The Role of microRNA in the Prognosis and Diagnosis of Ovarian Cancer
Abstract
1. Introduction
2. The Role of miRNAs in the Prognosis of OC
3. The Role of miRNAs in the Diagnosis of OC
4. Summary and Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Torre, L.A.; Trabert, B.; DeSantis, C.E.; Miller, K.D.; Samimi, G.; Runowicz, C.D.; Gaudet, M.M.; Jemal, A.; Siegel, R.L. Ovarian cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 284–296. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Hinchcliff, E.; Westin, S.N.; Herzog, T.J. State of the science: Contemporary front-line treatment of advanced ovarian cancer. Gynecol. Oncol. 2022, 166, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Ergin, K.; Çetinkaya, R. Regulation of MicroRNAs. Methods Mol. Biol. 2022, 2257, 1–32. [Google Scholar] [CrossRef]
- Gregory, R.I.; Yan, K.-P.; Amuthan, G.; Chendrimada, T.; Doratotaj, B.; Cooch, N.; Shiekhattar, R. The Microprocessor complex mediates the genesis of microRNAs. Nature 2004, 432, 235–240. [Google Scholar] [CrossRef]
- Nguyen, T.A.; Jo, M.H.; Choi, Y.-G.; Park, J.; Kwon, S.C.; Hohng, S.; Kim, V.N.; Woo, J.-S. Functional anatomy of the human microprocessor. Cell 2015, 161, 1374–1387. [Google Scholar] [CrossRef] [PubMed]
- Lund, E.; Güttinger, S.; Calado, A.; Dahlberg, J.E.; Kutay, U. Nuclear Export of MicroRNA Precursors. Science 2004, 303, 95–98. [Google Scholar] [CrossRef]
- Bernstein, E.; Caudy, A.A.; Hammond, S.M.; Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001, 409, 363–366. [Google Scholar] [CrossRef]
- Song, J.-J.; Smith, S.K.; Hannon, G.J.; Joshua-Tor, L. Crystal structure of argonaute and its implications for RISC slicer activity. Science 2004, 305, 1434–1437. [Google Scholar] [CrossRef]
- Schwarz, D.S.; Hutvágner, G.; Du, T.; Xu, Z.; Aronin, N.; Zamore, P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell 2003, 115, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Lewis, B.P.; Shih, I.-H.; Jones-Rhoades, M.W.; Bartel, D.P.; Burge, C.B. Prediction of Mammalian MicroRNA Targets. Cell 2003, 115, 787–798. [Google Scholar] [CrossRef] [PubMed]
- Lai, E.C. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat. Genet. 2002, 30, 363–364. [Google Scholar] [CrossRef]
- Ulitsky, I. Interactions between short and long noncoding RNAs. FEBS Lett. 2018, 592, 2874–2883. [Google Scholar] [CrossRef] [PubMed]
- Boussios, S.; Devo, P.; Goodall, I.C.A.; Sirlantzis, K.; Ghose, A.; Shinde, S.D.; Papadopoulos, V.; Sanchez, E.; Rassy, E.; Ovsepian, S.V. Exosomes in the Diagnosis and Treatment of Renal Cell Cancer. Int. J. Mol. Sci. 2023, 24, 14356. [Google Scholar] [CrossRef]
- MacFarlane, L.-A.; Murphy, P.R. MicroRNA: Biogenesis, Function and Role in Cancer. Curr. Genom. 2010, 11, 537–561. [Google Scholar] [CrossRef]
- Zhang, B.; Li, Y.; Li, Y.; Zhao, H.; An, R. High Expression of MicroRNA-200a/b Indicates Potential Diagnostic and Prognostic Biomarkers in Epithelial Ovarian Cancer. Dis. Markers 2022, 2022, 2751696. [Google Scholar] [CrossRef]
- Záveský, L.; Jandáková, E.; Weinberger, V.; Minář, L.; Hanzíková, V.; Dušková, D.; Drábková, L.Z.; Svobodová, I.; Hořínek, A. Ascites-Derived Extracellular microRNAs as Potential Biomarkers for Ovarian Cancer. Reprod. Sci. 2019, 26, 510–522. [Google Scholar] [CrossRef]
- Meng, X.; Müller, V.; Milde-Langosch, K.; Trillsch, F.; Pantel, K.; Schwarzenbach, H. Diagnostic and prognostic relevance of circulating exosomal miR-373, miR-200a, miR-200b and miR-200c in patients with epithelial ovarian cancer. Oncotarget 2016, 7, 16923–16935. [Google Scholar] [CrossRef]
- Gao, Y.-C.; Wu, J. MicroRNA-200c and microRNA-141 as potential diagnostic and prognostic biomarkers for ovarian cancer. Tumor Biol. 2015, 36, 4843–4850. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Jiang, Z.; Xie, G.; Lu, Y. Serum microRNA-145 as a novel biomarker in human ovarian cancer. Tumor Biol. 2015, 36, 5305–5313. [Google Scholar] [CrossRef]
