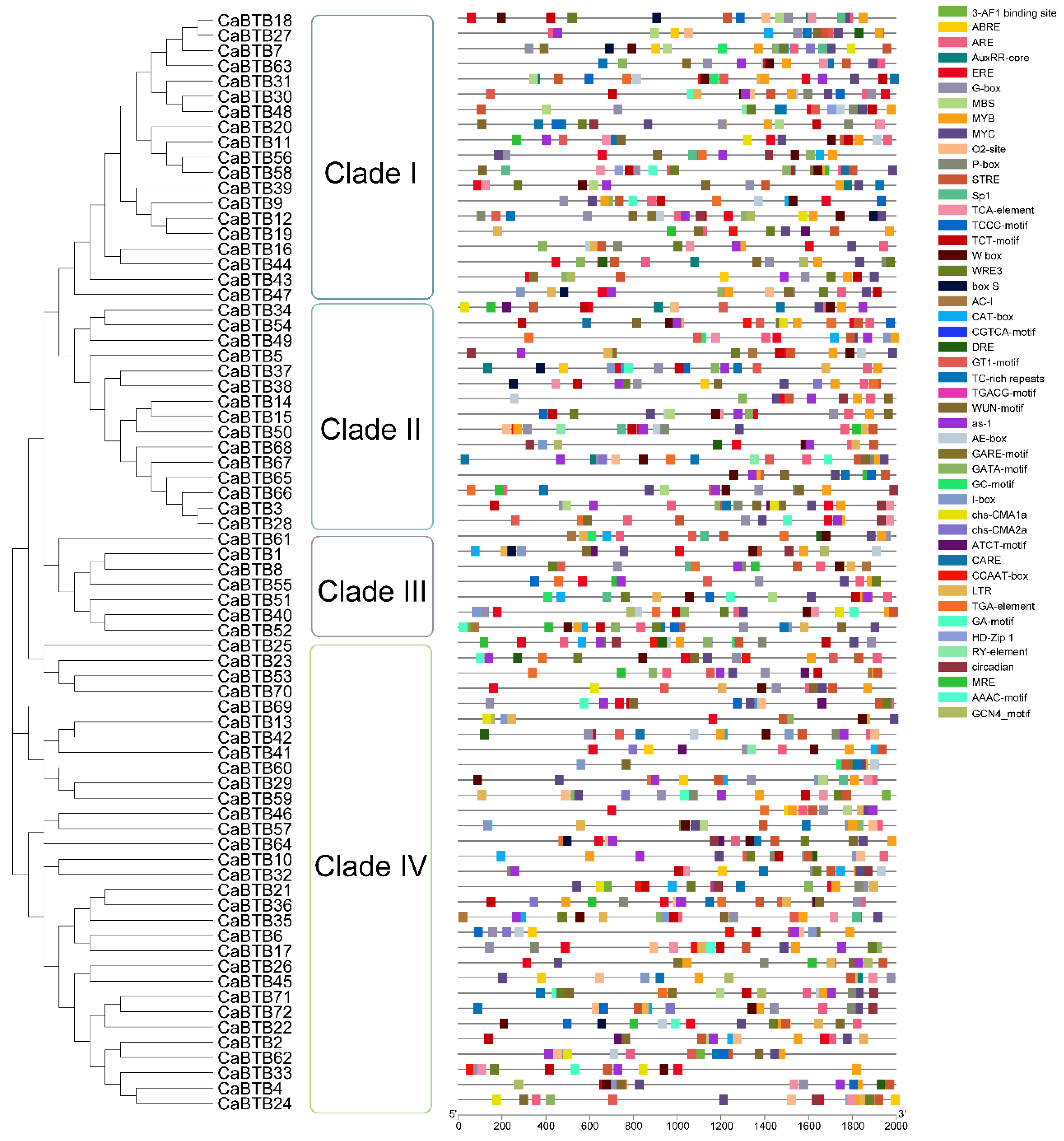
2.5. Cis-Acting Element Prediction of CaBTB Promoters
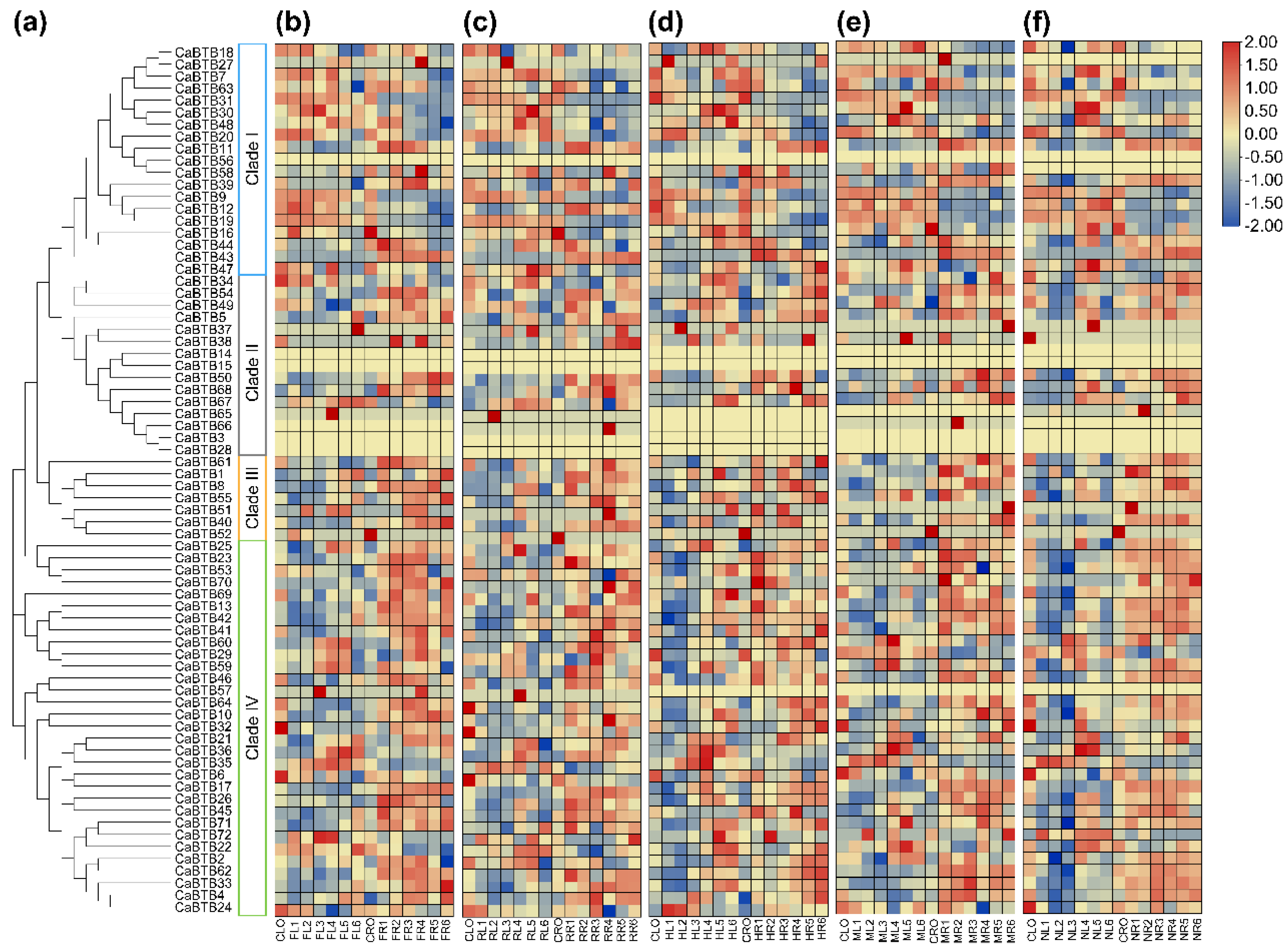
2.6. Expression Pattern Analysis of CaBTB Genes in Various Tissues
2.7. Expression Profiles of CaBTB Genes in Response to Exogenous Hormones
2.8. Expression Profiles of CaBTB Genes Under Multiple Abiotic Stresses
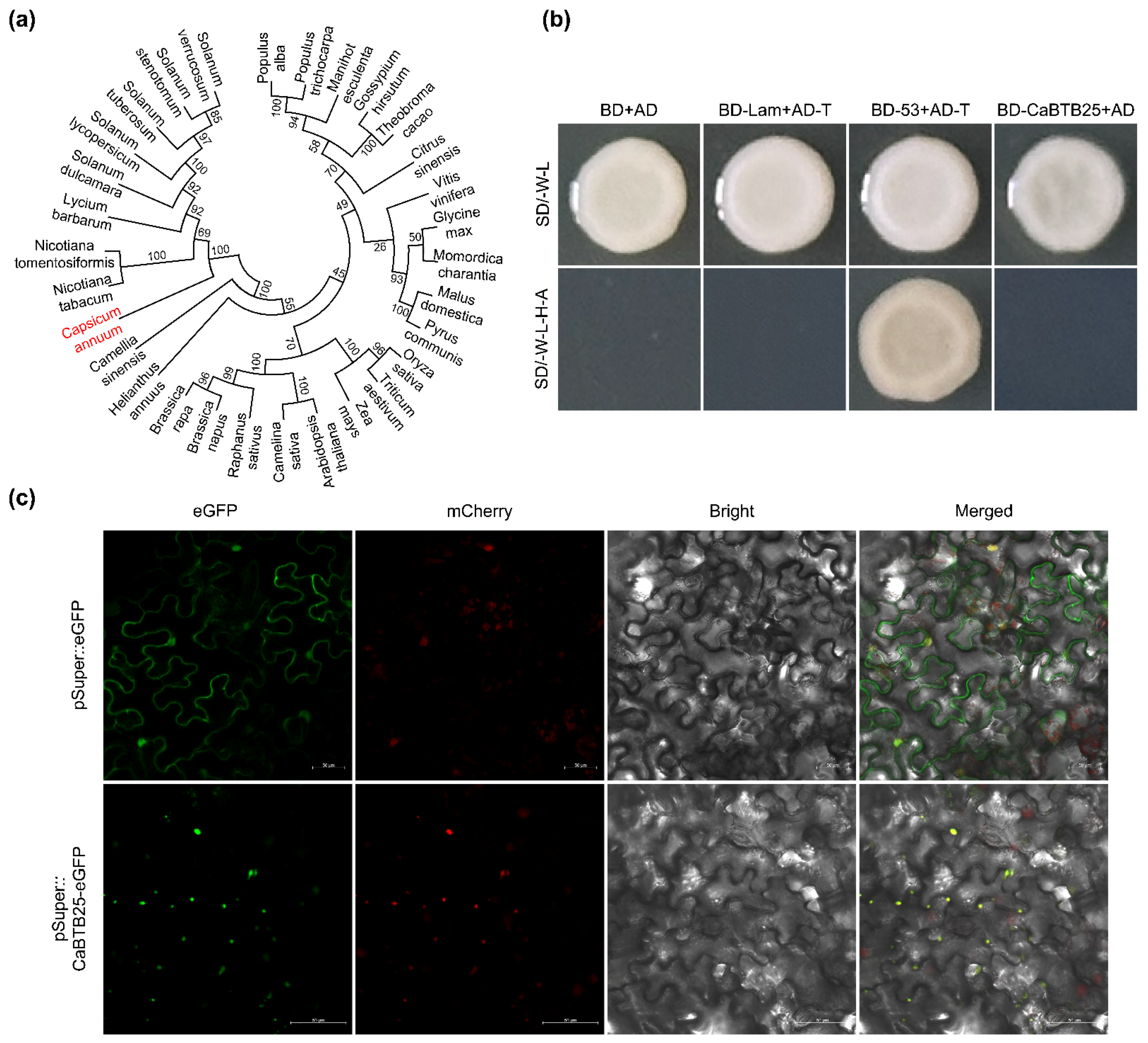
2.9. Homology, Transcription Activation, and Subcellular Localization of CaBTB25
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Identification of Pepper Genes Encoding BTB Domain Proteins
4.3. Phylogenetic Analysis of the BTB Proteins in Pepper
4.4. Gene Structure and Significant Motifs Analysis
4.5. Chromosomal Distribution and Detection of Gene Duplication
4.6. Prediction of the Cis-Acting Elements in the CaBTB Promoters
4.7. Expression Pattern Analysis of Pepper CaBTB Genes
4.8. RNA Isolation and Reverse Transcription Quantitative PCR (RT-qPCR)
4.9. Subcellular Localization
4.10. Transcriptional Activation Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Correction Statement
References
- Chaharbakhshi, E.; Jemc, J.C. Broad-complex, tramtrack, and bric-a-brac (BTB) proteins: Critical regulators of development. Genesis 2016, 54, 505–518. [Google Scholar] [CrossRef] [PubMed]
- Stogios, P.; Downs, G.; Jauhal, J.; Nandra, S.; Privé, G. Sequence and structural analysis of BTB domain proteins. Genome Biol. 2005, 6, R82. [Google Scholar] [CrossRef]
- Weber, H.; Bernhardt, A.; Dieterle, M.; Hano, P.; Mutlu, A.; Estelle, M.; Genschik, P.; Hanjo Hellmann, H. Arabidopsis AtCUL3a and AtCUL3b form complexes with members of the BTB/POZ-MATH protein family. Plant Physiol. 2005, 137, 83–93. [Google Scholar] [CrossRef] [PubMed]
- Cheng, D.; Qian, W.; Meng, M.; Wang, Y.; Peng, J.; Xia, Q. Identification and expression profiling of the BTB domain-containing protein gene family in the silkworm, Bombyx mori. Int. J. Genom. 2014, 2014, 865065. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.F.; Zhang, K.; Li, Y.Q.; Zhou, F.; Bai, H.; Zang, J.P.; Cao, H.Z.; Xing, J.H.; Dong, J.G. Identification and expression analysis of BTB family genes in Zea mays. Acta Phytopathol. Sin. 2021, 51, 268–281. [Google Scholar]
