Transcriptomic Analysis of Muscle Satellite Cell Regulation on Intramuscular Preadipocyte Differentiation in Tan Sheep
Abstract
1. Introduction
2. Results
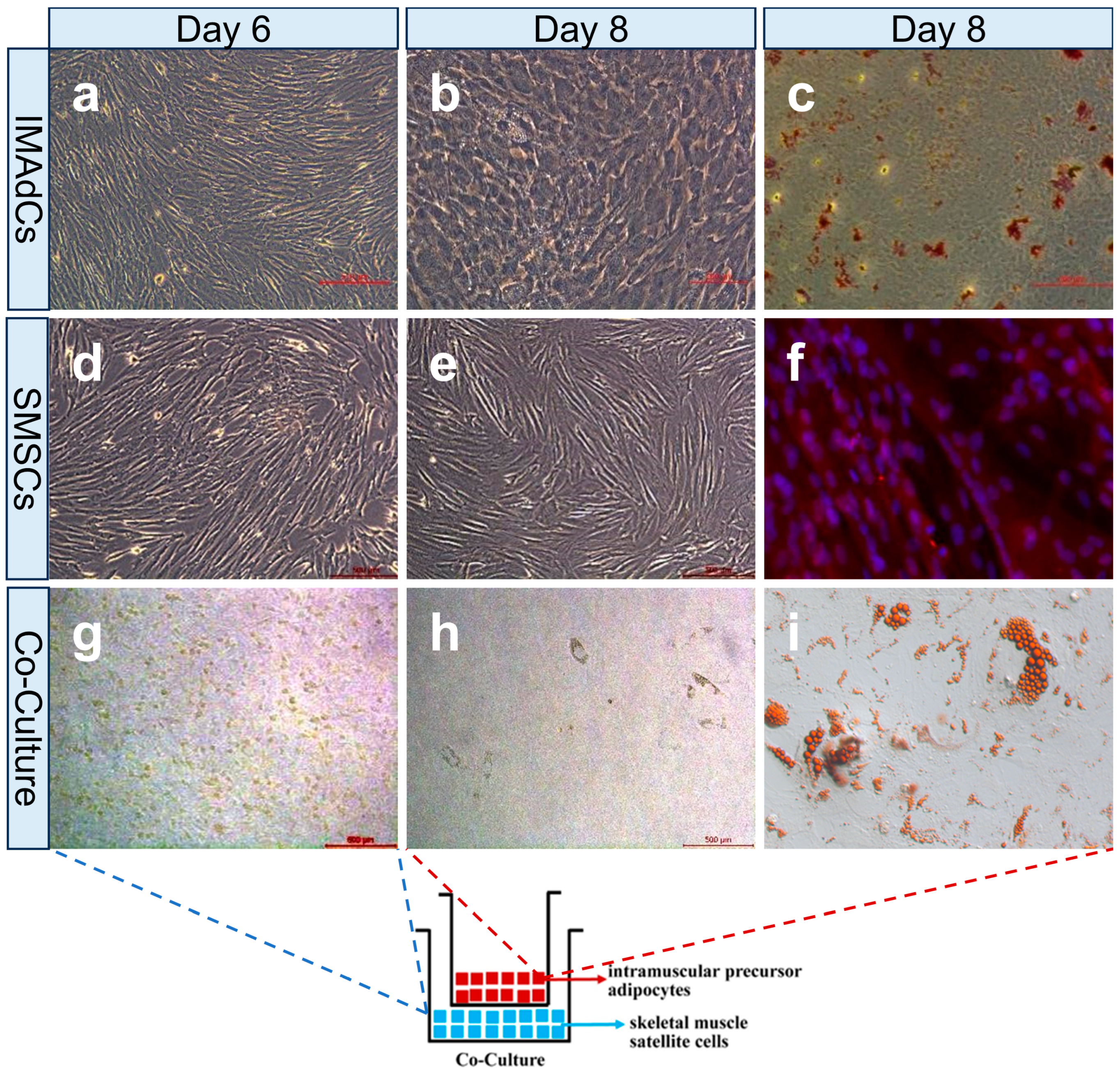
2.1. Establishment of a Co-Culture System with Intramuscular Preadipocytes and Skeletal Muscle Satellite Cells from Tan Sheep
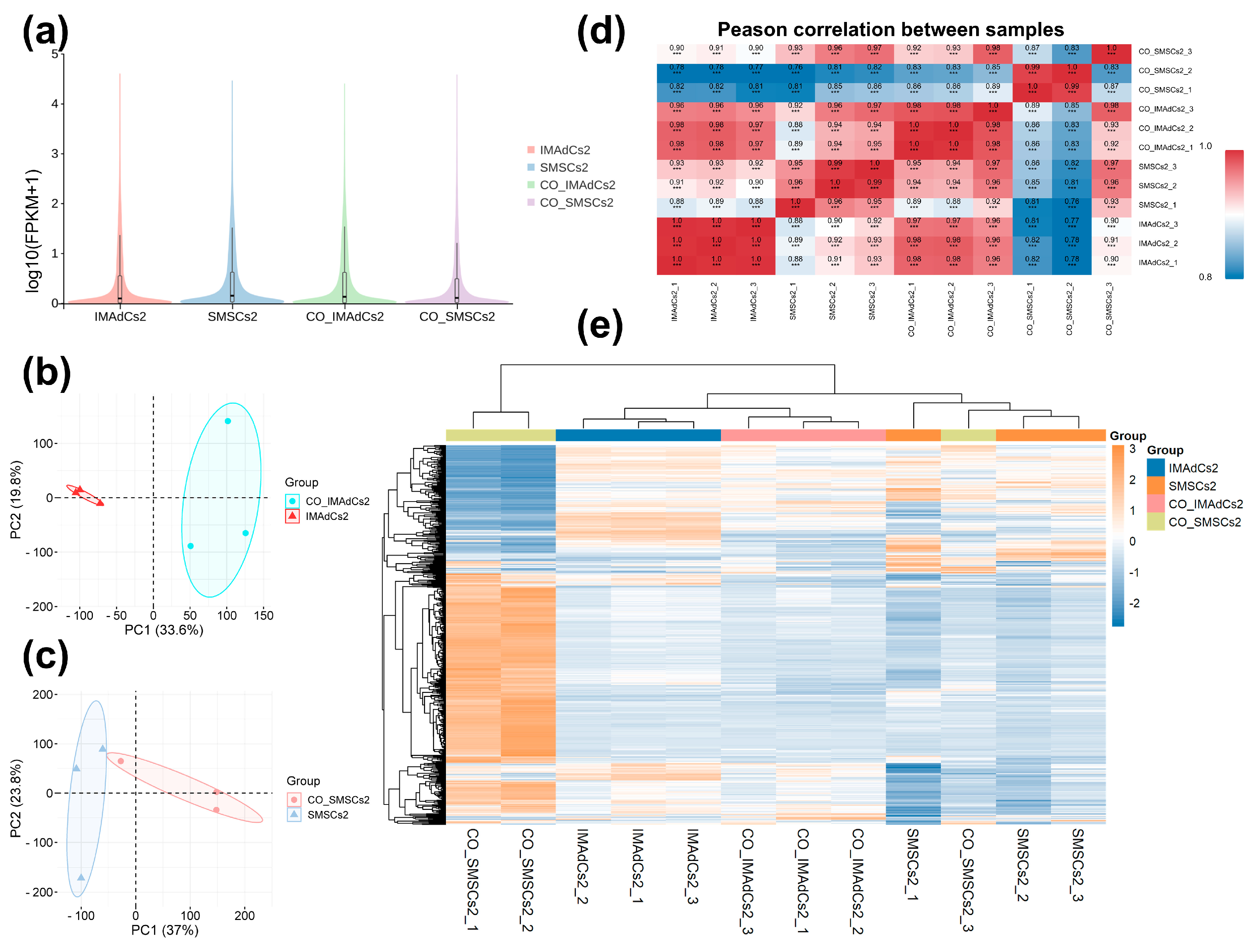
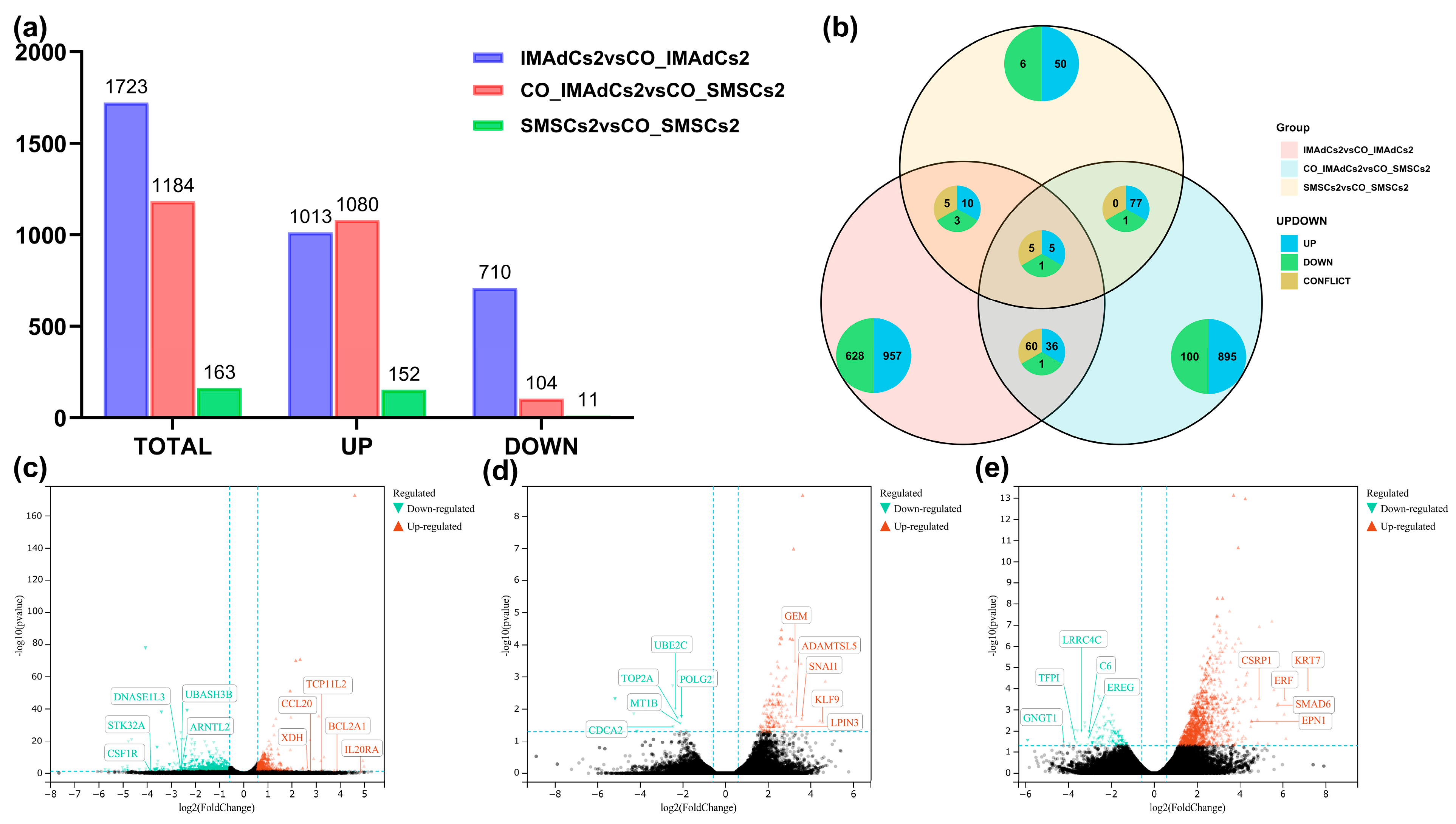
2.2. Quantitative Analysis of the Sequencing Data of Tan Sheep and Identification of Differentially Expressed Genes (DEGs)
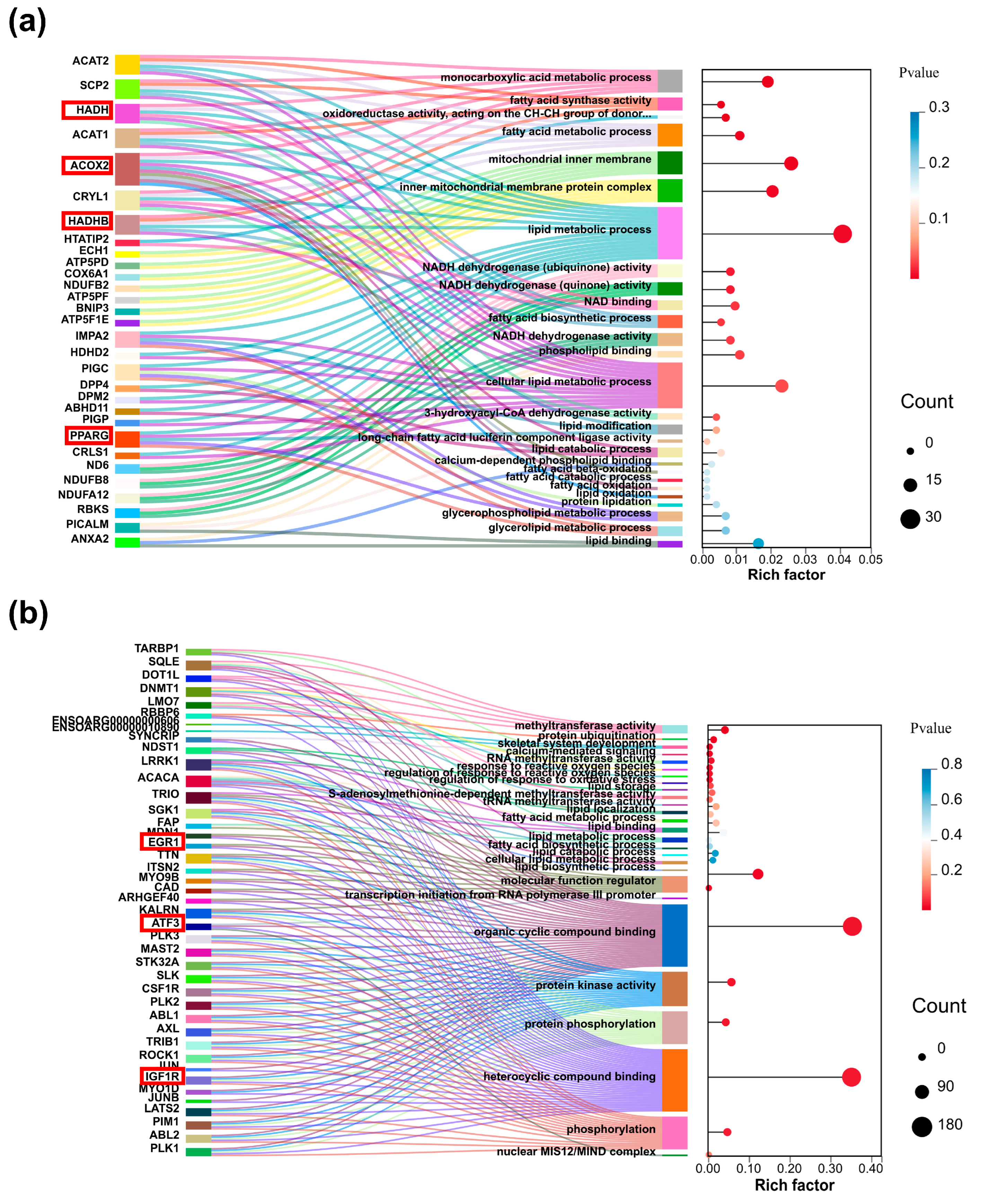
2.3. Functional Enrichment Analysis of Differentially Expressed Genes
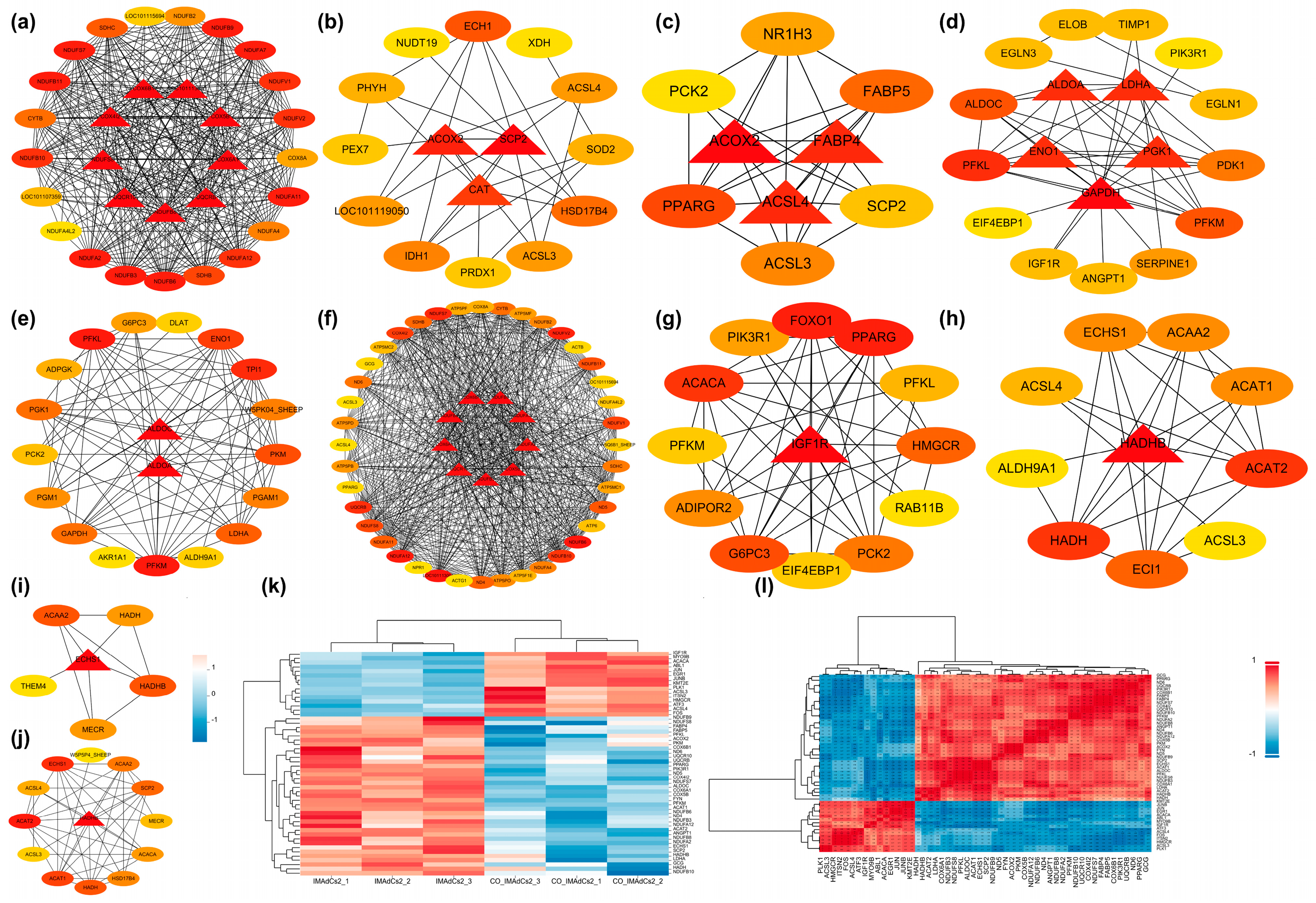
2.4. Protein–Protein Interaction (PPI) Network and Module Analysis
2.5. Identification of Ligands and Receptors Affected by Co-Culture and Construction of Ligand–Receptor Pairs Mediating Autocrine and Paracrine Crosstalk in Homologous and Heterologous Cell Types
2.6. qRT-PCR to Verify the Differentially Expressed Genes
3. Discussion
4. Materials and Methods
4.1. Culture and Differentiation Identification of Intramuscular Preadipocytes and Skeletal Muscle Satellite Cells
4.2. Co-Culture of Intramuscular Preadipocytes and Skeletal Muscle Satellite Cells
4.3. Cell Total RNA Extraction and Detection
4.4. Library Construction and High-Throughput Sequencing
4.5. Sequencing Data Processing
4.6. Quantitative Analysis
4.7. Screening of Differentially Expressed Genes
4.8. GO Function and KEGG Pathway Enrichment Analysis of Differentially Expressed Genes
4.9. mRNA-Protein Network Interaction Analysis in Adipose Tissue
4.10. Identification of Ligands and Receptors Affected by Co-Culture and Construction of Ligand–Receptor Pairs Mediating Autocrine and Paracrine Crosstalk in Homologous and Heterologous Cell Types
4.11. qRT-PCR Verification
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Realini, C.E.; Pavan, E.; Johnson, P.L.; Font-i-Furnols, M.; Jacob, N.; Agnew, M.; Craigie, C.R.; Moon, C.D. Consumer liking of M. longissimus lumborum from New Zealand pasture-finished lamb is influenced by intramuscular fat. Meat Sci. 2021, 173, 108380. [Google Scholar] [CrossRef]
- Jung, U.; Kim, M.; Wang, T.; Lee, J.-S.; Seo, S.; Lee, H.-G. Identification of candidate proteins regulated by long-term caloric restriction and feed efficiency in longissimus dorsi muscle in Korean native steer. J. Anim. Sci. Technol. 2022, 64, 330. [Google Scholar] [CrossRef] [PubMed]
- Yi, W.; Huang, Q.; Wang, Y.; Shan, T. Lipo-nutritional quality of pork: The lipid composition, regulation, and molecular mechanisms of fatty acid deposition. Anim. Nutr. 2023, 13, 373–385. [Google Scholar] [CrossRef]
- Ran, H.; He, Q.; Han, Y.; Wang, J.; Wang, H.; Yue, B.; Zhang, M.; Chai, Z.; Cai, X.; Zhong, J.; et al. Functional study and epigenetic targets analyses of SIRT1 in intramuscular preadipocytes via ChIP-seq and mRNA-seq. Epigenetics 2023, 18, 2135194. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.H.; Chung, K.Y.; Johnson, B.J.; Go, G.W.; Kim, K.H.; Choi, C.W.; Smith, S.B. Co-culture of bovine muscle satellite cells with preadipocytes increases PPARgamma and C/EBPbeta gene expression in differentiated myoblasts and increases GPR43 gene expression in adipocytes. J. Nutr. Biochem. 2013, 24, 539–543. [Google Scholar] [CrossRef] [PubMed]