- Kim, T.H.; Song, J.-Y.; Park, H.; Jeong, J.-Y.; Kwon, A.-Y.; Heo, J.H.; Kang, H.; Kim, G.; An, H.J. miR-145, targeting high-mobility group A2, is a powerful predictor of patient outcome in ovarian carcinoma. Cancer Lett. 2015, 356, 937–945. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Liu, Z.; Chen, L.; Zhou, L.; Yao, Y. MicroRNA-23b is an independent prognostic marker and suppresses ovarian cancer progression by targeting runt-related transcription factor-2. FEBS Lett. 2014, 588, 1608–1615. [Google Scholar] [CrossRef] [PubMed]
- Su, L.; Liu, M. Correlation analysis on the expression levels of microRNA-23a and microRNA-23b and the incidence and prognosis of ovarian cancer. Oncol. Lett. 2018, 16, 262–266. [Google Scholar] [CrossRef]
- Li, J.; Yue, H.; Li, W.; Zhu, G.; Zhu, T.; Chen, R.; Lu, X. Bevacizumab confers significant improvements in survival for ovarian cancer patients with low miR-25 expression and high miR-142 expression. J. Ovarian Res. 2021, 14, 166. [Google Scholar] [CrossRef]
- Wang, X.; Meng, X.; Li, H.; Liu, W.; Shen, S.; Gao, Z. MicroRNA-25 expression level is an independent prognostic factor in epithelial ovarian cancer. Clin. Transl. Oncol. 2014, 16, 954–958. [Google Scholar] [CrossRef]
- Biegała, Ł.; Kołat, D.; Gajek, A.; Płuciennik, E.; Marczak, A.; Śliwińska, A.; Mikula, M.; Rogalska, A. Uncovering miRNA–mRNA Regulatory Networks Related to Olaparib Resistance and Resensitization of BRCA2MUT Ovarian Cancer PEO1-OR Cells with the ATR/CHK1 Pathway Inhibitors. Cells 2024, 13, 867. [Google Scholar] [CrossRef]
- Wilczyński, M.; Żytko, E.; Szymańska, B.; Dzieniecka, M.; Nowak, M.; Danielska, J.; Stachowiak, G.; Wilczyński, J.R. Expression of miR-146a in patients with ovarian cancer and its clinical significance. Oncol. Lett. 2017, 14, 3207–3214. [Google Scholar] [CrossRef]
- Kovač, M.P.; Tadić, V.; Kralj, J.; Periša, M.M.; Orešković, S.; Babić, I.; Banović, V.; Zhang, W.; Culig, Z.; Brozovic, A. MiRNA-mRNA integrative analysis reveals epigenetically regulated and prognostic miR-103a with a role in migration and invasion of carboplatin-resistant ovarian cancer cells that acquired mesenchymal-like phenotype. Biomed. Pharmacother. 2023, 166, 115349. [Google Scholar] [CrossRef]
- Zhang, R.; Guo, X.; Zhao, L.; He, T.; Feng, W.; Ren, S. Abnormal expressions of PURPL, miR-363-3p and ADAM10 predicted poor prognosis for patients with ovarian serous cystadenocarcinoma. J. Cancer 2023, 14, 2908–2918. [Google Scholar] [CrossRef] [PubMed]
- Gahlawat, A.W.; Witte, T.; Haarhuis, L.; Schott, S. A novel circulating miRNA panel for non-invasive ovarian cancer diagnosis and prognosis. Br. J. Cancer 2022, 127, 1550–1556. [Google Scholar] [CrossRef] [PubMed]
- Lopacinska-Jørgensen, J.; Oliveira, D.V.N.P.; Novotny, G.W.; Høgdall, C.K.; Høgdall, E.V. Integrated microRNA and mRNA signatures associated with overall survival in epithelial ovarian cancer. PLoS ONE 2021, 16, e0255142. [Google Scholar] [CrossRef]
- Zhang, R.; Shi, H.; Ren, F.; Liu, Z.; Ji, P.; Zhang, W.; Wang, W. Down-regulation of miR-338-3p and up-regulation of MACC1 indicated poor prognosis of epithelial ovarian cancer patients. J. Cancer 2019, 10, 1385–1392. [Google Scholar] [CrossRef]
- Zhang, R.; He, T.; Shi, H.; Yuan, C.; Wei, F.; Liu, Z.; Wang, W. Disregulations of PURPL and MiR-338-3p could serve as prognosis biomarkers for epithelial ovarian cancer. J. Cancer 2021, 12, 5674–5680. [Google Scholar] [CrossRef]
- Minareci, Y.; Ak, N.; Sozen, H.; Tosun, O.A.; Kucukgergin, C.; Aydin, F.; Bingul, I.; Salihoglu, M.Y.; Topuz, S. The evaluation of miR-1181 and miR-4314 as serum microRNA biomarkers for epithelial ovarian cancer diagnosis and prognosis. Mol. Biol. Rep. 2024, 51, 515. [Google Scholar] [CrossRef]
- Flores-Colón, M.; Rivera-Serrano, M.; Reyes-Burgos, V.G.; Rolón, J.G.; Pérez-Santiago, J.; Marcos-Martínez, M.J.; Valiyeva, F.; Vivas-Mejía, P.E. MicroRNA Expression Profiles in Human Samples and Cell Lines Revealed Nine miRNAs Associated with Cisplatin Resistance in High-Grade Serous Ovarian Cancer. Int. J. Mol. Sci. 2024, 25, 3793. [Google Scholar] [CrossRef]
