Single-Nucleotide Polymorphisms Related to Multiple Myeloma Risk: A Systematic Review and Meta-Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Protocol and Registration
2.2. Search Strategy
2.3. Study Selection
2.4. Data Extraction
2.5. Quality Assessment
2.6. Data Analysis
3. Results
3.1. Search Results
3.2. Characteristics of Studies, Genes/SNPs, and Participants
3.3. Quality Assessment
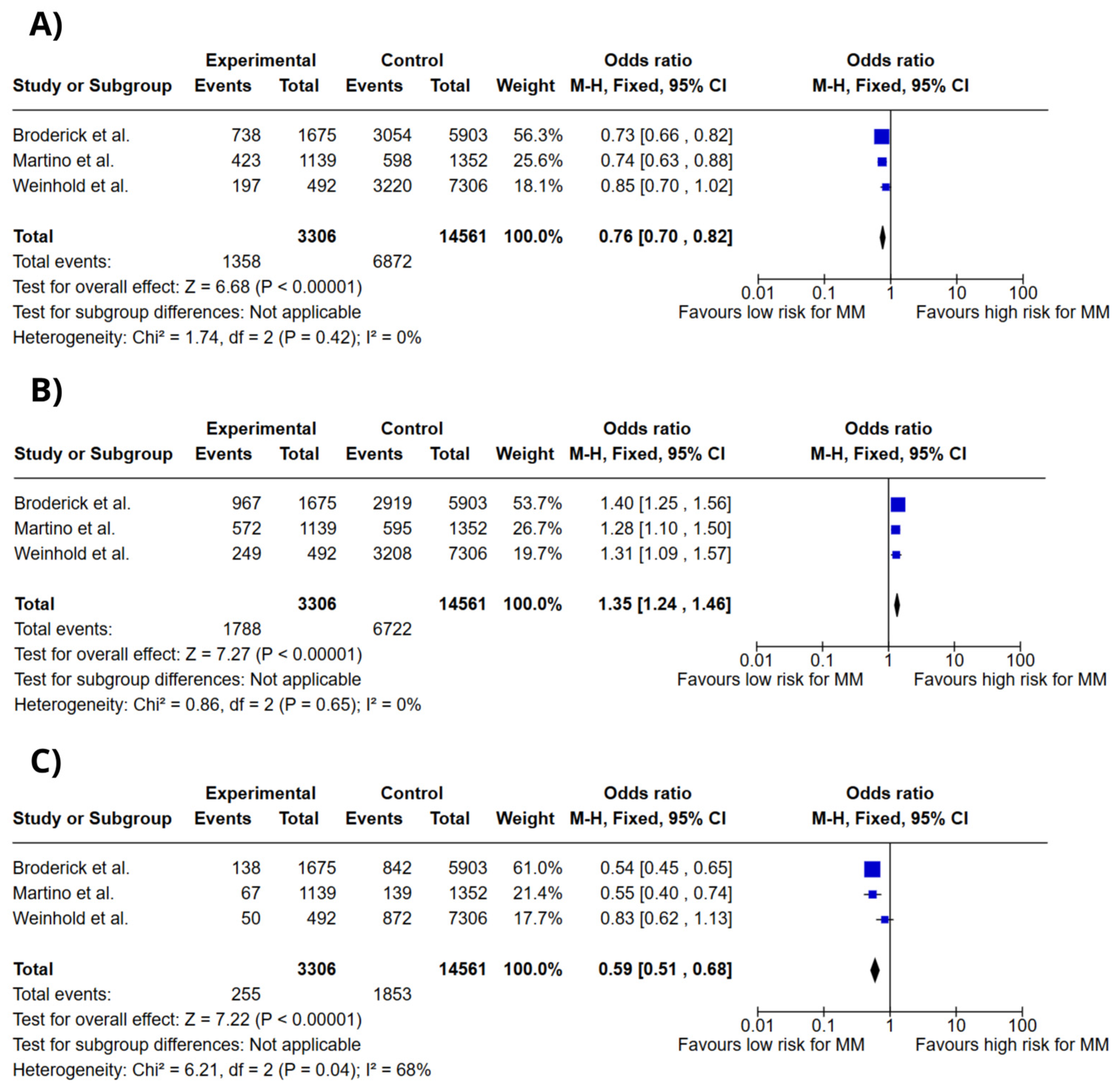
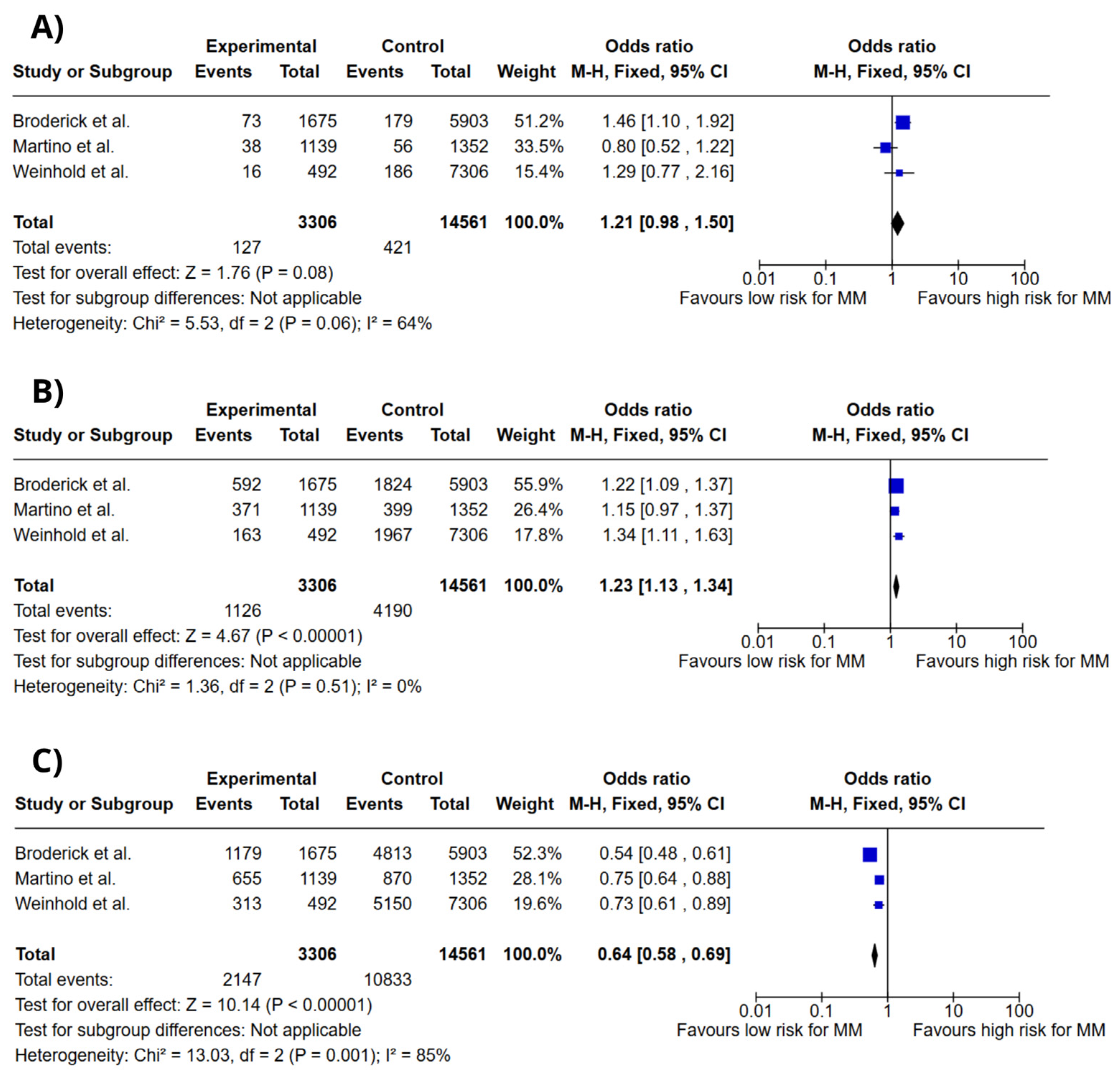
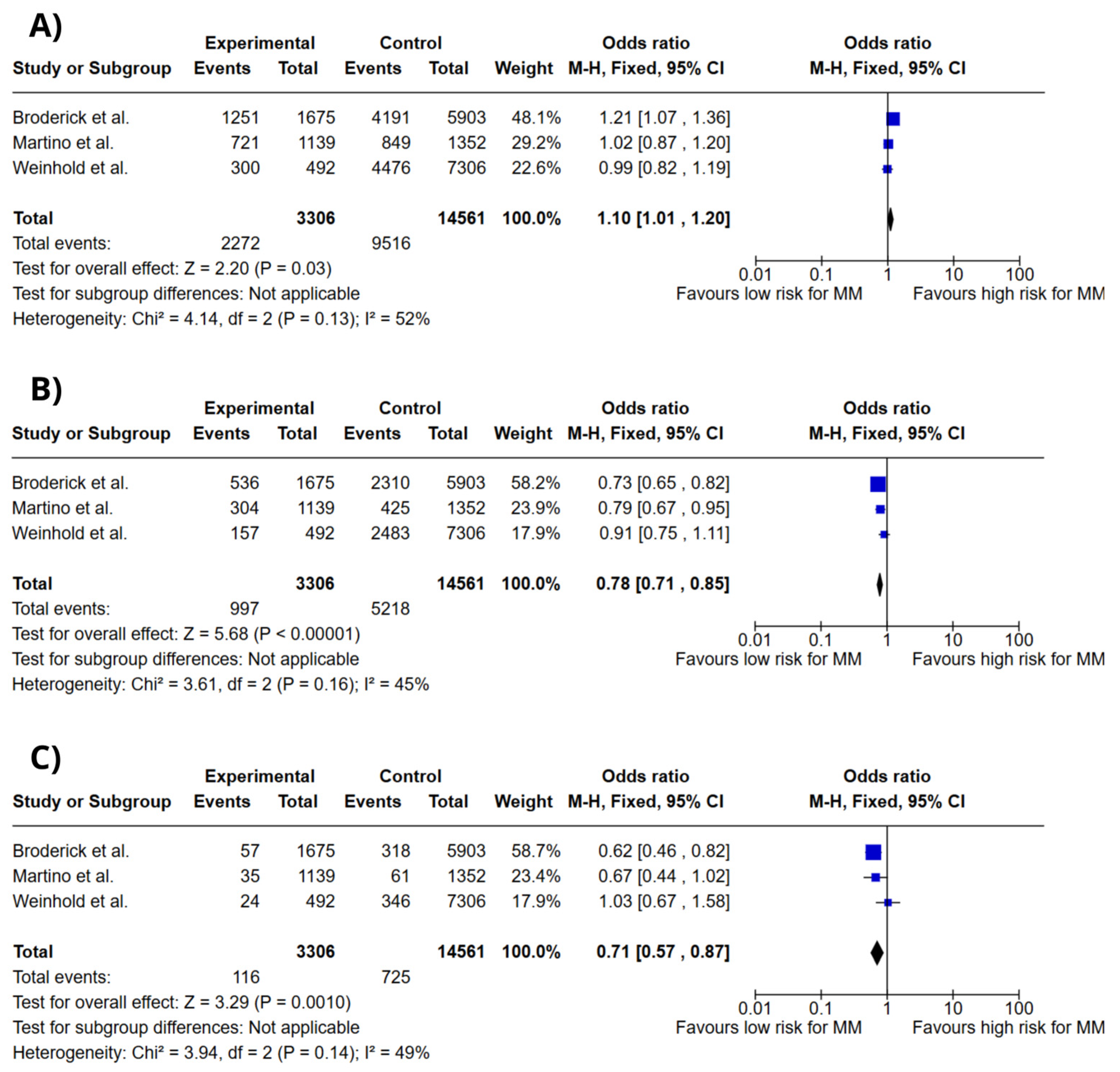
3.4. Meta-Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MM | Multiple myeloma |
| SNPs | Single-nucleotide polymorphisms |
| NR | Not reported |
References
- Jurczyszyn, A.; Hutch, R.; Waszczuk-Gajda, A.; Suska, A.; Krzanowska, K.; Vesole, D.H. Monoclonal gammopathies of undetermined significance and smoldering myeloma. Acta Haematol. Pol. 2020, 51, 193–202. [Google Scholar] [CrossRef]
- International Agency for Research on Cancer. GLOBOCAN 2022: Estimated Cancer Incidence, Mortality and Prevalence Worldwide; IARCP: Lyon, France, 2022. [Google Scholar]
- Cowan, A.J.; Green, D.J.; Kwok, M.; Lee, S.; Coffey, D.G.; Holmberg, L.A.; Tuazon, S.; Gopal, A.K.; Libby, E.N. Diagnosis and management of multiple myeloma: A review. JAMA 2022, 327, 464–477. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, H.; Lan, H.; Wu, J.; Xiao, Y. CAR-T cell therapy in multiple myeloma: Current limitations and potential strategies. Front. Immunol. 2023, 14, 1101495. [Google Scholar] [CrossRef]