- Cheng, Y.F.; Qin, G.J.; Dai, X.H.; Zhao, Y.D. NPY1, a BTB-NPH3-like protein, plays a critical role in auxin-regulated organogenesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 18825–18829. [Google Scholar] [CrossRef]
- Shalman, A.; Huang, Y.B.; Chen, Y.B.; Muhammad, I.; Li, B.B.; Ullah, U.; Jing, X.Q.; Bhanbhro, N.; Liu, W.T.; Li, W.Q.; et al. The highly interactive BTB domain targeting other functional domains to diversify the function of BTB proteins in rice growth and development. Int. J. Biol. Macromol. 2021, 192, 1311–1324. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, Q.; Wang, X.D.; Zhao, C.L.; Wang, X.; Li, Y.L.; Jia, H.L.; Ding, G.Z.; Chen, L. Mining and bioinformatics analysis of BTB protein family in Sugar beet. Sugar Crops China 2021, 43, 9–16. [Google Scholar]
- Gu, S.Y.; Lo, W.S.; Wu, S.J.; Wang, L.C. Dimerization of the ETO1 family proteins plays a crucial role in regulating ethylene biosynthesis in Arabidopsis thaliana. Plant J. 2021, 105, 1293–1308. [Google Scholar] [CrossRef]
- Mosavi, L.; Cammett, T.; Desrosiers, D.; Peng, Z.Y. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 2004, 13, 1435–1448. [Google Scholar] [CrossRef]
- Bomont, P.; Cavalier, L.; Blondeau, F.; Ben, H.C.; Belal, S.; Tazir, M.; Demir, E.; Topaloglu, H.; Korinthenberg, R.; Tüysüz, B.; et al. The gene encoding gigaxonin, a new member of the cytoskeletal BTB/kelch repeat family, is mutated in giant axonal neuropathy. Nat. Genet. 2000, 26, 370–374. [Google Scholar] [PubMed]
- Collins, T.; Stone, J.R.; Williams, A.J. All in the family: The BTB/POZ, KRAB, and SCAN domains. Mol. Cell. Biol. 2001, 21, 3609–3915. [Google Scholar] [PubMed]
- Ban, Z.; Estelle, M. CUL3 E3 ligases in plant development and environmental response. Nat. Plants 2021, 7, 6–16. [Google Scholar] [CrossRef] [PubMed]