- Liu, L.; Han, Q.; Cai, J.; Xiao, M.; Huang, D.; Cao, J. The clinical validity of miR-126 as a prognostic marker in epithelial ovarian cancer. Medicine 2023, 102, e33085. [Google Scholar] [CrossRef]
- Zhou, M.; Wu, T.; Yuan, Y.; Dong, S.-J.; Zhang, Z.-M.; Wang, Y.; Wang, J. A risk score system based on a six-microRNA signature predicts the overall survival of patients with ovarian cancer. J. Ovarian Res. 2022, 15, 54. [Google Scholar] [CrossRef]
- Chen, B.; Jin, X.; Wang, H.; Zhou, Q.; Li, G.; Lu, X. Expression, clinical significance, and prospective pathway signaling of miR-501-3p in ovarian cancer based on database and informatics analysis. Int. J. Gen. Med. 2021, 14, 5193–5201. [Google Scholar] [CrossRef]
- Teng, C.; Zheng, H. Low expression of microRNA-1908 predicts a poor prognosis for patients with ovarian cancer. Oncol. Lett. 2017, 14, 4277–4281. [Google Scholar] [CrossRef] [PubMed]
- Fukagawa, S.; Miyata, K.; Yotsumoto, F.; Kiyoshima, C.; Nam, S.O.; Anan, H.; Katsuda, T.; Miyahara, D.; Murata, M.; Yagi, H.; et al. MicroRNA-135a-3p as a promising biomarker and nucleic acid therapeutic agent for ovarian cancer. Cancer Sci. 2017, 108, 886–896. [Google Scholar] [CrossRef]
- Zhu, T.; Gao, W.; Chen, X.; Zhang, Y.; Wu, M.; Zhang, P.; Wang, S. A Pilot Study of Circulating MicroRNA-125b as a Diagnostic and Prognostic Biomarker for Epithelial Ovarian Cancer. Int. J. Gynecol. Cancer 2017, 27, 3–10. [Google Scholar] [CrossRef]
- Gong, L.; Wang, C.; Gao, Y.; Wang, J. Decreased expression of microRNA-148a predicts poor prognosis in ovarian cancer and associates with tumor growth and metastasis. Biomed. Pharmacother. 2016, 83, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Zuberi, M.; Khan, I.; Gandhi, G.; Ray, P.C.; Saxena, A. The conglomeration of diagnostic, prognostic and therapeutic potential of serum miR-199a and its association with clinicopathological features in epithelial ovarian cancer. Tumor Biol. 2016, 37, 11259–11266. [Google Scholar] [CrossRef]
- Chen, H.; Zhang, L.; Zhang, L.; Du, J.; Wang, H.; Wang, B. MicroRNA-183 correlates cancer prognosis, regulates cancer proliferation and bufalin sensitivity in epithelial ovarian caner. Am. J. Transl. Res. 2016, 8, 1748–1755. [Google Scholar]
- Cong, J.; Liu, R.; Wang, X.; Wang, J.; Wang, H.; Hou, J. Low miR-498 expression levels are associated with poor prognosis in ovarian cancer. Eur. Rev. Med. Pharmacol. Sci. 2015, 19, 4762–4765. [Google Scholar] [PubMed]
- Zhang, X.; Guo, G.; Wang, G.; Zhao, J.; Wang, B.; Yu, X.; Ding, Y. Profile of differentially expressed miRNAs in high-grade serous carcinoma and clear cell ovarian carcinoma, and the expression of miR-510 in ovarian carcinoma. Mol. Med. Rep. 2015, 12, 8021–8031. [Google Scholar] [CrossRef]
- Meng, X.; A Joosse, S.; Müller, V.; Trillsch, F.; Milde-Langosch, K.; Mahner, S.; Geffken, M.; Pantel, K.; Schwarzenbach, H. Diagnostic and prognostic potential of serum miR-7, miR-16, miR-25, miR-93, miR-182, miR-376a and miR-429 in ovarian cancer patients. Br. J. Cancer 2015, 113, 1358–1366. [Google Scholar] [CrossRef]
- Qin, C.Z.; Lou, X.Y.; Lv, Q.L.; Cheng, L.; Wu, N.Y.; Hu, L.; Zhou, H.H. MicroRNA-184 acts as a potential diagnostic and prognostic marker in epithelial ovarian cancer and reg-ulates cell proliferation, apoptosis and inflammation. Die Pharm. -Int. J. Pharm. Sci. 2015, 70, 668–673. [Google Scholar] [CrossRef]
- Fan, Y.; Fan, J.; Huang, L.; Ye, M.; Huang, Z.; Wang, Y.; Li, Q.; Huang, J. Increased expression of microRNA-196a predicts poor prognosis in human ovarian carcinoma. Int. J. Clin. Exp. Pathol. 2015, 8, 4132–4137. [Google Scholar] [PubMed]
- Ling, S.; Ruiqin, M.; Guohong, Z.; Ying, W. Expression and prognostic significance of microRNA-451 in human epithelial ovarian cancer. Eur. J. Gynaecol. Oncol. 2015, 36, 463–468. [Google Scholar]
