Association of Intergenic and Intragenic MGMT Enhancer Methylation with MGMT Promoter Methylation, MGMT Protein Expression and Clinical and Demographic Parameters in Glioblastoma
Abstract
1. Introduction
2. Results
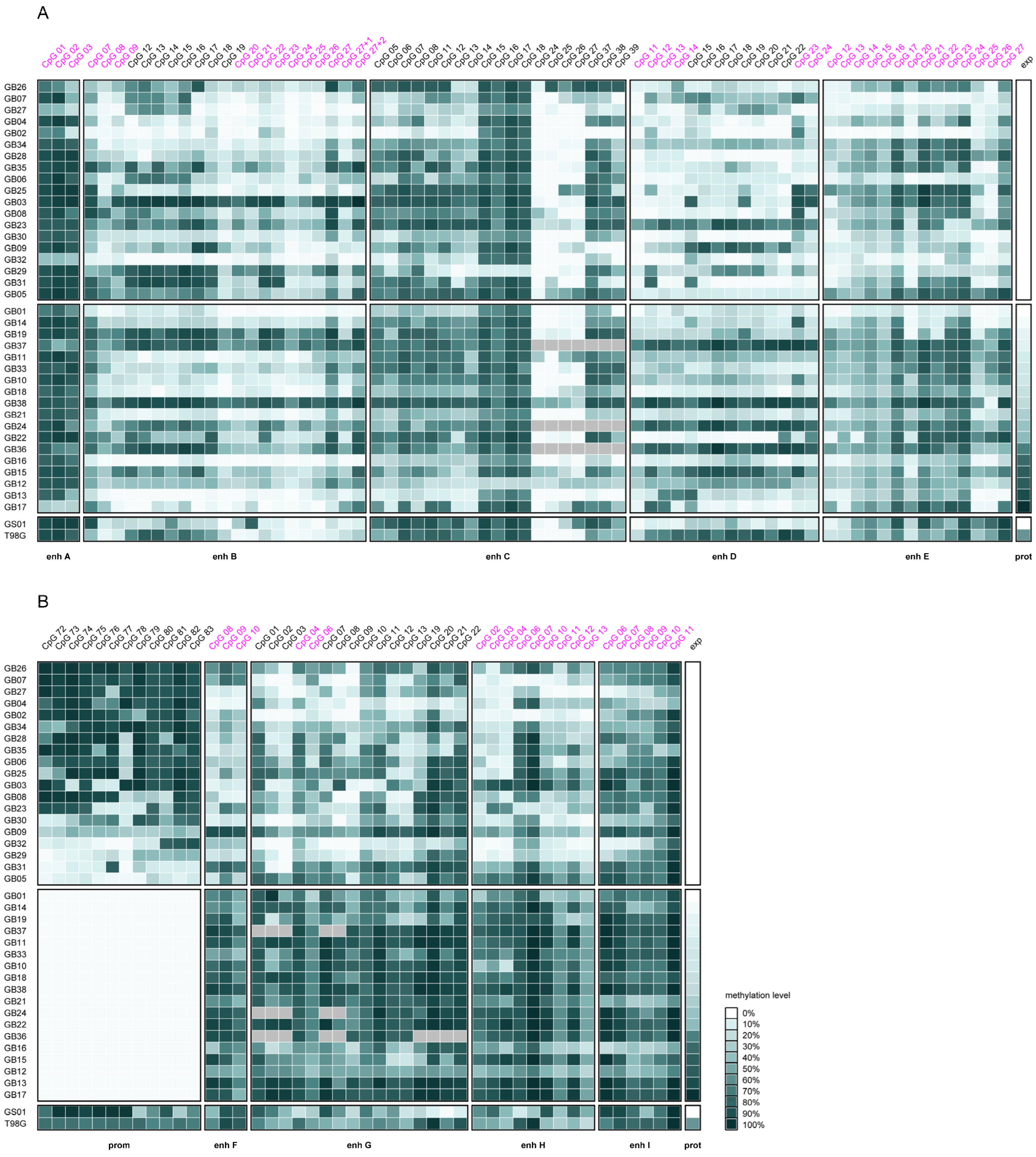
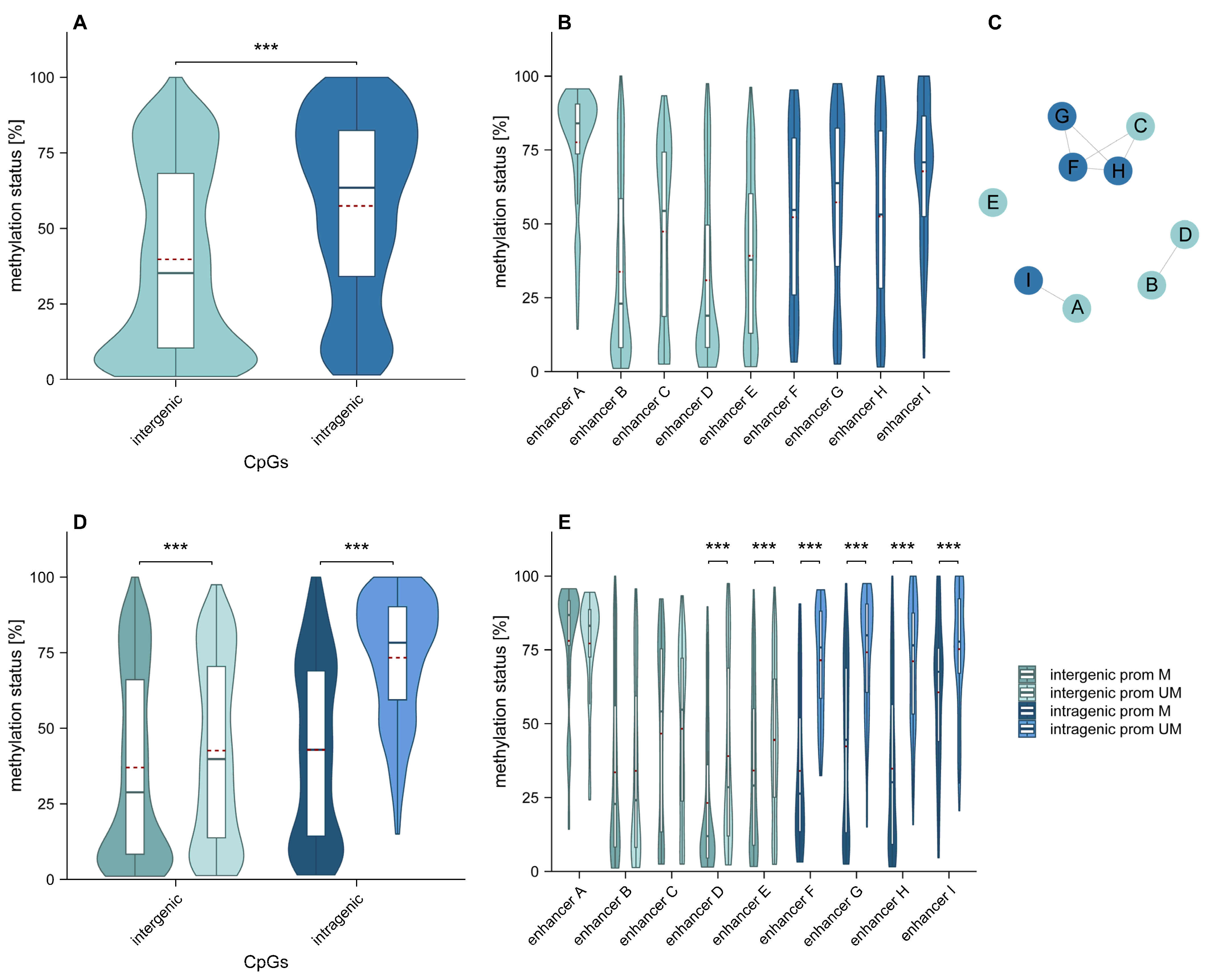
2.1. Overall Intergenic and Intragenic Enhancer Methylation
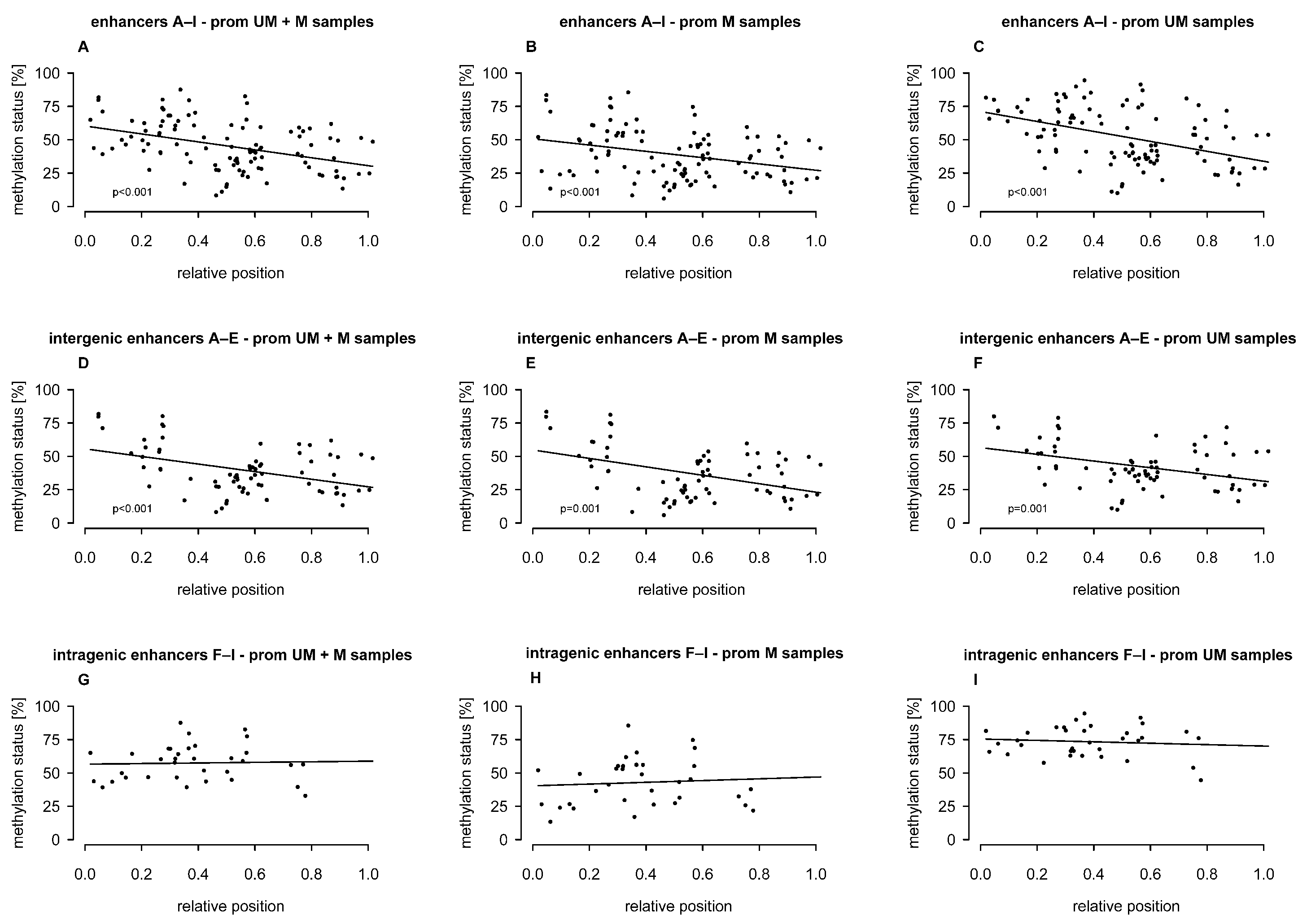
2.2. Association of Intergenic and Intragenic Enhancer Methylation with the Relative Position of CpGs
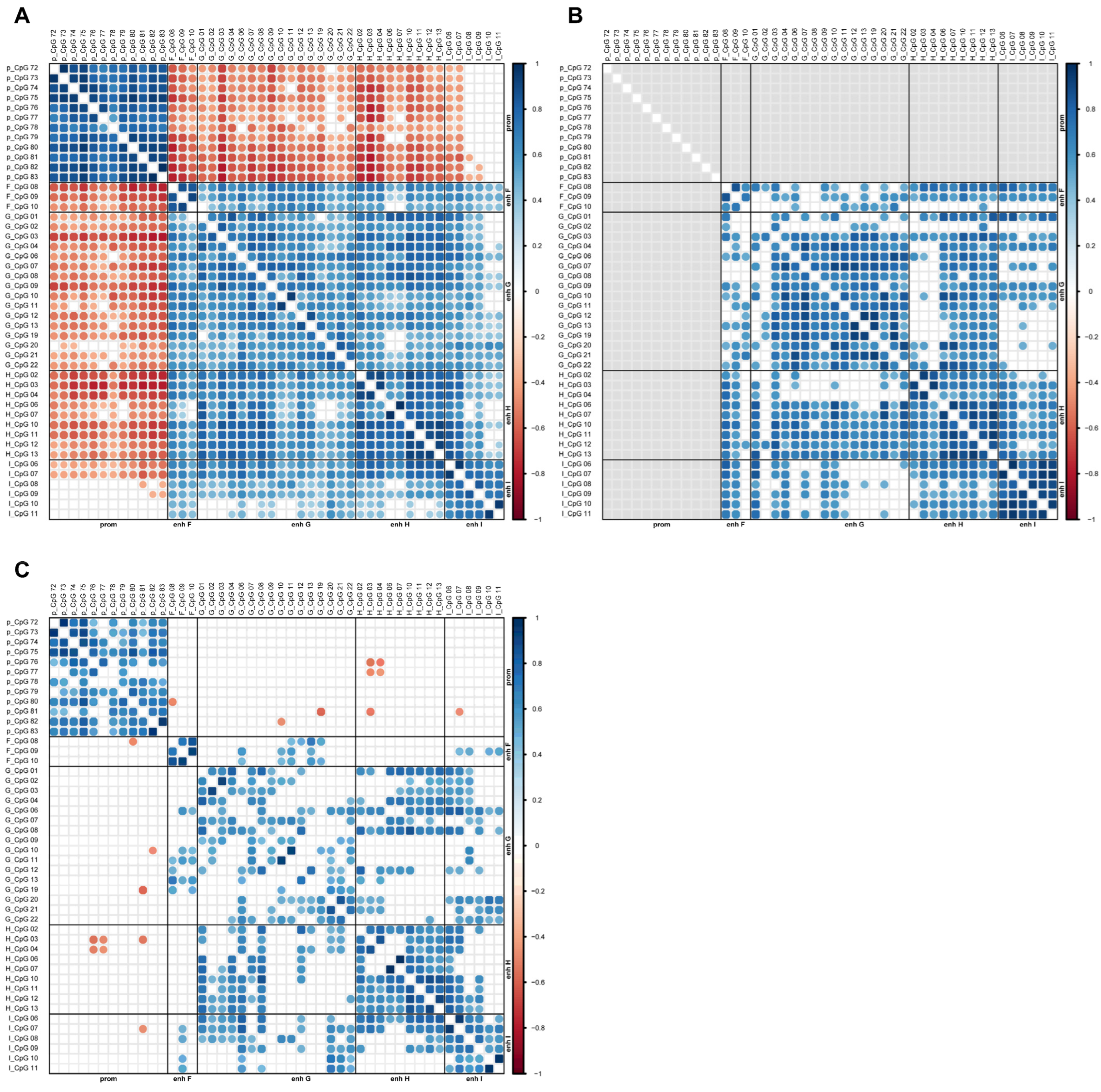
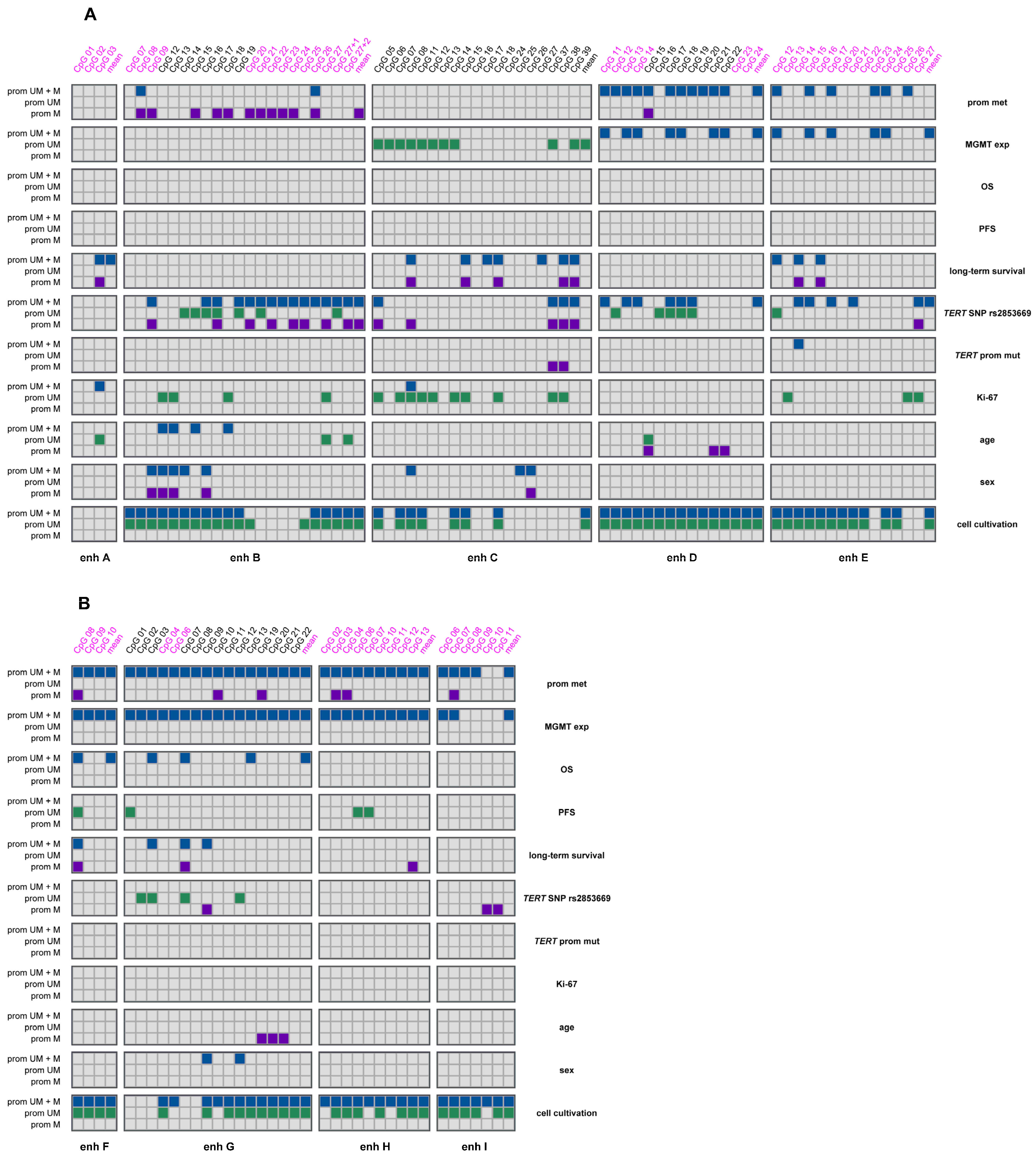
2.3. Association Between MGMT Promoter and Enhancer Methylation
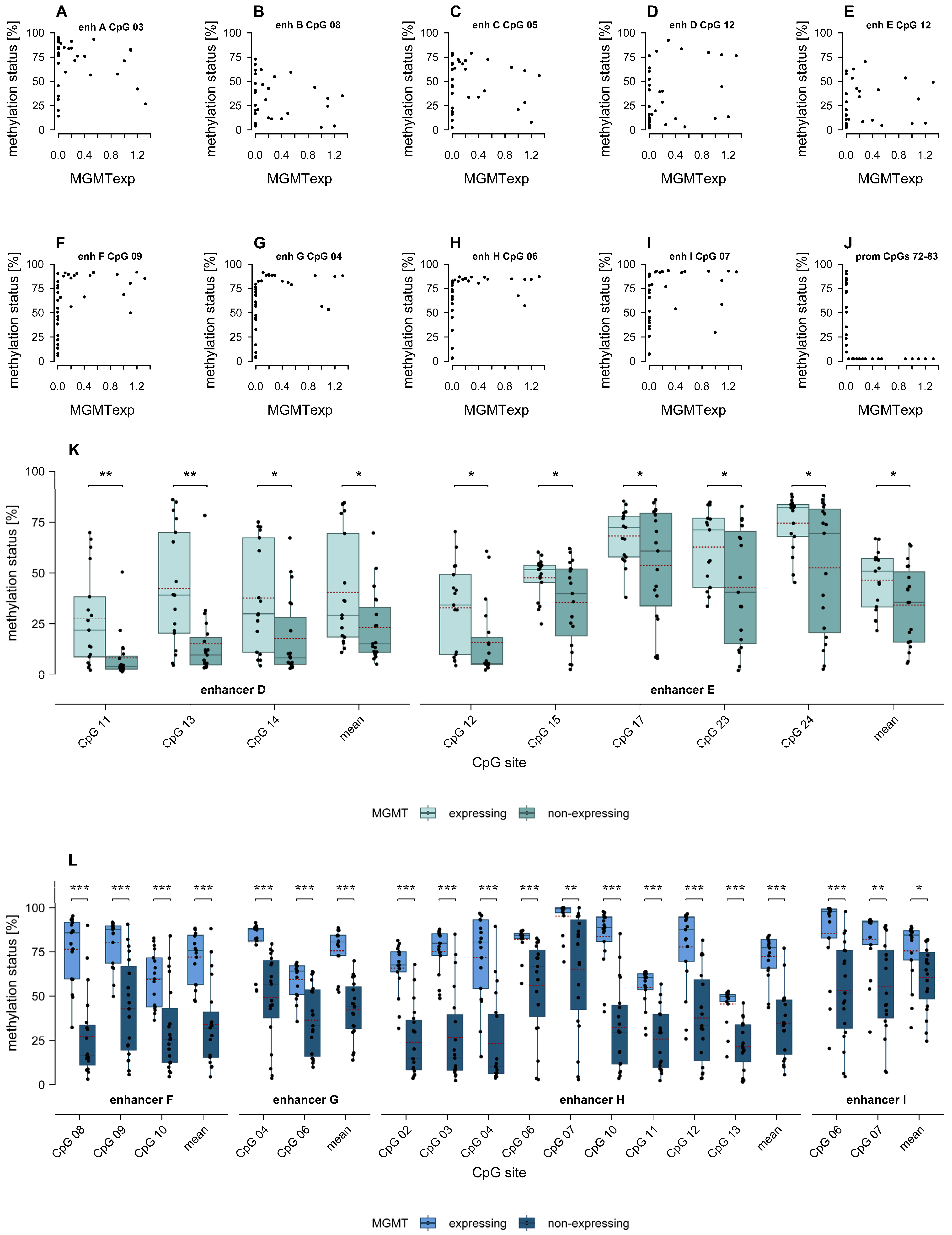
2.4. Association of MGMT Promoter and Enhancer Methylation with MGMT Protein Expression
2.5. Association of MGMT Enhancer Methylation with Clinical and Demographic Parameters
2.6. Association of MGMT Enhancer Methylation with Overall Survival, Progression-Free Survival and Long-Term Survival
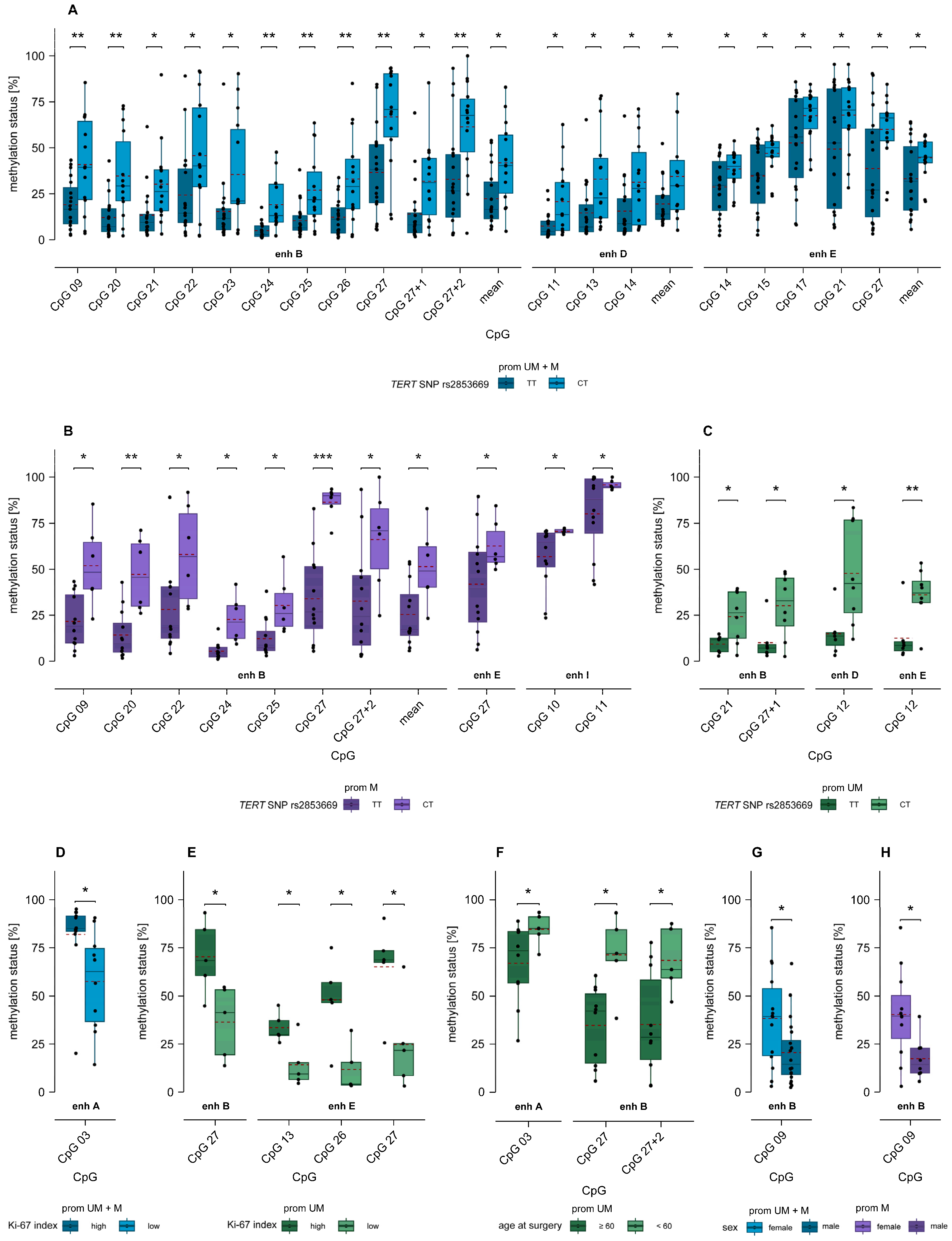
2.7. Association of MGMT Enhancer Methylation with TERT SNP rs2853669 and TERT Promoter Mutations C228T and C250T
2.8. Association of MGMT Enhancer Methylation with Ki-67 Index
2.9. Association of MGMT Enhancer Methylation with Age and Sex
3. Discussion
4. Materials and Methods
4.1. Samples and Cell Culturing
4.2. Determination of MGMT Protein Expression, Ki-67 Index, Genetic Variants, and Clinical Parameters
4.3. DNA Extraction and Bisulfite Conversion
4.4. Primer Design and PCR Conditions for DNA Methylation Analysis
4.5. PSQ of PCR Products
4.6. Data Analysis and Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CpG | CpG dinucleotide |
| EGFR | epidermal growth factor receptor |
| GB | Glioblastoma |
| GS | Gliosarcoma |
| HRM | High-resolution melting |
| IDH | Isocitrate dehydrogenase |
| KPS | Karnofsky Performance Score |
| MGMT | O6-methylguanine-DNA methyltransferase |
| NCBI | National Center for Biotechnology Information |
| OS | Overall survival |
| PCR | Polymerase chain reaction |
| PDCs | Patient-derived cells |
| PSQ | Pyrosequencing |
| TERT | Telomerase reverse transcriptase |
| TMZ | Temozolomide |
| WHO | World Health Organization |
References
- Gerson, S.L. MGMT: Its role in cancer aetiology and cancer therapeutics. Nat. Rev. Cancer 2004, 4, 296–307. [Google Scholar] [CrossRef] [PubMed]