- Ullah, U.; Mao, W.L.; Abbas, W.; Alharthi, B.; Bhanbhro, N.; Xiong, M.; Gul, N.; Shalmani, A. OsMBTB32, a MATH-BTB domain-containing protein that interacts with OsCUL1s to regulate salt tolerance in rice. Funct. Integr. Genom. 2023, 23, 139. [Google Scholar]
- Zhang, Q.Y.; Ma, C.N.; Gu, K.D.; Wang, J.H.; Yu, J.Q.; Liu, B.; Wang, Y.; He, J.X.; Hu, D.G.; Sun, Q. The BTB-BACK-TAZ domain protein MdBT2 reduces drought resistance by weakening the positive regulatory effect of MdHDZ27 on apple drought tolerance via ubiquitination. Plant J. 2024, 119, 283–299. [Google Scholar]
- Du, L.; Guan, Z.J.; Liu, Y.L.; Hu, D.G.; Gao, J.P.; Sun, C.H. Scaffold protein BTB/TAZ domain-containing genes (CmBTs) play a negative role in root development of chrysanthemum. Plant Sci. 2024, 341, 111997. [Google Scholar]
- Shalmani, A.; Ullah, U.; Tai, L.; Zhang, R.; Jing, X.Q.; Muhammad, I.; Bhanbhro, N.; Liu, W.T.; Li, W.Q.; Chen, K.M. OsBBX19-OsBTB97/OsBBX11 module regulates spikelet development and yield production in rice. Plant Sci. 2023, 334, 111779. [Google Scholar]
- Fincher, G.B. Molecular and cellular biology associated with endo sperm mobilization in germinating cereal grains. Annu. Rev. Plant Biol. 2003, 40, 305–346. [Google Scholar] [CrossRef]
- Woodger, F.J.; Jacobsen, J.V.; Gubler, F. GMPOZ, a BTB/POZ domain nuclear protein, is a regulator of hormone responsive gene expression in barley aleurone. Plant Cell Physiol. 2004, 45, 945–950. [Google Scholar]
- Kim, S.H.; Seo, D.H.; Chung, S.L.; Kim, S.W.; Lee, J.S.; Kim, W.T.; Lee, J.H. ABA-HYPERSENSITIVE BTB/POZ PROTEIN 1 functions as a negative regulator in ABA-mediated inhibition of germination in Arabidopsis. Plant Mol. Biol. 2016, 90, 303–315. [Google Scholar] [CrossRef]
- Zhou, Y.Y.; Zhai, H.; He, S.Z.; Zhu, H.; Gao, S.P.; Xing, S.H.; Wei, Z.H.; Zhao, N.; Liu, Q.C. The sweet potato BTB-TAZ protein gene, IbBT4, enhances drought tolerance in transgenic Arabidopsis. Front. Plant Sci. 2020, 11, 877. [Google Scholar] [CrossRef]
- Yue, J.; Dai, X.R.; Li, Q.Z.; Wei, M.K. Genome-wide characterization of the BTB gene family in poplar and expression analysis in response to hormones and biotic/abiotic stresses. Int. J. Mol. Sci. 2024, 25, 9048. [Google Scholar] [CrossRef] [PubMed]