- Uezumi, A.; Fukada, S.-I.; Yamamoto, N.; Takeda, S.I.; Tsuchida, K.J. Mesenchymal progenitors distinct from satellite cells contribute to ectopic fat cell formation in skeletal muscle. Nat. Cell Biol. 2010, 12, 143–152. [Google Scholar] [CrossRef]
- Li, Y.; Li, F.; Lin, B.; Kong, X.; Tang, Y.; Yin, Y. Myokine IL-15 regulates the crosstalk of co-cultured porcine skeletal muscle satellite cells and preadipocytes. Mol. Biol. Rep. 2014, 41, 7543–7553. [Google Scholar] [CrossRef]
- Li, F.-F.; Zhang, H.; Li, J.-J.; Cao, Y.-N.; Dong, X.; Gao, C. Interaction with adipocytes induces lung adenocarcinoma A549 cell migration and tumor growth. Mol. Med. Rep. 2018, 18, 1973–1980. [Google Scholar] [CrossRef]
- Vaughn, M.A.; Lancaster, P.A.; Roden, K.C.; Sharman, E.D.; Krehbiel, C.R.; Horn, G.W.; Starkey, J.D. Effect of stocker management program on beef cattle skeletal muscle growth characteristics, satellite cell activity, and paracrine signaling impact on preadipocyte differentiation. J. Anim. Sci. Technol. 2019, 61, 260. [Google Scholar] [CrossRef]
- Xu, X.; Zhao, R.; Ma, W.; Zhao, Q.; Zhang, G. Comparison of lipid deposition of intramuscular preadipocytes in Tan sheep co-cultured with satellite cells or alone. J. Anim. Physiol. Anim. Nutr. 2022, 106, 733–741. [Google Scholar] [CrossRef]
- Liu, K.; Zhang, X.; Wei, W.; Liu, X.; Tian, Y.; Han, H.; Zhang, L.; Wu, W.; Chen, J. Myostatin/SMAD4 signaling-mediated regulation of miR-124-3p represses glucocorticoid receptor expression and inhibits adipocyte differentiation. Am. J. Physiol. Endocrinol. Metab. 2019, 316, E635–E645. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, C.; Kong, Y.; Li, F.; Yue, X. Effects of intramuscular fat on meat quality and its regulation mechanism in Tan sheep. Front. Nutr. 2022, 9, 908355. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, H.; Liu, M.; Zhao, X.; Luo, H. Effect of Breed on the Volatile Compound Precursors and Odor Profile Attributes of Lamb Meat. Foods 2020, 9, 1178. [Google Scholar] [CrossRef] [PubMed]
- Jia, W.; Wu, X.; Li, R.; Liu, S.; Shi, L. Effect of nisin and potassium sorbate additions on lipids and nutritional quality of Tan sheep meat. Food Chem. 2021, 365, 130535. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genom. Biol. 2014, 15, 550. [Google Scholar]
- Hwang, Y.-H.; Lee, S.-J.; Lee, E.-Y.; Joo, S.-T. Effects of carcass weight increase on meat quality and sensory properties of pork loin. J. Anim. Sci. Technol. 2020, 62, 753. [Google Scholar] [CrossRef] [PubMed]
- Blanchard, P.J.; Willis, M.B.; Warkup, C.C.; Ellis, M. The influence of carcass backfat and intramuscular fat level on pork eating quality. J. Sci. Food Agric. 2000, 80, 145–151. [Google Scholar]
- Wang, Y.; Ma, C.; Sun, Y.; Li, Y.; Kang, L.; Jiang, Y. Dynamic transcriptome and DNA methylome analyses on longissimus dorsi to identify genes underlying intramuscular fat content in pigs. BMC Genom. 2017, 18, 780. [Google Scholar] [CrossRef]