- Wan, W.-N.; Zhang, Y.-Q.; Wang, X.-M.; Liu, Y.-J.; Zhang, Y.-X.; Que, Y.-H.; Zhao, W.-J.; Li, P. Down-regulated miR-22 as predictive biomarkers for prognosis of epithelial ovarian cancer. Diagn. Pathol. 2014, 9, 178. [Google Scholar] [CrossRef]
- Jin, M.; Yang, Z.; Ye, W.; Xu, H.; Hua, X. MicroRNA-150 Predicts a favorable prognosis in patients with epithelial ovarian cancer, and inhibits cell invasion and metastasis by suppressing transcriptional repressor ZEB1. PLoS ONE 2014, 9, e103965. [Google Scholar] [CrossRef]
- Oliveira, D.N.P.; Carlsen, A.L.; Heegaard, N.H.H.; Prahm, K.P.; Christensen, I.J.; Høgdall, C.K.; Høgdall, E.V. Diagnostic plasma miRNA-profiles for ovarian cancer in patients with pelvic mass. PLoS ONE 2019, 14, e0225249. [Google Scholar] [CrossRef]
- Záveský, L.; Jandáková, E.; Weinberger, V.; Minář, L.; Hanzíková, V.; Dušková, D.; Drábková, L.Z.; Hořínek, A. Ovarian Cancer: Differentially Expressed microRNAs in Tumor Tissue and Cell-Free Ascitic Fluid as Potential Novel Biomarkers. Cancer Investig. 2019, 37, 440–452. [Google Scholar] [CrossRef]
- Zuberi, M.; Mir, R.; Khan, I.; Javid, J.; Guru, S.A.; Bhat, M.; Sumi, M.P.; Ahmad, I.; Masroor, M.; Yadav, P.; et al. The Promising Signatures of Circulating microRNA-145 in Epithelial Ovarian Cancer Patients. MicroRNA 2019, 9, 49–57. [Google Scholar] [CrossRef]
- Wang, W.; Yin, Y.; Shan, X.; Zhou, X.; Liu, P.; Cao, Q.; Zhu, D.; Zhang, J.; Zhu, W. The Value of Plasma-Based MicroRNAs as Diagnostic Biomarkers for Ovarian Cancer. Am. J. Med. Sci. 2019, 358, 256–267. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wu, L.-R.; Li, C.; Zhou, X.; Liu, P.; Jia, X.; Chen, Y.; Zhu, W. Five serum microRNAs for detection and predicting of ovarian cancer. Eur. J. Obstet. Gynecol. Reprod. Biol. 2019, 3, 100017. [Google Scholar] [CrossRef]
- Zuberi, M.; Khan, I.; Mir, R.; Gandhi, G.; Ray, P.C.; Saxena, A. Utility of serum miR-125b as a diagnostic and prognostic indicator and its alliance with a panel of tumor suppressor genes in epithelial ovarian cancer. PLoS ONE 2016, 11, e0153902. [Google Scholar] [CrossRef]
- Yokoi, A.; Matsuzaki, J.; Yamamoto, Y.; Yoneoka, Y.; Takahashi, K.; Shimizu, H.; Uehara, T.; Ishikawa, M.; Ikeda, S.-I.; Sonoda, T.; et al. Integrated extracellular microRNA profiling for ovarian cancer screening. Nat. Commun. 2018, 9, 4319. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Gong, G.; Tan, H.; Dai, F.; Zhu, X.; Chen, Y.; Wang, J.; Liu, Y.; Chen, P.; Wu, X.; et al. Urinary microRNA-30a-5p is a potential biomarker for ovarian serous adenocarcinoma. Oncol. Rep. 2015, 33, 2915–2923. [Google Scholar] [CrossRef] [PubMed]
- Kumari, P.; Sharma, I.; Saha, S.C.; Srinivasan, R.; Minhas, P. Diagnostic potential of differentially regulated microRNAs among endometriosis, endometrioid ovarian cancer, and endometrial cancer. J. Cancer Res. Ther. 2021, 17, 1003–1011. [Google Scholar] [CrossRef]
- Kumar, V.; Pandey, A.; Arora, A.; Gautam, P.; Bisht, D.; Gupta, S.; Chaurasia, A.; Sachan, M. Diagnostics and Therapeutic Potential of miR-205 and miR-34a in Ovarian Cancer Management: A miRNA-Target-Based Analysis. DNA Cell Biol. 2023, 42, 151–162. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhu, M.-J.; Ren, A.-M.; Wu, H.-F.; Han, W.-M.; Tan, R.-Y.; Tu, R.-Q. A Ten-MicroRNA signature identified from a genome-wide MicroRNA expression profiling in human epithelial ovarian cancer. PLoS ONE 2014, 9, e96472. [Google Scholar] [CrossRef]
- Choi, P.-W.; Bahrampour, A.; Liu, S.K.; Qiu, W.; Xie, F.; Kuo, W.P.; Kwong, J.; Hales, K.H.; Hales, D.B.; Wong, K.-K.; et al. Characterization of miR-200 family members as blood biomarkers for human and laying hen ovarian cancer. Sci. Rep. 2020, 10, 20071. [Google Scholar] [CrossRef]
- Delek, F.S.P.; Tunçer, Ş.B.; Ödemiş, D.A.; Erciyas, S.K.; Erdoğan, Ö.Ş.; Saip, P.; Yazıcı, H. miR-3653-3p Expression in PBMCs: Unveiling the Diagnostic Potential for Ovarian Cancer. Biochem. Genet. 2024, 1–18. [Google Scholar] [CrossRef]