- Elsanosi, H.A.; Zhang, J.H.; Mostafa, S.; Geng, X.Y.; Zhou, G.S.; Awdelseid, A.H.M.; Song, L. Genome-wide identification, structural and gene expression analysis of BTB gene family in soybean. BMC Plant Biol. 2024, 24, 663. [Google Scholar]
- He, Y.M.; Liu, K.K.; Zhang, H.X.; Cheng, G.X.; Ali, M.; Haq, S.U.; Wei, A.M.; Gong, Z.H. Contribution of CaBPM4, a BTB domain-containing gene, to the response of pepper to phytophthora capsici infection and abiotic stresses. Agronomy 2019, 9, 417. [Google Scholar] [CrossRef]
- Lu, M.C.; Ji, J.N.; Lv, Y.F.; Zhao, J.; Liu, Y.T.; Jiao, Q.; Liu, T.; Mou, Y.; You, Q.D.; Jiang, Z.Y. Bivalent inhibitors of the BTB E3 ligase KEAP1 enable instant NRF2 activation to suppress acute inflammatory response. Cell Chem. Biol. 2024, 31, 1188–1202. [Google Scholar]
- Li, P.F.; Liu, P.; Zang, D.S.; Li, C.C.; Wang, C.; Zhu, Y.Z.; Liu, M.Q.; Lu, L.L.; Wu, X.B.; Nie, H.T. Genome-wide identification and expression analysis of the BTB gene superfamily provides insight into sex determination and early gonadal development of alligator sinensis. Int. J. Mol. Sci. 2024, 25, 10771. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zhou, N.; Fu, S.N.; Wang, L.L.; Liu, Y.D.; Fu, C.F.; Xu, F.; Guo, W.Y.; Wu, Y.H.; Cheng, J.; et al. Endothelial cell-derived exosomes trigger a positive feedback loop in osteogenesis-angiogenesis coupling via up-regulating zinc finger and BTB domain containing 16 in bone marrow mesenchymal stem cell. J. Nanobiotechnol. 2024, 22, 721. [Google Scholar] [CrossRef]
- Gingerich, D.J.; Hanada, K.; Shiu, S.H.; Vierstra, R.D. Large-scale, lineage-specific expansion of a bric-a-brac/tramtrack/broad complex ubiquitin-ligase gene family in rice. Plant Cell 2007, 19, 2329–2348. [Google Scholar]
- Li, J.; Su, X.; Wang, Y.; Yang, W.; Pan, Y.; Su, C.; Zhang, X. Genome-wide identification and expression analysis of the BTB domain-containing protein gene family in tomato. Genes Genom. 2018, 40, 1–15. [Google Scholar]