- Cheng, F.; Liang, J.; Yang, L.; Lan, G.; Wang, L.; Wang, L. Systematic Identification and Comparison of the Expressed Profiles of lncRNAs, miRNAs, circRNAs, and mRNAs with Associated Co-Expression Networks in Pigs with Low and High Intramuscular Fat. Animals 2021, 11, 3212. [Google Scholar] [CrossRef]
- Santos, G.M.; Neves, F.d.A.R.; Amato, A.A. Thermogenesis in white adipose tissue: An unfinished story about PPARγ. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2015, 1850, 691–695. [Google Scholar]
- Gill, J.; La Merrill, M.A. An emerging role for epigenetic regulation of Pgc-1α expression in environmentally stimulated brown adipose thermogenesis. Environ. Epigenet. 2017, 3, dvx009. [Google Scholar] [CrossRef]
- Konno, T.; Sasaki, K.; Kobayashi, K.; Murata, T.J. Indirubin promotes adipocyte differentiation and reduces lipid accumulation in 3T3-L1 cells via peroxisome proliferator-activated receptor γ activation. Mol. Med. Rep. 2020, 21, 1552–1560. [Google Scholar] [PubMed]
- Huang, C.-J.; Choo, K.B.; Chen, C.-F. The MicroRNA-signaling-peroxisome proliferator-activated receptor gamma connection in the modulation of adipogenesis: Bioinformatics projection on chicken. Poult. Sci. 2022, 101, 101950. [Google Scholar] [PubMed]
- Nishida, N.; Yada, N.; Hagiwara, S.; Sakurai, T.; Kitano, M.; Kudo, M. Unique features associated with hepatic oxidative DNA damage and DNA methylation in non-alcoholic fatty liver disease. J. Gastroenterol. Hepatol. 2016, 31, 1646–1653. [Google Scholar] [PubMed]
- Liu, L.; Cui, H.; Fu, R.; Zheng, M.; Liu, R.; Zhao, G.; Wen, J. The regulation of IMF deposition in pectoralis major of fast-and slow-growing chickens at hatching. J. Anim. Sci. Biotechnol. 2017, 8, 77. [Google Scholar]
- Cursons, J.; Leuchowius, K.-J.; Waltham, M.; Tomaskovic-Crook, E.; Foroutan, M.; Bracken, C.P.; Redfern, A.; Crampin, E.J.; Street, I.; Davis, M.J. Stimulus-dependent differences in signalling regulate epithelial-mesenchymal plasticity and change the effects of drugs in breast cancer cell lines. Cell Commun. Signal. 2015, 13, 26. [Google Scholar]
- Chen, M.; Wang, J.; Wang, Y.; Wu, Y.; Fu, J.; Liu, J.-F. Genome-wide detection of selection signatures in Chinese indigenous Laiwu pigs revealed candidate genes regulating fat deposition in muscle. BMC Genet. 2018, 19, 31. [Google Scholar]
- Seo, J.; Jeong, D.-W.; Park, J.-W.; Lee, K.-W.; Fukuda, J.; Chun, Y.-S. Fatty-acid-induced FABP5/HIF-1 reprograms lipid metabolism and enhances the proliferation of liver cancer cells. Commun. Biol. 2020, 3, 638. [Google Scholar]
- Zhang, C.; Liao, Y.; Liu, P.; Du, Q.; Liang, Y.; Ooi, S.; Qin, S.; He, S.; Yao, S.; Wang, W. FABP5 promotes lymph node metastasis in cervical cancer by reprogramming fatty acid metabolism. Theranostics 2020, 10, 6561. [Google Scholar]
- Xu, B.; Chen, L.; Zhan, Y.; Marquez, K.N.S.; Zhuo, L.; Qi, S.; Zhu, J.; He, Y.; Chen, X.; Zhang, H. The biological functions and regulatory mechanisms of fatty acid binding protein 5 in various diseases. Front. Cell Dev. Biol. 2022, 10, 857919. [Google Scholar]