- Ralser, D.J.; Condic, M.; Egger, E.; Koensgen, D.; Mustea, A.; Stope, M.B. Evaluation of the Diagnostic Potential of Circulating MicroRNAs miR-1 and miR-21 in Patients With Ovarian Cancer. Anticancer. Res. 2022, 42, 5839–5845. [Google Scholar] [CrossRef]
- Ali, F.T.; Soliman, R.M.; Hassan, N.S.; Ibrahim, A.M.; El-Gizawy, M.M.; Mandoh, A.A.Y.; Ibrahim, E.A. Sensitivity and specificity of microRNA-204, CA125, and CA19.9 as biomarkers for diagnosis of ovarian cancer. PLoS ONE 2022, 17, e0272308. [Google Scholar] [CrossRef]
- Kumar, V.; Gupta, S.; Varma, K.; Chaurasia, A.; Sachan, M. Diagnostic performance of microRNA-34a, let-7f and microRNA-31 in epithelial ovarian cancer prediction. J. Gynecol. Oncol. 2022, 33, e49. [Google Scholar] [CrossRef]
- Jeon, H.; Seo, S.M.; Kim, T.W.; Ryu, J.; Kong, H.; Jang, S.H.; Jang, Y.S.; Kim, K.S.; Kim, J.H.; Ryu, S.; et al. Circulating Exosomal miR-1290 for Diagnosis of Epithelial Ovarian Cancer. Curr. Issues Mol. Biol. 2022, 44, 288–300. [Google Scholar] [CrossRef] [PubMed]
- Ghafour, A.A.; Odemis, D.A.; Tuncer, S.B.; Kurt, B.; Saral, M.A.; Erciyas, S.K.; Erdogan, O.S.; Celik, B.; Saip, P.; Yazici, H. High expression level of miR-1260 family in the peripheral blood of patients with ovarian carcinoma. J. Ovarian Res. 2021, 14, 131. [Google Scholar] [CrossRef] [PubMed]
- El-Shal, A.S.; Matboli, M.; Abdelaziz, A.M.; Morsy, A.A.; Abdelbary, E.H. Role of a novel circulatory RNA-based biomarker panel expression in ovarian cancer. IUBMB Life 2019, 71, 2031–2047. [Google Scholar] [CrossRef] [PubMed]
- Günel, T.; Gumusoglu, E.; Dogan, B.; Ertem, F.B.; Hosseini, M.K.; Cevik, N.; Senol, T.; Topuz, S.; Aydinli, K. Potential biomarker of circulating hsa-miR-1273g-3p level for detection of recurrent epithelial ovarian cancer. Arch. Gynecol. Obstet. 2018, 298, 1173–1180. [Google Scholar] [CrossRef]

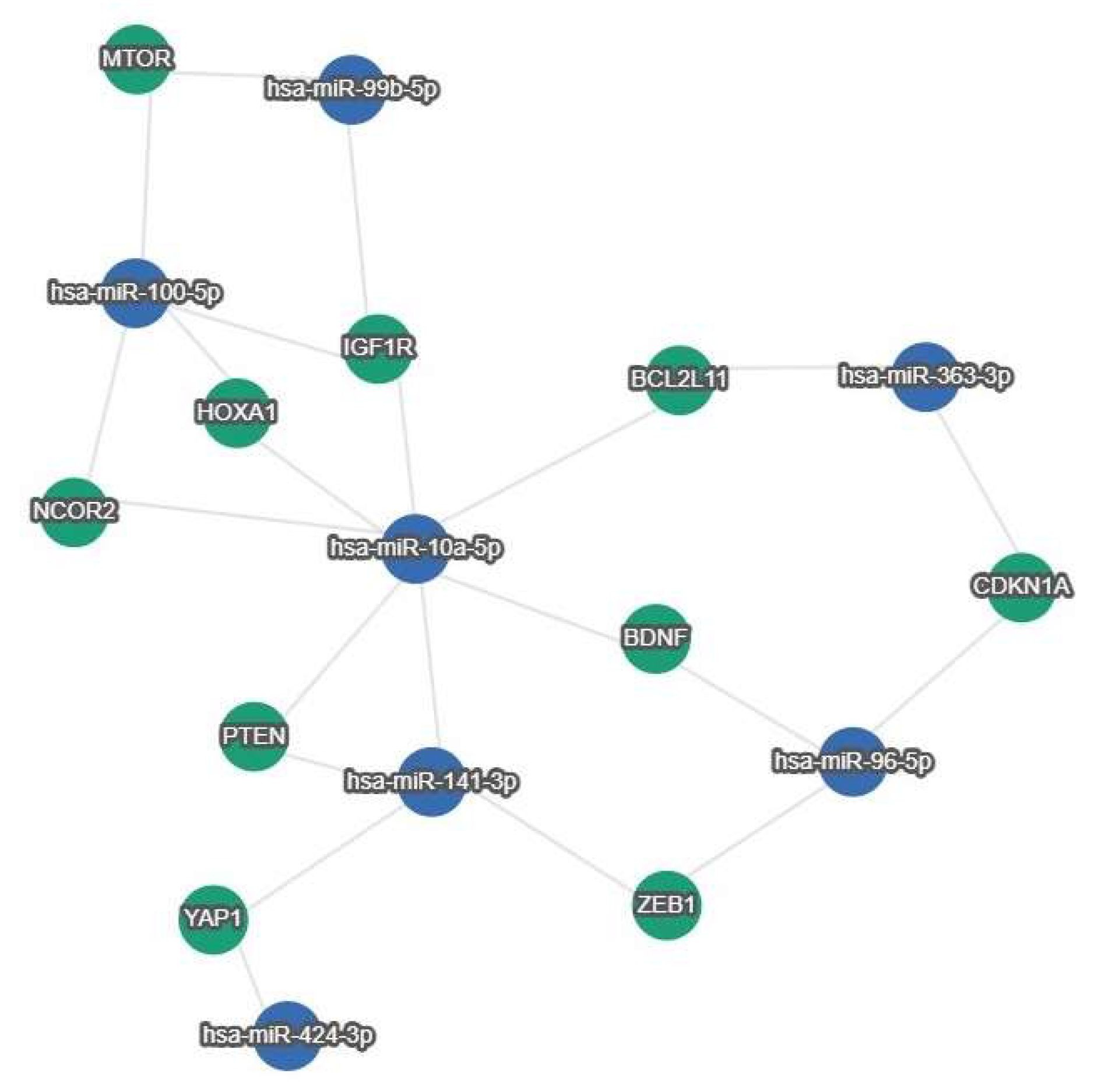
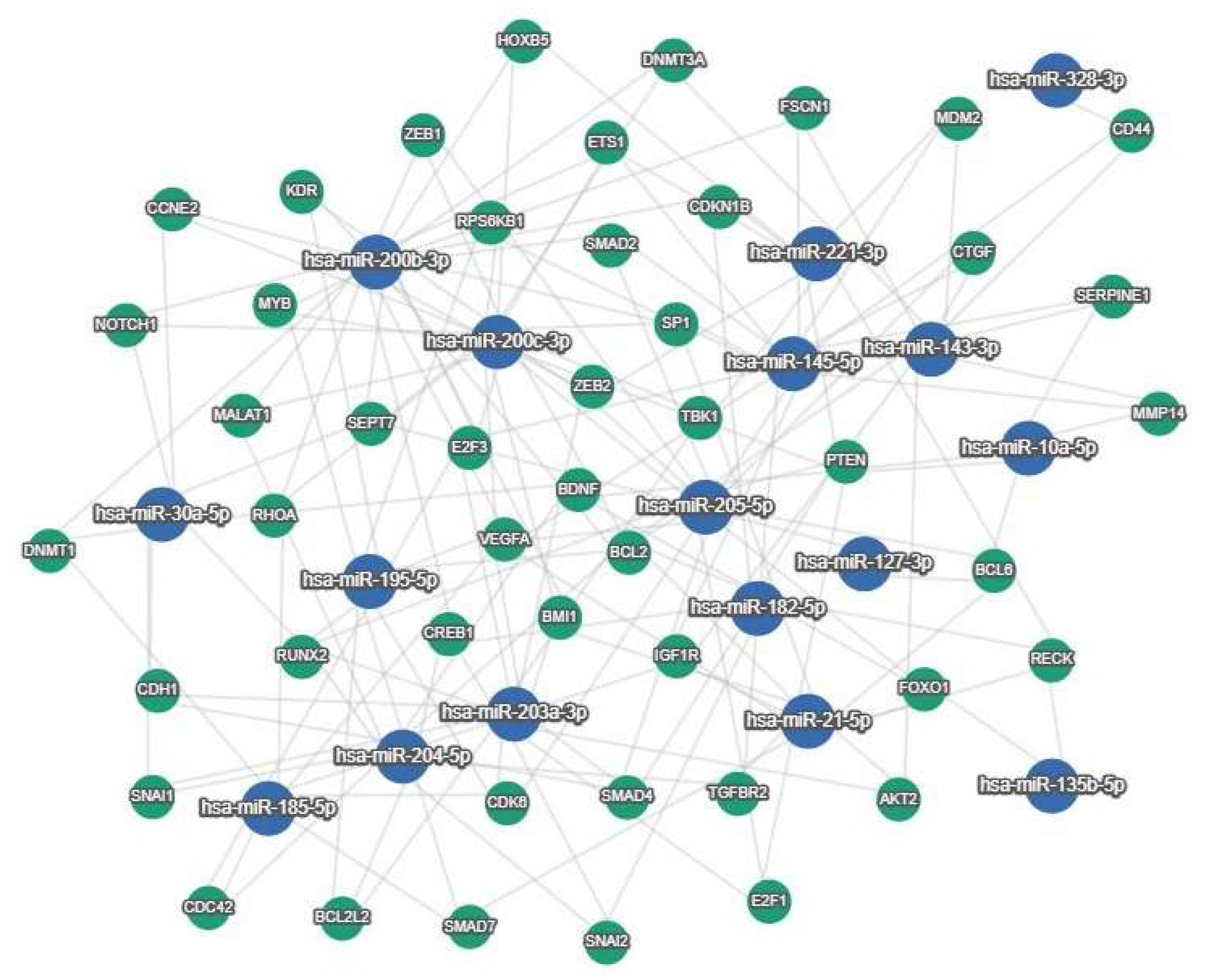
| miRNAs | Survival | Cohort | Compared Expression Groups | Reference |
|---|---|---|---|---|
| miR-99b-5p | OS: better OS, p = 0.011 PFI: better PFI, p = 0.0025 | Serous ovarian cancer | High vs. low | [28] |
| miR-100-5p | PFI: shorter PFI, p = 0.0088 | Serous ovarian cancer | High vs. low | [28] |
| miR-125a-3p | OS: shorter OS, p = 0.039 | Serous ovarian cancer | High vs. low | [28] |
| miR-505-5p | OS: better OS, p = 0.0009 | Serous ovarian cancer | High vs. low | [28] |
| miR-424-3p | OS: better OS, p = 0.021 PFI: better PFI, p = 0.044 | Serous ovarian cancer | High vs. low | [28] |
| miR-324-5p | OS: better OS, p = 0.0048 | Serous ovarian cancer | High vs. low | [28] |
| miR-4314 | OS: shorter OS, p = 0.007 DFS: shorter DFS, p < 0.004 | Epithelial ovarian cancer | High vs. low | [36] |
| miR-1181 | OS: shorter OS, p < 0.001 DFS: shorter DFS, p < 0.001 | Epithelial ovarian cancer | High vs. low | [36] |
| miR-1206 | OS: shorter OS, p = 7.1 × 10−7 | Ovarian cancer | High vs. low | [37] |
| miR-96-5p | OS: shorter OS, p = 0.0026 | Ovarian cancer | High vs. low | [37] |
| miR-10a-5p | OS: shorter OS, p = 0.021 | Ovarian cancer | High vs. low | [37] |
| miR-141-3p | OS: shorter OS, p = 0.046 | Ovarian cancer | High vs. low | [37] |
| miR-103a–3p | OS: better OS, p = 0.015 | Ovarian serous cystadenocarcinoma—MSC: enriched | High vs. low | [30] |
| miR-107 | OS: better OS, p = 0.0039 | Ovarian serous cystadenocarcinoma—all patients | High vs. low | [30] |
| miR-107 | OS: better OS, p = 0.0067 | Ovarian serous cystadenocarcinoma—MSC: enriched | High vs. low | [30] |
| miR-363-3p | OS: better OS, p = 0.0060 PFS: better PFS, p = 0.0284 | Ovarian serous cystadenocarcinoma | High vs. low | [31] |
| miR-126 | OS: shorter OS, p = 0.006 RFS: shorter RFS, p = 0.007 | Epithelial ovarian cancer | High vs. low | [38] |
| miR-6509-5p | OS: better OS, p = 0.006 | Ovarian cancer | High vs. low | [39] |