- Goyal, N.; Bhuria, M.; Verma, D.; Garewal, N.; Singh, K. Genome-wide identification of BTB domain-containing gene family in grapevine (Vitis vinifera L.). Agriculture 2023, 13, 252. [Google Scholar] [CrossRef]
- Zhu, P.P.; Fan, Y.J.; Xu, P.L.; Fan, G.Q. Bioinformatic analysis of the BTB gene family in paulownia fortunei and functional characterization in response to abiotic and biotic stresses. Plants 2023, 12, 4144. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.; Lian, X.D.; Cheng, J.; Zeng, W.F.; Zheng, X.B.; Wang, W.; Ye, X.; Li, J.D.; Li, Z.Q.; Zhang, L.L.; et al. Genome-wide identification and transcriptome profiling reveal that E3 ubiquitin ligase genes relevant to ethylene, auxin and abscisic acid are differentially expressed in the fruits of melting flesh and stony hard peach varieties. BMC Genom. 2019, 20, 892. [Google Scholar]
- Weber, H.; Hellmann, H. Arabidopsis thaliana BTB/POZ-MATH proteins interact with members of the ERF/AP2 transcription factor family. FEBS J. 2009, 276, 6624–6635. [Google Scholar] [CrossRef]
- Jeffares, D.C.; Penkett, C.J.; Bähler, J. Rapidly regulated genes are intron poor. Trends Genet. 2008, 24, 375–378. [Google Scholar]
- Heyn, P.; Kalinka, A.T.; Tomancak, P.; Neugebauer, K.M. Introns and gene expression: Cellular constraints, transcriptional regulation, and evolutionary consequences. Bioessays 2015, 37, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Perez-Torrado, R.; Yamada, D.; Defossez, P.A. Born to bind: The BTB protein-protein interaction domain. Bioessays 2006, 28, 1194–1202. [Google Scholar]
- Teufel, A.I.; Johnson, M.M.; Laurent, J.M.; Kachroo, A.H.; Marcotte, E.M.; Wilke, C.O. The many nuanced evolutionary consequences of duplicated genes. Mol. Biol. Evol. 2019, 36, 304–314. [Google Scholar] [PubMed]
- Juranić, M.; Dresselhaus, T. Phylogenetic analysis of the expansion of the MATH-BTB gene family in the grasses. Plant Signal. Behav. 2014, 9, e28242. [Google Scholar]
- Villao-Uzho, L.; Chávez-Navarrete, T.; Pacheco-Coello, R.; Eduardo Sánchez-Timm, E.; Santos-Ordóñez, E. Plant promoters: Their identification, characterization, and role in gene regulation. Genes 2023, 14, 1226. [Google Scholar] [CrossRef]