- Hsiao, Y.-H.; Chen, N.-C.; Koh, Y.-C.; Nagabhushanam, K.; Ho, C.-T.; Pan, M.-H. Pterostilbene inhibits adipocyte conditioned-medium-induced colorectal cancer cell migration through targeting FABP5-related signaling pathway. J. Agric. Food Chem. 2019, 67, 10321–10329. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Ren, X.; Han, P.; Hu, S.; Wang, J.; Yin, J. SiRNA against Fabp5 induces 3T3-L1 cells apoptosis during adipocytic induction. Mol. Biol. Rep. 2010, 37, 4003–4011. [Google Scholar] [CrossRef]
- Yamamoto, T.; Furuhashi, M.; Sugaya, T.; Oikawa, T.; Matsumoto, M.; Funahashi, Y.; Matsukawa, Y.; Gotoh, M.; Miura, T. Transcriptome and metabolome analyses in exogenous FABP4-and FABP5-treated adipose-derived stem cells. PLoS ONE 2016, 11, e0167825. [Google Scholar] [CrossRef] [PubMed]
- Yamada, E.; Pessin, J.E.; Kurland, I.J.; Schwartz, G.J.; Bastie, C.C. Fyn-dependent regulation of energy expenditure and body weight is mediated by tyrosine phosphorylation of LKB1. Cell Metab. 2010, 11, 113–124. [Google Scholar] [CrossRef]
- Lee, T.-W.A.; Kwon, H.; Zong, H.; Yamada, E.; Vatish, M.; Pessin, J.E.; Bastie, C.C. Fyn deficiency promotes a preferential increase in subcutaneous adipose tissue mass and decreased visceral adipose tissue inflammation. Diabetes 2013, 62, 1537–1546. [Google Scholar] [CrossRef]
- Tarabra, E.; Lee, T.-W.A.; Zammit, V.A.; Vatish, M.; Yamada, E.; Pessin, J.E.; Bastie, C.C. Differential activation of Fyn kinase distinguishes saturated and unsaturated fats in mouse macrophages. Oncotarget 2017, 8, 86634. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ma, Z.; Wang, Y.; Shen, J.; Yan, W.; Cao, C.; Feng, M.; Zhu, J.; Pang, W. Overexpression of ALDOC Promotes Porcine Intramuscular and Intermuscular Fat Deposition by Activating the AKT-mTORC1 Signaling Pathway. J. Agric. Food Chem. 2024, 72, 23893–23907. [Google Scholar] [CrossRef]
- Votava, J.A.; John, S.V.; Li, Z.; Chen, S.; Fan, J.; Parks, B.W. Mining cholesterol genes from thousands of mouse livers identifies aldolase C as a regulator of cholesterol biosynthesis. J. Lipid Res. 2024, 65, 100525. [Google Scholar] [CrossRef]
- Zhou, M.; Sun, X.; Wang, C.; Wang, F.; Fang, C.; Hu, Z. PFKM inhibits doxorubicin-induced cardiotoxicity by enhancing oxidative phosphorylation and glycolysis. Sci. Rep. 2022, 12, 11684. [Google Scholar] [CrossRef]
- Meng, Y.; Guo, D.; Lin, L.; Zhao, H.; Xu, W.; Luo, S.; Jiang, X.; Li, S.; He, X.; Zhu, R.; et al. Glycolytic enzyme PFKL governs lipolysis by promoting lipid droplet-mitochondria tethering to enhance beta-oxidation and tumor cell proliferation. Nat. Metab. 2024, 6, 1092–1107. [Google Scholar] [CrossRef]
- Pan, M.; Luo, M.; Liu, L.; Chen, Y.; Cheng, Z.; Wang, K.; Huang, L.; Tang, N.; Qiu, J.; Huang, A.; et al. EGR1 suppresses HCC growth and aerobic glycolysis by transcriptionally downregulating PFKL. J. Exp. Clin. Cancer Res. 2024, 43, 35. [Google Scholar] [CrossRef] [PubMed]
- Sekine, Y.; Yamamoto, K.; Kurata, M.; Honda, A.; Onishi, I.; Kinowaki, Y.; Kawade, G.; Watabe, S.; Nomura, S.; Fukuda, S.; et al. HADHB, a fatty acid beta-oxidation enzyme, is a potential prognostic predictor in malignant lymphoma. Pathology 2022, 54, 286–293. [Google Scholar] [CrossRef]
- Aredes, D.S.; De Paula, I.F.; Santos-Araujo, S.; Gondim, K.C. Silencing of Mitochondrial Trifunctional Protein A Subunit (HADHA) Increases Lipid Stores, and Reduces Oviposition and Flight Capacity in the Vector Insect Rhodnius prolixus. Front. Insect Sci. 2022, 2, 885172. [Google Scholar] [CrossRef]