| miR-342-5p | OS: better OS, p = 0.032 | Ovarian cancer | High vs. low | [39] |
| miR-3074-5p | OS: better OS, p = 0.015 | Ovarian cancer | High vs. low | [39] |
| miR-877-5p | OS: better OS, p = 0.021 | Ovarian cancer | High vs. low | [39] |
| miR-760 | OS: better OS, p = 0.020 | Ovarian cancer | High vs. low | [39] |
| miR-758-3p | OS: shorter OS, p < 0.001 | Ovarian cancer | High vs. low | [39] |
| miR-200a | OS: shorter OS, p = 0.0047 DFS: shorter DFS, p = 0.0187 | Epithelial ovarian cancer | High vs. low | [18] |
| miR-200b | OS: shorter OS, p = 0.0232 DFS: shorter DFS, p = 0.0364 | Epithelial ovarian cancer | High vs. low | [18] |
| miR-25 | OS: better OS, p = 0.004 PFS: better PFS, p = 0.005 | Ovarian cancer | High vs. low | [26] |
| miR-142 | OS: shorter OS, p = 0.049 | Ovarian cancer | High vs. low | [26] |
| miR-501-3p | OS: better OS, p = 0.02 DSS: better DSS, p = 0.038 | Ovarian cancer | High vs. low | [40] |
| miR-200b | OS: shorter OS, p = 0.019 | Ovarian cancer | High vs. low | [19] |
| miR-23a | OS: shorter OS, p < 0.01 | Ovarian epithelial cancer | High vs. low | [25] |
| miR-23b | OS: better OS, p < 0.01 | Ovarian epithelial cancer | High vs. low | [25] |
| miR-1908 | OS: better OS, p = 0.004 DFS: better DFS, p < 0.001 | Ovarian cancer | High vs. low | [41] |
| miR-146a | Survival: better survival, p = 0.003 | Advanced serous ovarian cancer | High vs. low | [29] |
| miR-135a-3p | PFS: better PFS, p = 0.0494 | Ovarian cancer | High vs. low | [42] |
| miR-125b | PFS: better PFS, p = 0.035 | Epithelial ovarian cancer | High vs. low | [43] |
| miR-148a | OS: better OS, p = 0.002 | Ovarian cancer | High vs. low | [44] |
| miR-199a | OS: better OS, p = 0.03 | Epithelial ovarian cancer | High vs. low | [45] |
| miR-183 | OS: shorter OS, p < 0.05 | Epithelial ovarian cancer | High vs. low | [46] |
| miR-373 | OS: shorter OS, p = 0.033 | Epithelial ovarian cancer | High vs. low | [20] |
| miR-200b | OS: shorter OS, p = 0.007 | Epithelial ovarian cancer | High vs. low | [20] |
| miR-200c | OS: shorter OS, p = 0.017 DFS: shorter DFS, p = 0.019 | Epithelial ovarian cancer | High vs. low | [20] |
| miR-498 | OS: better OS, p = 0.0056 PFS: better PFS, p = 0.003 | Ovarian cancer | High vs. low | [47] |
| miR-129-3p | OS: better OS, p = 0.039 | Epithelial ovarian cancer | High vs. low | [48] |
| miR-510 | OS: better OS, p = 0.048 | Epithelial ovarian cancer | High vs. low | [48] |
| miR-429 | OS: shorter, p = 0.011 | Epithelial ovarian cancer | High vs. low | [49] |
| miR-184 | OS: better OS, p < 0.001 | Epithelial ovarian cancer | High vs. low | [50] |
| miR-145 | OS: better OS, p = 0.023 | Malignant ovarian cancer | High vs. low | [22] |
| miR-200c | OS: better OS, p < 0.001 | Ovarian cancer | High vs. low | [21] |
| miR-141 | OS: shorter OS, p = 0.049 | Ovarian cancer | High vs. low | [21] |
| miR-196a | OS: shorter OS, p < 0.001 recurrent-free survival: shorter recurrent-free survival, p = 0.003 | Ovarian carcinoma | High vs. low | [51] |
| miR-145 | OS: better OS, p = 0.003 | High-grade ovarian serous carcinoma | High vs. low | [23] |
| miR-451 | OS: better OS, p < 0.001 | Epithelial ovarian cancer | High vs. low | [52] |
| miR-25 | OS: shorter OS, p = 0.001 | Epithelial ovarian cancer | High vs. low | [27] |
| miR-22 | OS: better OS, p = 0.005 PFS: better PFS, p = 0.004 | Epithelial ovarian cancer | High vs. low | [53] |
| miR-150 | OS: better OS, p < 0.001 PFS: better PFS, p < 0.001 | Epithelial ovarian cancer | High vs. low | [54] |
| miR-23b | OS: better OS, p < 0.001 PFS: better PFS, p < 0.001 | Epithelial ovarian cancer | High vs. low | [24] |
| miRNAs | AUC (95% CI) | Compared Cohorts | Test Sample | Reference |
|---|---|---|---|---|
| miR-3653-3p | 0.833 (0.779–0.887) | Ovarian cancer vs. healthy controls | PBMCs | [67] |
| miR-4314 | 0.78 (0.69–0.85) | Epithelial ovarian cancer vs. healthy controls | Serum | [36] |
| miR-1181 | 0.76 (0.67-0.86) | Epithelial ovarian cancer vs. healthy controls | Serum | [36] |