- Nanjareddy, K.; Arthikala, M.K.; Aguirre, A.L.; Gómez, B.M.; Lara, M. Plant promoter analysis: Identification and characterization of root nodule specific promoter in the common bean. J. Vis. Exp. 2017, 130, 56140. [Google Scholar]
- Kang, H.; Zhang, T.T.; Li, Y.Y.; Wang, K.L.; Espley, R.V.; Du, Y.P.; Guan, Q.M.; Ma, F.W.; Hao, Y.J.; You, C.X.; et al. The apple BTB protein MdBT2 positively regulates MdCOP1 abundance to repress anthocyanin biosynthesis. Plant Physiol. 2022, 190, 305–318. [Google Scholar] [PubMed]
- Xu, G.; Park, S.J.; Eck, J.V.; Lippman, Z.B. Control of inflorescence architecture in tomato by BTB/POZ transcriptional regulators. Genes Dev. 2016, 30, 2048–2061. [Google Scholar] [PubMed]
- An, J.P.; Wang, X.F.; Zhang, X.W.; You, C.X.; Yu-Jin Hao, Y.J. Apple BT2 protein negatively regulates jasmonic acid-triggered leaf senescence by modulating the stability of MYC2 and JAZ2. Plant Cell Environ. 2021, 44, 216–233. [Google Scholar]
- Ding, Y.; Sun, T.J.; Ao, K.; Peng, Y.J.; Zhang, Y.X.; Li, X.; Zhang, Y.L. Opposite roles of salicylic acid receptors NPR1 and NPR3/NPR4 in transcriptional regulation of plant immunity. Cell 2018, 173, 1454–1467.e15. [Google Scholar] [CrossRef]
- Silva, K.J.P.; Brunings, A.; Peres, N.A.; Mou, Z.L.; Folta, K.M. The Arabidopsis NPR1 gene confers broad-spectrum disease resistance in strawberry. Transgenic Res. 2015, 24, 693–704. [Google Scholar]
- Saputro, T.B.; Jakada, B.H.; Chutimanukul, P.; Comai, L.; Buaboocha, T.; Chadchawan, S. OsBTBZ1 confers salt stress tolerance in Arabidopsis thaliana. Int. J. Mol. Sci. 2023, 24, 14483. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.T.; Lin, Y.; Lin, Y.; Luo, J.Z.; Ding, J.H. Regeneration and Agrobacterium-mediated genetic transformation of twelve Eucalyptus species. For. Res. 2022, 2, 15. [Google Scholar]
- Xu, H.; Liu, Q.; Yao, T.; Fu, X.D. Shedding light on integrative GA signaling. Curr. Opin. Plant Biol. 2014, 21, 89–95. [Google Scholar] [PubMed]