- Fu, X.; Zheng, F.; Zhang, Y.; Bao, X.; Wang, S.; Yang, Y.; Xiong, H.J. Mitochondrial trifunctional protein deficiency due to HADHB gene mutation in a Chinese family. Mol. Genet. Metab. Rep. 2015, 5, 80–84. [Google Scholar] [CrossRef]
- Yi, X.; Feng, M.; Zhu, J.; Yu, H.; He, Z.; Zhang, Z.; Zhao, T.; Zhang, Q.; Pang, W.J. Adipocyte Progenitor Pools Composition and Cellular Niches Affect Adipogenesis Divergence in Porcine Subcutaneous and Intramuscular Fat. J. Agric. Food Chem. 2024, 72, 14044–14056. [Google Scholar]
- Yu, W.; Yao, Y.; Ye, N.; Zhao, Y.; Ye, Z.; Wei, W.; Zhang, L.; Chen, J. The myokine CCL5 recruits subcutaneous preadipocytes and promotes intramuscular fat deposition in obese mice. Am. J. Physiol. Physiol. 2024, 326, C1320–C1333. [Google Scholar]
- Lin, A.; He, W. LINC01705 derived from adipocyte exosomes regulates hepatocyte lipid accumulation via an miR-552-3p/LXR axis. J. Diabetes Investig. 2023, 14, 1160–1171. [Google Scholar] [CrossRef]
- Ambrosio, M.R.; Adriaens, M.; Derks, K.; Migliaccio, T.; Costa, V.; Liguoro, D.; Cataldi, S.; D’Esposito, V.; Maneli, G.; Bassolino, R.; et al. Glucose impacts onto the reciprocal reprogramming between mammary adipocytes and cancer cells. Sci. Rep. 2024, 14, 24674. [Google Scholar] [CrossRef]
- Guo, L.; Cui, H.; Zhao, G.; Liu, R.; Li, Q.; Zheng, M.; Guo, Y.; Wen, J. Intramuscular preadipocytes impede differentiation and promote lipid deposition of muscle satellite cells in chickens. BMC Genom. 2018, 19, 838. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, W.; Kong, P.; Feng, K.; Liu, C.; Sun, T.; Sang, Y.; Duan, X.; Tao, Z.; Liu, W. New insights into lipid metabolism and prostate cancer. Int. J. Oncol. 2023, 62, 74. [Google Scholar]
- Doll, S.; Proneth, B.; Tyurina, Y.Y.; Panzilius, E.; Kobayashi, S.; Ingold, I.; Irmler, M.; Beckers, J.; Aichler, M.; Walch, A.; et al. ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat. Chem. Biol. 2017, 13, 91–98. [Google Scholar]
- Zhang, H.; Sun, Q.; Dong, H.; Jin, Z.; Li, M.; Jin, S.; Zeng, X.; Fan, J.; Kong, Y. Long-chain acyl-CoA synthetase-4 regulates endometrial decidualization through a fatty acid β-oxidation pathway rather than lipid droplet accumulation. Mol. Metab. 2024, 84, 101953. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Li, X.; Ding, N.; Teng, J.; Zhang, S.; Zhang, Q.; Tang, H. miR-34a regulates adipogenesis in porcine intramuscular adipocytes by targeting ACSL4. BMC Genet. 2020, 21, 33. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.-F.; Ku, H.-C.; Cheng, J.-J.; Chao, S.-W.; Li, H.-F.; Lai, P.-F.; Chang, C.-C.; Don, M.-J.; Chen, H.-H.; Lin, H. Adipocyte browning and resistance to obesity in mice is induced by expression of ATF3. Commun. Biol. 2019, 2, 389. [Google Scholar]
- Jang, M.K.; Kim, C.H.; Seong, J.K.; Jung, M.H. ATF3 inhibits adipocyte differentiation of 3T3-L1 cells. Biochem. Biophys. Res. Commun. 2012, 421, 38–43. [Google Scholar]