- Christmann, M.; Verbeek, B.; Roos, W.P.; Kaina, B. O6-Methylguanine-DNA methyltransferase (MGMT) in normal tissues and tumors: Enzyme activity, promoter methylation and immunohistochemistry. BBA Rev. Cancer 2011, 1816, 179–190. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Salehi, F.; Scheithauer, B.W.; Rotondo, F.; Syro, L.V.; Kovacs, K. Role of MGMT in tumor development, progression, diagnosis, treatment and prognosis. Anticancer Res. 2009, 29, 3759–3768. [Google Scholar]
- Gerson, S.L. Clinical relevance of MGMT in the treatment of cancer. J. Clin. Oncol. 2002, 20, 2388–2399. [Google Scholar] [CrossRef]
- Bai, P.; Fan, T.; Sun, G.; Wang, X.; Zhao, L.; Zhong, R. The dual role of DNA repair protein MGMT in cancer prevention and treatment. DNA Repair 2023, 123, 103449. [Google Scholar] [CrossRef]
- Stupp, R.; Mason, W.P.; Van Den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.B.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO classification of tumors of the central nervous system: A summary. Neuro-Oncology 2021, 23, 1231–1251. [Google Scholar] [CrossRef]
- Ceccarelli, M.; Barthel, F.P.; Malta, T.M.; Sabedot, T.S.; Salama, S.R.; Murray, B.A.; Morozova, O.; Newton, Y.; Radenbaugh, A.; Pagnotta, S.M.; et al. Molecular Profiling Reveals Biologically Discrete Subsets and Pathways of Progression in Diffuse Glioma. Cell 2016, 164, 550–563. [Google Scholar] [CrossRef]
- Vinel, C.; Boot, J.; Jin, W.; Pomella, N.; Hadaway, A.; Mein, C.; Zabet, N.R.; Marino, S. Mapping chromatin remodelling in glioblastoma identifies epigenetic regulation of key molecular pathways and novel druggable targets. BMC Biol. 2025, 23, 26. [Google Scholar] [CrossRef]
- Prakash, V.; Gabrani, R. Epigenetic dysregulation in glioblastoma: Potential pathways to precision medicine. Neurogenetics 2025, 26, 5. [Google Scholar] [CrossRef]
- Mansouri, A.; Hachem, L.D.; Mansouri, S.; Nassiri, F.; Laperriere, N.J.; Xia, D.; Lindeman, N.I.; Wen, P.Y.; Chakravarti, A.; Mehta, M.P.; et al. MGMT promoter methylation status testing to guide therapy for glioblastoma: Refining the approach based on emerging evidence and current challenges. Neuro-Oncology 2019, 21, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Hegi, M.E.; Diserens, A.C.; Gorlia, T.; Hamou, M.F.; De Tribolet, N.; Weller, M.; Kros, J.M.; Hainfellner, J.A.; Mason, W.; Mariani, L.; et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 2005, 352, 997–1003. [Google Scholar] [CrossRef] [PubMed]