- Rajkumar, S.V. Multiple myeloma: 2024 update on diagnosis, risk-stratification, and management. Am. J. Hematol. 2024, 99, 1802–1824. [Google Scholar] [CrossRef] [PubMed]
- Karunarathna, I.; Gunawardana, K.; Aluthge, P.; Gunasena, P.; Gunathilake, S. Advances in Understanding and Managing Multiple Myeloma: A Comprehensive Review. Uva Clin. Med. 2024, 97, 1–15. [Google Scholar]
- Li, Z.; Zhao, H.; Li, Z.; He, Y. Correlation analysis of laboratory indicators, genetic abnormalities and staging in patients with newly diagnosed multiple myeloma. Medicine 2024, 103, e40710. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wells, G.A.; Shea, B.; O’Connell, D.; Peterson, J.; Welch, V.; Losos, M.; Tugwell, P. The Newcastle-Ottawa Scale (NOS) for Assessing the Quality of Nonrandomised Studies in Meta-Analyses; Ottawa Health Research Institute: Ottawa, ON, Canada, 2014. [Google Scholar]
- Little, J.; Higgins, J.; Ioannidis, J.; Moher, D.; Gagnon, F.; Vandenbroucke, J.; Zeggini, E. Strengthening the reporting of genetic association studies (STREGA)—An extension of the STROBE statement. Genet. Epidemiol. 2009, 33, 1061–1074. [Google Scholar] [CrossRef]
- 1000 Genomes Project Consortium. A Global Reference for Human Genetic Variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Landrum, M.J.; Lee, J.M.; Benson, M.; Brown, G.R.; Chao, C.; Chitipiralla, S.; Gu, B.; Hart, J.; Hoffman, D.; Jang, W.; et al. ClinVar: Improving Access to Variant Interpretations and Supporting Evidence. Nucleic Acids Res. 2018, 46, D1062–D1067. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Broderick, P.; Chubb, D.; Johnson, D.C.; Weinhold, N.; Försti, A.; Lloyd, A.; Olver, B.; Ma, Y.; Dobbins, S.E.; Walker, B.A.; et al. Common variation at 3p22.1 and 7p15.3 influences multiple myeloma risk. Nat. Genet. 2011, 44, 58–61. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Rui, H.; Liu, Y.; Lin, M.; Zheng, X. Vitamin D receptor gene polymorphism is associated with multiple myeloma. J. Cell Biochem. 2019, 120, 10147–10153. [Google Scholar] [CrossRef]
- Kumar, R.; Himani Gupta, N.; Singh, V.; Kumar, V.; Haq, A.; Mirza, A.A.; Sharma, A. Unveiling molecular associations of polymorphic variants of VDR gene (FokI, BsmI and ApaI) in multiple myeloma patients of Indian population. J. Steroid Biochem. Mol. Biol. 2020, 199, 105588. [Google Scholar] [CrossRef] [PubMed]
- Martino, A.; Campa, D.; Jamroziak, K.; Reis, R.M.; Sainz, J.; Buda, G.; García-Sanz, R.; Lesueur, F.; Marques, H.; Moreno, V.; et al. Impact of polymorphic variation at 7p15.3, 3p22.1 and 2p23.3 loci on risk of multiple myeloma. Br. J. Haematol. 2012, 158, 805–809. [Google Scholar] [CrossRef] [PubMed]
- Weinhold, N.; Johnson, D.C.; Rawstron, A.C.; Försti, A.; Doughty, C.; Vijayakrishnan, J.; Broderick, P.; Dahir, N.B.; Begum, D.B.; Hosking, F.J.; et al. Inherited genetic susceptibility to monoclonal gammopathy of unknown significance. Blood 2014, 123, 2513–2517, quiz 2593. [Google Scholar] [CrossRef] [PubMed]