- Ku, H.-C.; Chan, T.-Y.; Chung, J.-F.; Kao, Y.-H.; Cheng, C.-F. The ATF3 inducer protects against diet-induced obesity via suppressing adipocyte adipogenesis and promoting lipolysis and browning. Biomed. Pharmacother. 2022, 145, 112440. [Google Scholar] [CrossRef]
- Bléher, M.; Meshko, B.; Cacciapuoti, I.; Gergondey, R.; Kovacs, Y.; Duprez, D.; L’honoré, A.; Havis, E. Egr1 loss-of-function promotes beige adipocyte differentiation and activation specifically in inguinal subcutaneous white adipose tissue. Sci. Rep. 2020, 10, 15842. [Google Scholar]
- Boyle, K.B.; Hadaschik, D.; Virtue, S.; Cawthorn, W.P.; Ridley, S.H.; O’Rahilly, S.; Siddle, K. The transcription factors Egr1 and Egr2 have opposing influences on adipocyte differentiation. Cell Death Differ. 2009, 16, 782–789. [Google Scholar] [CrossRef]
- Milet, C.; Bléher, M.; Allbright, K.; Orgeur, M.; Coulpier, F.; Duprez, D.; Havis, E. Egr1 deficiency induces browning of inguinal subcutaneous white adipose tissue in mice. Sci. Rep. 2017, 7, 16153. [Google Scholar]
- Muñoz, M.; García-Casco, J.M.; Caraballo, C.; Fernández-Barroso, M.Á.; Sánchez-Esquiliche, F.; Gómez, F.; Rodríguez, M.d.C.; Silió, L. Identification of candidate genes and regulatory factors underlying intramuscular fat content through longissimus dorsi transcriptome analyses in heavy Iberian pigs. Front. Genet. 2018, 9, 608. [Google Scholar] [CrossRef]
- Han, H.; Wei, W.; Chu, W.; Liu, K.; Tian, Y.; Jiang, Z.; Chen, J. Muscle conditional medium reduces intramuscular adipocyte differentiation and lipid accumulation through regulating insulin signaling. Int. J. Mol. Sci. 2017, 18, 1799. [Google Scholar] [CrossRef]
- Ahmad, S.S.; Ahmad, K.; Lee, E.J.; Lee, Y.-H.; Choi, I. Implications of insulin-like growth factor-1 in skeletal muscle and various diseases. Cells 2020, 9, 1773. [Google Scholar] [CrossRef] [PubMed]
- Kloting, N.; Koch, L.; Wunderlich, T.; Kern, M.; Ruschke, K.; Krone, W.; Bruning, J.C.; Bluher, M. Autocrine IGF-1 action in adipocytes controls systemic IGF-1 concentrations and growth. Diabetes 2008, 57, 2074–2082. [Google Scholar] [PubMed]
- Zhang, L.H.; Zhuo, H.Q.; Hou, J.J.; Zhou, Y.; Cheng, J.; Cai, J.C. Proteomic signatures of infiltrative gastric cancer by proteomic and bioinformatic analysis. World J. Gastrointest. Oncol. 2022, 14, 2097–2107. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Kang, Y.; Wang, R.; Deng, J.; Yu, Y.; Yu, J.; Wang, J. Emerging Roles of NDUFS8 Located in Mitochondrial Complex I in Different Diseases. Molecules 2022, 27, 8754. [Google Scholar] [CrossRef]
- Han, J.; Ma, S.; Liang, B.; Bai, T.; Zhao, Y.; Ma, Y.; MacHugh, D.E.; Ma, L.; Jiang, L. Transcriptome Profiling of Developing Ovine Fat Tail Tissue Reveals an Important Role for MTFP1 in Regulation of Adipogenesis. Front. Cell Dev. Biol. 2022, 10, 839731. [Google Scholar] [CrossRef]
- Manning, B.D.; Dibble, C.C. Growth Signaling Networks Orchestrate Cancer Metabolic Networks. Cold Spring Harb. Perspect. Med. 2024, 14, a041543. [Google Scholar] [CrossRef]
- Kostenis, E.; Bravo, S.; Gomeza, J.J. Giving ERK a jERK from the endosome. Trends Pharmacol. Sci. 2023, 44, 131–133. [Google Scholar] [CrossRef]
- Stansbury, C.M.; Dotson, G.A.; Pugh, H.; Rehemtulla, A.; Rajapakse, I.; Muir, L.A. A lipid-associated macrophage lineage rewires the spatial landscape of adipose tissue in early obesity. J. Clin. Investig. 2023, 8, e171701. [Google Scholar]