- Reifenberger, G.; Hentschel, B.; Felsberg, J.; Schackert, G.; Simon, M.; Schnell, O.; Westphal, M.; Wick, W.; Pietsch, T.; Loeffler, M.; et al. Predictive impact of MGMT promoter methylation in glioblastoma of the elderly. Int. J. Cancer 2012, 131, 1342–1350. [Google Scholar] [CrossRef] [PubMed]
- Spiegl-Kreinecker, S.; Pirker, C.; Filipits, M.; Lötsch, D.; Buchroithner, J.; Pichler, J.; Silye, R.; Weis, S.; Micksche, M.; Fischer, J.; et al. O6-Methylguanine DNA methyltransferase protein expression in tumor cells predicts outcome of temozolomide therapy in glioblastoma patients. Neuro-Oncology 2010, 12, 28–36. [Google Scholar] [CrossRef]
- Zappe, K.; Pühringer, K.; Pflug, S.; Berger, D.; Böhm, A.; Spiegl-Kreinecker, S.; Cichna-Markl, M. Association between MGMT Enhancer Methylation and MGMT Promoter Methylation, MGMT Protein Expression, and Overall Survival in Glioblastoma. Cells 2023, 12, 1639. [Google Scholar] [CrossRef]
- Charlet, J.; Duymich, C.E.; Lay, F.D.; Mundbjerg, K.; Dalsgaard Sørensen, K.; Liang, G.; Jones, P.A. Bivalent Regions of Cytosine Methylation and H3K27 Acetylation Suggest an Active Role for DNA Methylation at Enhancers. Mol. Cell 2016, 62, 422–431. [Google Scholar] [CrossRef]
- Bell, R.E.; Golan, T.; Sheinboim, D.; Malcov, H.; Amar, D.; Salamon, A.; Liron, T.; Gelfman, S.; Gabet, Y.; Shamir, R.; et al. Enhancer methylation dynamics contribute to cancer plasticity and patient mortality. Genome Res. 2016, 26, 601–611. [Google Scholar] [CrossRef]
- Calo, E.; Wysocka, J. Modification of Enhancer Chromatin: What, How, and Why? Mol. Cell 2013, 49, 825–837. [Google Scholar] [CrossRef]
- Ong, C.T.; Corces, V.G. Enhancer function: New insights into the regulation of tissue-specific gene expression. Nat. Rev. Genet. 2011, 12, 283–293. [Google Scholar] [CrossRef]
- Spitz, F. Gene regulation at a distance: From remote enhancers to 3D regulatory ensembles. Semin. Cell Dev. Biol. 2016, 57, 57–67. [Google Scholar] [CrossRef]
- Angeloni, A.; Bogdanovic, O. Enhancer DNA methylation: Implications for gene regulation. Essays Biochem. 2019, 63, 707–715. [Google Scholar] [CrossRef] [PubMed]