- Davière, J.M.; Achard, P. A pivotal role of DELLAs in regulating multiple hormone signals. Mol. Plant 2016, 9, 10–20. [Google Scholar]
- Ren, Y.R.; Zhao, Q.; Yang, Y.Y.; Zhang, R.; Wang, X.F.; Zhang, T.E.; You, C.X.L.; Huo, H.Q.; Hao, Y.J. Interaction of BTB-TAZ protein MdBT2 and DELLA protein MdRGL3a regulates nitrate-mediated plant growth. Plant Physiol. 2021, 6, 750–766. [Google Scholar]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Qin, C.; Yu, C.S.; Shen, Y.; Fang, X.D.; Chen, L.; Min, J.M.; Cheng, J.W.; Zhao, S.C.; Xu, M.; Luo, Y.; et al. Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc. Natl. Acad. Sci. USA 2014, 111, 5135–5140. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Yu, H.Y.; Deng, Y.T.; Zheng, J.Y.; Liu, M.L.; Ou, L.J.; Yang, B.Z.; Dai, X.Z.; Ma, Y.Q.; Feng, S.Y.; et al. PepperHub, an informatics hub for the chili pepper research community. Mol. Plant 2017, 10, 1129–1132. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, Q.; Wang, J.; Liu, F.; Dai, X.; Zhu, F.; Zou, X.; Xiong, C. Genome-Wide Identification of the BTB Domain-Containing Protein Gene Family in Pepper (Capsicum annuum L.). Int. J. Mol. Sci. 2025, 26, 3429. https://doi.org/10.3390/ijms26073429
Yuan Q, Wang J, Liu F, Dai X, Zhu F, Zou X, Xiong C. Genome-Wide Identification of the BTB Domain-Containing Protein Gene Family in Pepper (Capsicum annuum L.). International Journal of Molecular Sciences. 2025; 26(7):3429. https://doi.org/10.3390/ijms26073429
Chicago/Turabian StyleYuan, Qiaoling, Jin Wang, Feng Liu, Xiongze Dai, Fan Zhu, Xuexiao Zou, and Cheng Xiong. 2025. "Genome-Wide Identification of the BTB Domain-Containing Protein Gene Family in Pepper (Capsicum annuum L.)" International Journal of Molecular Sciences 26, no. 7: 3429. https://doi.org/10.3390/ijms26073429
APA StyleYuan, Q., Wang, J., Liu, F., Dai, X., Zhu, F., Zou, X., & Xiong, C. (2025). Genome-Wide Identification of the BTB Domain-Containing Protein Gene Family in Pepper (Capsicum annuum L.). International Journal of Molecular Sciences, 26(7), 3429. https://doi.org/10.3390/ijms26073429