- Zhu, Y.-Z.; Liu, J.-K.; Li, X.-E.; Yu, Z.-P.; Yang, L.-Q.; Wan, Q.; Zhao, Y.; Saeed, M.; Wu, A.-D.; Tian, X.-L. Genome-wide search links senescence-associated secretory proteins with susceptibility for coronary artery disease in mouse and human. J. Gerontol. Ser. A 2024, 79, glae070. [Google Scholar]
- Verkerke, A.R.; Kajimura, S. Oil does more than light the lamp: The multifaceted role of lipids in thermogenic fat. Dev. Cell 2021, 56, 1408–1416. [Google Scholar]
- Schmidt, M.; Morais, K.; de Almeida, M.; Iqbal, A.; Goldfeder, M.; Chudzinski-Tavassi, A. Amblyomin-X, a recombinant Kunitz-type inhibitor, regulates cell adhesion and migration of human tumor cells. Cell Adhes. Migr. 2018, 14, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Song, N.-J.; Lee, A.; Yasmeen, R.; Shen, Q.; Yang, K.; Kumar, S.B.; Muhanna, D.; Arnipalli, S.; Noria, S.F.; Needleman, B.J. Epiregulin as an alternative ligand for leptin receptor alleviates glucose intolerance without change in obesity. Cells 2022, 11, 425. [Google Scholar] [CrossRef]
- Guo, B.; Qu, X.; Chen, Z.; Yu, J.; Yan, L.; Zhu, H.J. Transcriptome analysis reveals transforming growth factor-β1 prevents extracellular matrix degradation and cell adhesion during the follicular-luteal transition in cows. J. Reprod. Dev. 2022, 68, 12–20. [Google Scholar]
- Chen, X.; Zhang, W.; Zhu, H.; Lin, F. Development and validation of a 5-gene autophagy-based prognostic index in endometrial carcinoma. Med. Sci. Monit. 2021, 27, e928949. [Google Scholar] [PubMed]
- Lucas, L.M.; Dwivedi, V.; Senfeld, J.I.; Cullum, R.L.; Mill, C.P.; Piazza, J.T.; Bryant, I.N.; Cook, L.J.; Miller, S.T.; Lott, J.H., IV. The yin and yang of ERBB4: Tumor suppressor and oncoprotein. Pharmacol. Rev. 2022, 74, 18–47. [Google Scholar] [PubMed]
- Byeon, S.-J.; Lee, H.S.; Kim, M.-A.; Lee, B.L.; Kim, W.H. Expression of the ERBB family of ligands and receptors in gastric cancer. Pathobiology 2017, 84, 210–217. [Google Scholar]
- Cushman, S.M.; Jiang, C.; Hatch, A.J.; Shterev, I.; Sibley, A.B.; Niedzwiecki, D.; Venook, A.P.; Owzar, K.; Hurwitz, H.I.; Nixon, A.B. Gene expression markers of efficacy and resistance to cetuximab treatment in metastatic colorectal cancer: Results from CALGB 80203 (Alliance). Clin. Cancer Res. 2015, 21, 1078–1086. [Google Scholar]
- Katoh, Y.; Katoh, M. Conserved POU-binding site linked to SP1-binding site within FZD5 promoter: Transcriptional mechanisms of FZD5 in undifferentiated human ES cells, fetal liver/spleen, adult colon, pancreatic islet, and diffuse-type gastric cancer. Int. J. Oncol. 2007, 30, 751–755. [Google Scholar]
- Katoh, M.; Katoh, M. Comparative integromics on non-canonical WNT or planar cell polarity signaling molecules: Transcriptional mechanism of PTK7 in colorectal cancer and that of SEMA6A in undifferentiated ES cells. Int. J. Mol. Med. 2007, 20, 405–409. [Google Scholar] [CrossRef]
- Katoh, M. WNT/PCP signaling pathway and human cancer. Oncol. Rep. 2005, 14, 1583–1588. [Google Scholar] [CrossRef]