- Benetatos, L.; Vartholomatos, G. Enhancer DNA methylation in acute myeloid leukemia and myelodysplastic syndromes. Cell. Mol. Life Sci. 2018, 75, 1999–2009. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.W.; Shim, H.S.; Lee, C.Y.; Park, S.Y.; Hong, M.H.; Lee, I.; Kim, H.R. The importance of enhancer methylation for epigenetic regulation of tumorigenesis in squamous lung cancer. Exp. Mol. Med. 2022, 54, 12–22. [Google Scholar] [CrossRef]
- Kron, K.J.; Bailey, S.D.; Lupien, M. Enhancer alterations in cancer: A source for a cell identity crisis. Genome Med. 2014, 6, 77. [Google Scholar] [CrossRef]
- Zappe, K.; Pirker, C.; Miedl, H.; Schreiber, M.; Heffeter, P.; Pfeiler, G.; Hacker, S.; Haslik, W.; Spiegl-Kreinecker, S.; Cichna-Markl, M. Discrimination between 34 of 36 possible combinations of three C>T SNP genotypes in the MGMT promoter by high resolution melting analysis coupled with pyrosequencing using a single primer set. Int. J. Mol. Sci. 2021, 22, 12527. [Google Scholar] [CrossRef]
- Stone, A.; Zotenko, E.; Locke, W.J.; Korbie, D.; Millar, E.K.A.; Pidsley, R.; Stirzaker, C.; Graham, P.; Trau, M.; Musgrove, E.A.; et al. DNA methylation of oestrogen-regulated enhancers defines endocrine sensitivity in breast cancer. Nat. Commun. 2015, 6, 7558. [Google Scholar] [CrossRef]
- Zappe, K.; Pühringer, K.; Pflug, S.; Berger, D.; Weis, S.; Spiegl-Kreinecker, S.; Cichna-Markl, M. Association of MGMT Promoter and Enhancer Methylation with Genetic Variants, Clinical Parameters, and Demographic Characteristics in Glioblastoma. Cancers 2023, 15, 5777. [Google Scholar] [CrossRef]
- Aran, D.; Sabato, S.; Hellman, A. DNA methylation of distal regulatory sites characterizes dysregulation of cancer genes. Genome Biol. 2013, 14, R21. [Google Scholar] [CrossRef]
- Visel, A.; Minovitsky, S.; Dubchak, I.; Pennacchio, L.A. VISTA Enhancer Browser—A database of tissue-specific human enhancers. Nucleic Acids Res. 2007, 35, D88–D92. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, M.; Gan, H.; Wang, H.; Lee, J.H.; Fang, D.; Kitange, G.J.; He, L.; Hu, Z.; Parney, I.F.; et al. A novel enhancer regulates MGMT expression and promotes temozolomide resistance in glioblastoma. Nat. Commun. 2018, 9, 2949. [Google Scholar] [CrossRef]
- Carullo, N.V.N.; Day, J.J. Genomic enhancers in brain health and disease. Genes 2019, 10, 43. [Google Scholar] [CrossRef] [PubMed]
- Kyrchanova, O.; Georgiev, P. Mechanisms of enhancer-promoter interactions in higher eukaryotes. Int. J. Mol. Sci. 2021, 22, 671. [Google Scholar] [CrossRef] [PubMed]