| miR-1 | 0.531 | Malignant vs. benign ovarian tumors | Serum | [68] |
| miR-21 | 0.648 | Malignant vs. benign ovarian tumors | Serum | [68] |
| miR-204 | 0.924 (0.866–0.982) | Early ovarian cancer | Serum | [69] |
| 0.942 (0.893–0.990) | Late ovarian cancer | |||
| miRNA-34a | 0.97 (0.932–1.008) | Advanced-stage epithelial ovarian cancer | Tissue | [70] |
| 0.92 (0.842–0.99) | Advanced-stage epithelial ovarian cancer | Serum | ||
| 0.969 (0.938–1.001) | Early-stage epithelial ovarian cancer | Tissue | ||
| 0.827 (0.628–0.95) | Early-stage epithelial ovarian cancer | Serum | ||
| miRNA-let-7f | 0.921 (0.853–0.989) | Advanced-stage epithelial ovarian cancer | Tissue | [70] |
| 0.879 (0.773–0.98) | Advanced-stage epithelial ovarian cancer | Serum | ||
| 0.871 (0.788–0.954) | Early-stage epithelial ovarian cancer | Tissue | ||
| 0.82 (0.677–0.96) | Early-stage epithelial ovarian cancer | Serum | ||
| miRNA-31 | 0.921 (0.725–0.949) | Advanced-stage epithelial ovarian cancer | Tissue | [70] |
| 0.856 (0.694–1.01) | Advanced-stage epithelial ovarian cancer | Serum | ||
| 0.866 (0.766–0.969) | Early-stage epithelial ovarian cancer | Tissue | ||
| 0.81 (0.642–0.97) | Early-stage epithelial ovarian cancer | Serum | ||
| miRNA-200a | 0.8088 (0.6749–0.9426) | Epithelial ovarian cancer vs. benign ovarian disease or healthy physical examination | Tissue | [18] |
| 0.8063 (0.6745–0.9380) | Epithelial ovarian cancer vs. benign ovarian disease or healthy physical examination | Serum | ||
| miRNA-200b | 0.8425 (0.7197–0.9653) | Epithelial ovarian cancer vs. benign ovarian disease or healthy physical examination | Tissue | [18] |
| 0.8625 (0.7459–0.9791) | Epithelial ovarian cancer vs. benign ovarian disease or healthy physical examination | Serum | ||
| miR-1290 | 0.988 | EOC vs. benign ovarian neoplasm | Tissue | [71] |
| 0.794 | EOC vs. benign ovarian neoplasm | Serum | ||
| miR-1260a | 0.660 (0.588–0.733) | Ovarian cancer vs. healthy control | Peripheral blood lymphocytes | [72] |
| miR-1260b | 0.704 (0.635–0.773) | Ovarian cancer vs. healthy control | Peripheral blood lymphocytes | [72] |
| miR-143 | 0.933 (0.842–1.000) | Endometrioid ovarian cancer vs. endometriosis and endometrioid endometrial cancer | Tissue | [63] |
| miR-145 | 0.928 (0.86–0.95) | Epithelial ovarian cancer vs. healthy controls | Serum | [57] |
| miR-361-3p | 0.838 | Ovarian cancer vs. control group and patients with benign mass | Serum | [73] |
| miR-200c-3p | 0.78 | Malignant pelvic mass vs. patients with a benign ovarian tumor | Plasma | [55] |
| miR-221-3p | 0.65 | Malignant pelvic mass vs. patients with a benign ovarian tumor | Plasma | [55] |
| miR-195-5p | 0.63 | Malignant pelvic mass vs. patients with a benign ovarian tumor | Plasma | [55] |
| miR-21-5p | 0.63 | Malignant pelvic mass vs. patients with a benign ovarian tumor | Plasma | [55] |
| miR-451a | 0.62 | Malignant pelvic mass vs. patients with a benign ovarian tumor | Plasma | [55] |
| miR-484 | 0.63 | Malignant pelvic mass vs. patients with a benign ovarian tumor | Plasma | [55] |
| miR-205-5p | 0.681 | Ovarian cancer vs. normal controls | Plasma | [58] |
| miR-145-5p | 0.702 | Ovarian cancer vs. normal controls | Plasma | [58] |
| miR-10a-5p | 0.680 | Ovarian cancer vs. normal controls | Plasma | [58] |
| miR-346 | 0.737 | Ovarian cancer vs. normal controls | Plasma | [58] |
| miR-328-3p | 0.700 | Ovarian cancer vs. normal controls | Plasma | [58] |
| miR-200c-3p | 0.726 | Ovarian cancer vs. normal controls | Serum | [59] |
| miR-346 | 0.693 | Ovarian cancer vs. normal controls | Serum | [59] |
| miR-127-3p | 0.698 | Ovarian cancer vs. normal controls | Serum | [59] |
| miR-143-3p | 0.687 | Ovarian cancer vs. normal controls | Serum | [59] |
| miR-205-5p | 0.689 | Ovarian cancer vs. normal controls | Serum | [59] |
| miR-200b-3p | 1.000 (0.877–1.000) | Ovarian cancer vs. normal ovary | Tissue | [56] |