- Erickson, S.W.; Raj, V.R.; Stephens, O.W.; Dhakal, I.; Chavan, S.S.; Sanathkumar, N.; Coleman, E.A.; Lee, J.Y.; Goodwin, J.A.; Apewokin, S.; et al. Genome-wide scan identifies variant in 2q12.3 associated with risk for multiple myeloma. Blood 2014, 124, 2001–2003. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hosgood III, H.D.; Baris, D.; Zhang, Y.; Zhu, Y.; Zheng, T.; Yeager, M.; Welch, R.; Zahm, S.; Chanock, S.; Rothman, N.; et al. Caspase polymorphisms and genetic susceptibility to multiple myeloma. Hematol. Oncol. 2008, 26, 148–151. [Google Scholar] [CrossRef] [PubMed]
- Karabon, L.; Pawlak-Adamska, E.; Tomkiewicz, A.; Jedynak, A.; Kielbinski, M.; Woszczyk, D.; Potoczek, S.; Jonkisz, A.; Kuliczkowski, K.; Frydecka, I. Variations in suppressor molecule CTLA-4 gene are related to susceptibility to multiple myeloma in a Polish population. Pathol. Oncol. Res. 2012, 18, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Huo, J.; Shi, J.; Yuan, Z.; Zhang, C.; Fu, W.; Jiang, H.; Yi, Q.; Hou, J. Polymorphisms of nuclear factor-κB family genes are associated with development of multiple myeloma and treatment outcome in patients receiving bortezomib-based regimens. Haematologica 2011, 96, 729–737. [Google Scholar] [CrossRef] [PubMed]
- Campa, D.; Martino, A.; Sainz, J.; Buda, G.; Jamroziak, K.; Weinhold, N.; Reis, R.M.V.; García-Sanz, R.; Jurado, M.; Ríos, R.; et al. Comprehensive investigation of genetic variation in the 8q24 region and multiple myeloma risk in the IMMEnSE consortium. Br. J. Haematol. 2012, 156, 507–514. [Google Scholar] [CrossRef] [PubMed]