- Anastasiadi, D.; Esteve-Codina, A.; Piferrer, F. Consistent inverse correlation between DNA methylation of the first intron and gene expression across tissues and species. Epigenet. Chromatin 2018, 11, 37. [Google Scholar] [CrossRef]
- Blattler, A.; Yao, L.; Witt, H.; Guo, Y.; Nicolet, C.M.; Berman, B.P.; Farnham, P.J. Global loss of DNA methylation uncovers intronic enhancers in genes showing expression changes. Genome Biol. 2014, 15, 469. [Google Scholar] [CrossRef]
- Wang, Q.; Xiong, F.; Wu, G.; Liu, W.; Chen, J.; Wang, B.; Chen, Y. Gene body methylation in cancer: Molecular mechanisms and clinical applications. Clin. Epigenet. 2022, 14, 154. [Google Scholar] [CrossRef]
- Ehrlich, K.C.; Paterson, H.L.; Lacey, M.; Ehrlich, M. DNA hypomethylation in intragenic and intergenic enhancer chromatin of muscle-specific genes usually correlates with their expression. Yale J. Biol. Med. 2016, 89, 441–455. [Google Scholar]
- Héberlé, É.; Bardet, A.F. Sensitivity of transcription factors to DNA methylation. Essays Biochem. 2019, 63, 727–741. [Google Scholar] [CrossRef]
- Farre, D. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 2003, 13, 3651–3653. [Google Scholar] [CrossRef]
- Madhugiri, V.S.; Moiyadi, A.V.; Shetty, P.; Gupta, T.; Epari, S.; Jalali, R.; Subeikshanan, V.; Dutt, A.; Sasidharan, G.M.; Roopesh Kumar, V.R.; et al. Analysis of Factors Associated with Long-Term Survival in Patients with Glioblastoma. World Neurosurg. 2021, 149, e758–e765. [Google Scholar] [CrossRef]
- Chehade, G.; Lawson, T.M.; Lelotte, J.; Daoud, L.; Di Perri, D.; Whenham, N.; Duprez, T.; Tajeddine, N.; Tissir, F.; Raftopoulos, C. Long-term survival in patients with IDH-wildtype glioblastoma: Clinical and molecular characteristics. Acta Neurochir. 2023, 165, 1075–1085. [Google Scholar] [CrossRef]
- Briceno, N.; Vera, E.; Komlodi-Pasztor, E.; Abdullaev, Z.; Choi, A.; Grajkowska, E.; Kunst, T.; Levine, J.; Lindsley, M.; Fernandez, K.; et al. Long-term survivors of glioblastoma: Tumor molecular, clinical, and imaging findings. Neuro-Oncol. Adv. 2024, 6, vdae019. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, T.; Wu, Z.; Zhang, K.; Li, W.; Yang, J.; Chen, C.; Chen, L.; Xing, J. Association between TERT rs2853669 polymorphism and cancer risk: A metaanalysis of 9, 157 cases and 11, 073 controls. PLoS ONE 2018, 13, e0191560. [Google Scholar] [CrossRef]