| miR-182-5p | 0.995 (0.867–1.000) | Ovarian cancer vs. normal ovary | Tissue | [56] |
| miR-135b-5p | 0.847 (0.661–0.954) | Ovarian cancer vs. normal ovary | Tissue | [56] |
| miR-451a | 0.974 (0.832–1.000) | Ovarian cancer vs. normal ovary | Tissue | [56] |
| miR-204-5p | 0.934 (0.772–0.993) | Ovarian cancer vs. normal ovary | Tissue | [56] |
| miR-185-5p | 0.811 (0.619–0.933) | Ovarian cancer vs. normal ovary | Tissue | [56] |
| miR-203a-3p | 0.765 (0.568–0.904) | Ovarian cancer vs. normal ovary | Tissue | [56] |
| miR-203a-3p | 1.000 (0.858–1.000) | Ovarian cancer vs. healthy controls | Ascitic fluid (ovarian cancer), plasma (healthy controls) | [56] |
| miR-204-5p | 1.000 (0.858–1.000) | Ovarian cancer vs. healthy controls | Ascitic fluid (ovarian cancer), plasma (healthy controls) | [56] |
| miR-135b-5p | 1.000 (0.858–1.000) | Ovarian cancer vs. healthy controls | Ascitic fluid (ovarian cancer), plasma (healthy controls) | [56] |
| miR-451a | 0.986 (0.833–1.000) | Ovarian cancer vs. healthy controls | Ascitic fluid (ovarian cancer), plasma (healthy controls) | [56] |
| miR-182-5p | 0.986 (0.833–1.000) | Ovarian cancer vs. healthy controls | Ascitic fluid (ovarian cancer), plasma (healthy controls) | [56] |
| miR-1273g-3p | 0.7 | Recurrent epithelial ovarian cancer vs. healthy controls | Serum | [74] |
| miR-320a | 0.96 (0.95–0.98) | Ovarian cancer vs. non-cancer | Serum | [61] |
| miR-665 | 0.86 (0.82–0.89) | Ovarian cancer vs. non-cancer | Serum | [61] |
| miR-3184-5p | 0.97 (0.96–0.98) | Ovarian cancer vs. non-cancer | Serum | [61] |
| miR-6717-5p | 0.73 (0.68–0.78) | Ovarian cancer vs. non-cancer | Serum | [61] |
| miR-4459 | 0.61 (0.56–0.65) | Ovarian cancer vs. non-cancer | Serum | [61] |
| miR-6076 | 0.56 (0.51–0.61) | Ovarian cancer vs. non-cancer | Serum | [61] |
| miR-3195 | 0.83 (0.79–0.87) | Ovarian cancer vs. non-cancer | Serum | [61] |
| miR-1275 | 0.87 (0.84–0.91) | Ovarian cancer vs. non-cancer | Serum | [61] |
| miR-3185 | 0.70 (0.65–0.75) | Ovarian cancer vs. non-cancer | Serum | [61] |
| miR-4640-5p | 0.54 (0.48–0.61) | Ovarian cancer vs. non-cancer | Serum | [61] |
| miR-125b | 0.737 | Epithelial ovarian cancer vs. benign ovarian tumor | Serum | [43] |
| miR-199a | 0.704 | Epithelial ovarian cancer vs. healthy controls | Serum | [45] |
| miR-125b | 0.728 (0.64–0.81) | Epithelial ovarian cancer vs. healthy controls | Serum | [60] |
| miR-145 | 0.82 (0.77–0.88) | Malignant ovarian cancer vs. healthy controls | Serum | [22] |
| miR-30a-5p | 0.862 (0.709–1.016) | Ovarian serous adenocarcinoma vs. healthy controls | Urine | [62] |
| miR-6076 | 0.693 (0.482–0.904) | Ovarian serous adenocarcinoma vs. healthy controls | Urine | [62] |
| miR-200c | 0.79 (0.71–0.87) | Ovarian cancer vs. healthy controls | Serum | [21] |
| miR-141 | 0.75 (0.67–0.83) | Ovarian cancer vs. healthy controls | Serum | [21] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kozłowski, M.; Borzyszkowska, D.; Golara, A.; Lubikowski, J.; Cymbaluk-Płoska, A. The Role of microRNA in the Prognosis and Diagnosis of Ovarian Cancer. Int. J. Mol. Sci. 2025, 26, 3413. https://doi.org/10.3390/ijms26073413
Kozłowski M, Borzyszkowska D, Golara A, Lubikowski J, Cymbaluk-Płoska A. The Role of microRNA in the Prognosis and Diagnosis of Ovarian Cancer. International Journal of Molecular Sciences. 2025; 26(7):3413. https://doi.org/10.3390/ijms26073413
Chicago/Turabian StyleKozłowski, Mateusz, Dominika Borzyszkowska, Anna Golara, Jerzy Lubikowski, and Aneta Cymbaluk-Płoska. 2025. "The Role of microRNA in the Prognosis and Diagnosis of Ovarian Cancer" International Journal of Molecular Sciences 26, no. 7: 3413. https://doi.org/10.3390/ijms26073413
APA StyleKozłowski, M., Borzyszkowska, D., Golara, A., Lubikowski, J., & Cymbaluk-Płoska, A. (2025). The Role of microRNA in the Prognosis and Diagnosis of Ovarian Cancer. International Journal of Molecular Sciences, 26(7), 3413. https://doi.org/10.3390/ijms26073413