- Martinez, S.; Scerbo, P.; Giordano, M.; Daulat, A.M.; Lhoumeau, A.-C.; Thomé, V.; Kodjabachian, L.; Borg, J.-P. The PTK7 and ROR2 protein receptors interact in the vertebrate WNT/planar cell polarity (PCP) pathway. J. Biol. Chem. 2015, 290, 30562–30572. [Google Scholar] [CrossRef] [PubMed]
- Raivola, J.; Dini, A.; Salokas, K.; Karvonen, H.; Niininen, W.; Piki, E.; Varjosalo, M.; Ungureanu, D. New insights into the molecular mechanisms of ROR1, ROR2, and PTK7 signaling from the proteomics and pharmacological modulation of ROR1 interactome. Cell. Mol. Life Sci. 2022, 79, 276. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Gingeras, T.R. Mapping RNA-seq reads with STAR. Curr. Protoc. Bioinform. 2015, 51, 11–14. [Google Scholar] [CrossRef]
- Shen, W.; Song, Z.; Zhong, X.; Huang, M.; Shen, D.; Gao, P.; Qian, X.; Wang, M.; He, X.; Wang, T.; et al. Sangerbox: A comprehensive, interaction-friendly clinical bioinformatics analysis platform. Imeta 2022, 1, e36. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L. ClusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Kawashima, M.; Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023, 51, D587–D592. [Google Scholar] [CrossRef]
- Demchak, B.; Hull, T.; Reich, M.; Liefeld, T.; Smoot, M.; Ideker, T.; Mesirov, J.P. Cytoscape: The network visualization tool for GenomeSpace workflows. F1000Research 2014, 3, 151. [Google Scholar] [CrossRef]


| Up/Down | Log2 FC | Primer Sequence (5′ → 3′) | |
|---|---|---|---|
| PPARγ | up | 1.0485 | F: TTCCGTTCCCAAGAGCTGAC |
| R: CGTGCACGCCGTATTTTAGG | |||
| FABP5 | up | 1.2384 | F: GGAAGATGGCGCTTAGTGGA |
| R: TGGCCATCGCACCTACTTTT | |||
| FYN | up | 0.9934 | F: AGACGTGGAAAAAGACCAGTCC |
| R: ACTTCCCCAAACTGCCCATT | |||
| EGR1 | down | −2.4717 | F: AGTGGAGACGAGTTACCCCA |
| R: CCGCTGACCAGACTGAAGAG | |||
| ATF3 | down | −3.0771 | F: CACAGTCGCACGCAGAGA |
| R: GCTCTGCAATGTTCCCTCCT | |||
| IGF1R | down | −0.4761 | F: ATCCGGATCGAGAAGAACGC |
| R: CGTAGTAGTAGTGGCGGCAG | |||
| GAPDH | / | / | F: AAGCTCATTTCCTGGTACGACA |
| R: GGAGGTCGGGAGATTCTCAG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, X.; Zhan, C.; Qiao, J.; Yang, Y.; Li, C.; Li, P.; Ma, S. Transcriptomic Analysis of Muscle Satellite Cell Regulation on Intramuscular Preadipocyte Differentiation in Tan Sheep. Int. J. Mol. Sci. 2025, 26, 3414. https://doi.org/10.3390/ijms26073414
Xu X, Zhan C, Qiao J, Yang Y, Li C, Li P, Ma S. Transcriptomic Analysis of Muscle Satellite Cell Regulation on Intramuscular Preadipocyte Differentiation in Tan Sheep. International Journal of Molecular Sciences. 2025; 26(7):3414. https://doi.org/10.3390/ijms26073414
Chicago/Turabian StyleXu, Xiaochun, Cong Zhan, Jiaqi Qiao, Yuxuan Yang, Changyuan Li, Pan Li, and Sen Ma. 2025. "Transcriptomic Analysis of Muscle Satellite Cell Regulation on Intramuscular Preadipocyte Differentiation in Tan Sheep" International Journal of Molecular Sciences 26, no. 7: 3414. https://doi.org/10.3390/ijms26073414
APA StyleXu, X., Zhan, C., Qiao, J., Yang, Y., Li, C., Li, P., & Ma, S. (2025). Transcriptomic Analysis of Muscle Satellite Cell Regulation on Intramuscular Preadipocyte Differentiation in Tan Sheep. International Journal of Molecular Sciences, 26(7), 3414. https://doi.org/10.3390/ijms26073414