- Bell, R.J.A.; Rube, H.T.; Xavier-Magalhães, A.; Costa, B.M.; Mancini, A.; Song, J.S.; Costello, J.F. Understanding TERT promoter mutations: A common path to immortality. Mol. Cancer Res. 2016, 14, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Heidenreich, B.; Rachakonda, P.S.; Hemminki, K.; Kumar, R. TERT promoter mutations in cancer development. Curr. Opin. Genet. Dev. 2014, 24, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Alkhaibary, A.; Alassiri, A.H.; AlSufiani, F.; Alharbi, M.A. Ki-67 labeling index in glioblastoma; does it really matter? Hematol./Oncol. Stem Cell Ther. 2019, 12, 82–88. [Google Scholar] [CrossRef]
- Sun, X.; Kaufman, P.D. Ki-67: More than a proliferation marker. Chromosoma 2018, 127, 175–186. [Google Scholar] [CrossRef]
- Armocida, D.; Frati, A.; Salvati, M.; Santoro, A.; Pesce, A. Is Ki-67 index overexpression in IDH wild type glioblastoma a predictor of shorter Progression Free survival? A clinical and Molecular analytic investigation. Clin. Neurol. Neurosurg. 2020, 198, 106126. [Google Scholar] [CrossRef]
- Franzen, J.; Georgomanolis, T.; Selich, A.; Kuo, C.C.; Stöger, R.; Brant, L.; Mulabdić, M.S.; Fernandez-Rebollo, E.; Grezella, C.; Ostrowska, A.; et al. DNA methylation changes during long-term in vitro cell culture are caused by epigenetic drift. Commun. Biol. 2021, 4, 598. [Google Scholar] [CrossRef]
- Bork, S.; Pfister, S.; Witt, H.; Horn, P.; Korn, B.; Ho, A.D.; Wagner, W. DNA methylation pattern changes upon long-term culture and aging of human mesenchymal stromal cells. Aging Cell 2010, 9, 54–63. [Google Scholar] [CrossRef]
- Kozak, K.R.; Mahadevan, A.; Moody, J.S. Adult gliosarcoma: Epidemiology, natural history, and factors associated with outcome. Neuro-Oncology 2009, 11, 183–191. [Google Scholar] [CrossRef]
- Kavouridis, V.K.; Ligon, K.L.; Wen, P.Y.; Iorgulescu, J.B. Survival outcomes associated with MGMT promoter methylation and temozolomide in gliosarcoma patients. J. Neurooncol. 2022, 158, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Spiegl-Kreinecker, S.; Pirker, C.; Marosi, C.; Buchroithner, J.; Pichler, J.; Silye, R.; Fischer, J.; Micksche, M.; Berger, W. Dynamics of chemosensitivity and chromosomal instability in recurrent glioblastoma. Br. J. Cancer 2007, 96, 960–969. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wesseling, P.; Capper, D. WHO 2016 Classification of gliomas. Neuropathol. Appl. Neurobiol. 2018, 44, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Spiegl-Kreinecker, S.; Lötsch, D.; Ghanim, B.; Pirker, C.; Mohr, T.; Laaber, M.; Weis, S.; Olschowski, A.; Webersinke, G.; Pichler, J.; et al. Prognostic quality of activating TERT promoter mutations in glioblastoma: Interaction with the rs2853669 polymorphism and patient age at diagnosis. Neuro-Oncology 2015, 17, 1231–1240. [Google Scholar] [CrossRef]
- Genbank. Available online: https://www.ncbi.nlm.nih.gov/nucleotide/ (accessed on 22 July 2024).
- Genome Data Viewer. Available online: https://www.ncbi.nlm.nih.gov/gdv (accessed on 22 July 2024).
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.D.A.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Wei, T.; Simko, V. R package ‘corrplot’: Visualization of a Correlation Matrix (Version 0.92). 2021. Available online: https://CRAN.R-project.org/package=corrplot (accessed on 19 September 2024).
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; Available online: https://ggplot2.tidyverse.org (accessed on 10 September 2024).
- Kassambra, A. ggpubr: ‘ggplot2’ Based Publication Ready Plots. R-package version 0.6.0. 2023. Available online: https://CRAN.R-project.org/package=ggpubr (accessed on 10 September 2024).
- Revelle, W. psych: Procedures for Psychological, Psychometric, and Personality Research. Northwestern University, Evanston, Illinois. R package version 2.4.6. 2024. Available online: https://CRAN.R-project.org/package=ggsignif (accessed on 19 September 2024).
- Auguie, B. gridExtra: Miscellaneous functions for “grid” graphics (Version 2.3). 2017. Available online: https://CRAN.R-project.org/package=gridExtra (accessed on 20 September 2024).
- Robinson, D.; Hayes, A.; Couch, S. broom: Convert Statistical Objects into Tidy Tibbles. R Package Version 1.0.6. 2022. Available online: https://CRAN.R-project.org/package=broom (accessed on 10 September 2024).
- Chang, W. extrafont: Tools for Using Fonts. R Package Version 0.19. 2023. Available online: https://CRAN.R-project.org/package=extrafont (accessed on 20 September 2024).
- Therneau, T. A Package for Survival Analysis in R. R Package Version 3.5-8. 2024. Available online: https://CRAN.R-project.org/package=survival (accessed on 11 September 2024).


| Patient | Age [y] | Sex | KPS [%] | Ki-67 [%] | OS [m] | PFS [m] | MGMT Exp | TERT Prom | TERT rs2853669 |
|---|---|---|---|---|---|---|---|---|---|
| GB01 | 64 | f | 60 | n.s. | 7.43 | 4.00 | 0.00 | C250T | TT |
| GB02 | 85 | m | 40 | ≤50 (30) | 3.00 | n.s. | 0.00 | wt | TT |
| GB03 | 53 | f | 100 | n.s. | 52.50 | 4.50 | 0.00 | C228T | CT |
| GB04 | 67 | f | 70 | >50 (90) | 46.63 | 0.53 | 0.00 | C228T | TT |
| GB05 | 57 | f | 100 | ≤50 (30) | 10.50 | 6.00 | 0.00 | C228T | TT |
| GB06 | 46 | m | 90 | n.s. | 30.50 | 4.00 | 0.00 | C228T | TT |
| GB07 | 50 | f | 90 | n.s. | 27.40 | n.s. | 0.00 | C250T | n.s. |
| GB08 | 74 | f | 70 | ≤50 (30) | 7.79 | n.s. | 0.00 | C228T | CT |
| GB09 | 48 | m | 60 | n.s. | 1.55 | n.s. | 0.00 | wt | TT |
| GB10 | 64 | m | 80 | >50 | 11.70 | n.s. | 0.20 | C228T | CT |
| GB11 | 73 | f | 60 | >50 | 10.60 | n.s. | 0.16 | C228T | TT |
| GB12 | 44 | m | 80 | >50 | 13.00 | 9.00 | 1.10 | C250T | CT |
| GB13 | 65 | m | 90 | ≤50 (15) | 9.27 | n.s. | 1.20 | C228T | TT |
| GB14 | 69 | m | 90 | ≤50 (40) | 8.00 | n.s. | 0.04 | C250T | TT |
| GB15 | 73 | f | 100 | n.s. | 7.00 | 2.63 | 1.10 | C250T | CT |
| GB16 | 83 | m | 90 | ≤50 | 9.57 | 9.00 | 1.00 | C228T | CT |
| GB17 | 74 | m | 90 | n.s. | 5.19 | n.s. | 1.32 | C250T | CT |
| GB18 | 44 | m | 100 | n.s. | 23.15 | n.s. | 0.25 | wt | TT |
| GB19 | 48 | m | 100 | >50 (60) | 18.67 | 4.54 | 0.09 | C228T | CT |
| GB21 | 60 | m | 90 | ≤50 (40) | 13.00 | 3.00 | 0.40 | C228T | TT |
| GB22 | 53 | m | 70 | >50 (60) | 0.89 | n.s. | 0.54 | C250T | TT |
| GB23 | 47 | f | 70 | >50 (70) | 37.25 | 8.00 | 0.00 | C250T | CT |
| GB24 | 64 | m | 90 | ≤50 (40) | 7.73 | n.s. | 0.49 | C228T | CT |
| GB25 | 67 | f | 70 | >50 | 16.80 | 10.00 | 0.00 | C228T | TT |
| GB26 | 75 | f | 70 | ≤50 (40) | 8.22 | n.s. | 0.00 | C228T | CT |
| GB27 | 52 | f | 90 | >50 (70) | n.s. | n.s. | 0.00 | C250T | TT |
| GB28 | 63 | m | n.s. | >50 | 21.80 | 8.00 | 0.00 | C250T | CT |
| GB29 | 79 | f | 40 | >50 | 1.31 | n.s. | 0.00 | C228T | CT |
| GB30 | 58 | m | 90 | >50 | 16.00 | 4.31 | 0.00 | C228T | TT |
| GB31 | 58 | f | n.s. | >50 | 13.30 | 4.00 | 0.00 | C228T | TT |
| GB32 | 71 | m | n.s. | ≤50 (20) | 1.25 | n.s. | 0.00 | C228T | TT |
| GB33 | 57 | m | n.s. | n.s. | 12.70 | 6.00 | 0.20 | C250T | CT |
| GB34 | 53 | m | n.s. | n.s. | 32.30 | 22.00 | 0.00 | C228T | TT |
| GB35 | 64 | m | 40 | n.s. | 10.60 | n.s. | 0.00 | C228T | TT |
| GB36 | 62 | m | 50 | ≤50 (25) | 1.48 | n.s. | 0.90 | wt | TT |
| GB37 | 63 | m | 80 | >50 (60) | 19.50 | 9.01 | 0.11 | wt | TT |
| GB38 | 76 | m | 30 | ≤50 (20) | 4.44 | n.s. | 0.29 | wt | CT |
| GS01 | 43 | f | 80 | ≤50 (30) | 9 | n.s. | 0.00 | C250T | TT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pühringer, K.; Czarda, P.; Iluca, S.; Zappe, K.; Weis, S.; Spiegl-Kreinecker, S.; Cichna-Markl, M. Association of Intergenic and Intragenic MGMT Enhancer Methylation with MGMT Promoter Methylation, MGMT Protein Expression and Clinical and Demographic Parameters in Glioblastoma. Int. J. Mol. Sci. 2025, 26, 3390. https://doi.org/10.3390/ijms26073390
Pühringer K, Czarda P, Iluca S, Zappe K, Weis S, Spiegl-Kreinecker S, Cichna-Markl M. Association of Intergenic and Intragenic MGMT Enhancer Methylation with MGMT Promoter Methylation, MGMT Protein Expression and Clinical and Demographic Parameters in Glioblastoma. International Journal of Molecular Sciences. 2025; 26(7):3390. https://doi.org/10.3390/ijms26073390
Chicago/Turabian StylePühringer, Katharina, Philipp Czarda, Sebastian Iluca, Katja Zappe, Serge Weis, Sabine Spiegl-Kreinecker, and Margit Cichna-Markl. 2025. "Association of Intergenic and Intragenic MGMT Enhancer Methylation with MGMT Promoter Methylation, MGMT Protein Expression and Clinical and Demographic Parameters in Glioblastoma" International Journal of Molecular Sciences 26, no. 7: 3390. https://doi.org/10.3390/ijms26073390
APA StylePühringer, K., Czarda, P., Iluca, S., Zappe, K., Weis, S., Spiegl-Kreinecker, S., & Cichna-Markl, M. (2025). Association of Intergenic and Intragenic MGMT Enhancer Methylation with MGMT Promoter Methylation, MGMT Protein Expression and Clinical and Demographic Parameters in Glioblastoma. International Journal of Molecular Sciences, 26(7), 3390. https://doi.org/10.3390/ijms26073390