- Chubb, D.; Weinhold, N.; Broderick, P.; Chen, B.; Johnson, D.C.; Försti, A.; Vijayakrishnan, J.; Migliorini, G.; Dobbins, S.E.; Holroyd, A.; et al. Common variation at 3q26.2, 6p21.33, 17p11.2, and 22q13.1 influences multiple myeloma risk. Nat. Genet. 2013, 45, 1221–1225. [Google Scholar] [CrossRef] [PubMed]
- Martino, A.; Campa, D.; Jurczyszyn, A.; Martínez-López, J.; Moreno, M.J.; Varkonyi, J.; Dumontet, C.; García-Sanz, R.; Gemignani, F.; Jamroziak, K.; et al. Genetic variants and multiple myeloma risk: IMMEnSE validation of the best reported associations—An extensive replication of the associations from the candidate gene era. Cancer Epidemiol. Biomarkers Prev. 2014, 23, 670–674. [Google Scholar] [CrossRef] [PubMed]
- Faber, E.W.; Lourenço, G.J.; Ortega, M.M.; Lorand-Metze, I.; De Souza, C.A.; Lima, C.S.P. Polymorphisms of VEGF, GSTM1 and GSTT1 genes in multiple myeloma risk. Hematol. Oncol. 2011, 29, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Vangsted, A.J.; Nielsen, K.R.; Klausen, T.W.; Haukaas, E.; Tjønneland, A.; Vogel, U. A functional polymorphism in the promoter region of the IL1B gene is associated with risk of multiple myeloma. Hematol. Oncol. 2012, 30, 179–183. [Google Scholar] [CrossRef] [PubMed]
- Scionti, F.; Agapito, G.; Caracciolo, D.; Riillo, C.; Grillone, K.; Cannataro, M.; Di Martino, M.T.; Tagliaferri, P.; Tassone, P.; Arbitrio, M. Risk Alleles for Multiple Myeloma Susceptibility in ADME Genes. Cells 2022, 11, 189. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wang, Q.; Wang, Y.; Wu, J.; Xie, X.; Qin, H.; Huang, C.; Li, Z.; Ling, Z.; Li, R. Association between BCL2 interacting protein 3 like (BNIP3L) genetic polymorphisms and the risk of multiple myeloma in China. Hematology 2024, 29, 2367918. [Google Scholar] [CrossRef] [PubMed]
- Niebudek, K.; Balcerczak, E.; Mirowski, M.; Żebrowska, M. Association of ABCB1 T-129C polymorphism and multiple myeloma risk in Polish population. Pol. J. Pathol. 2018, 69, 405–409. [Google Scholar] [CrossRef] [PubMed]
- Peng, M.; Zhao, G.; Yang, F.; Cheng, G.; Huang, J.; Qin, X.; Liu, Y.; Wang, Q.; Li, Y.; Qin, D. NCOA1 is a novel susceptibility gene for multiple myeloma in the Chinese population: A case-control study. PLoS ONE 2017, 12, e0173298. [Google Scholar] [CrossRef]
- Li, B.; Liu, C.; Cheng, G.; Peng, M.; Qin, X.; Liu, Y.; Li, Y.; Qin, D. LRP1B Polymorphisms Are Associated with Multiple Myeloma Risk in a Chinese Han Population. J. Cancer 2019, 10, 577–582. [Google Scholar] [CrossRef]
- Szemraj-Rogucka, Z.; Szemraj, J.; Grzybowska-Izydorczyk, O.; Robak, T.; Jamroziak, K. CD38 Gene Polymorphisms and Genetic Predisposition to Multiple Myeloma. Acta Haematol. Pol. 2013, 44, 58–62. [Google Scholar] [CrossRef]
- Brito, A.B.C.; Lourenço, G.J.; Oliveira, G.B.; De Souza, C.A.; Vassallo, J.; Lima, C.S.P. Associations of VEGF and VEGFR2 Polymorphisms with Increased Risk and Aggressiveness of Multiple Myeloma. Ann. Hematol. 2014, 93, 1363–1369. [Google Scholar] [CrossRef] [PubMed]
- Sodeifian, F.; Samieefar, N.; Shahkarami, S.; Rayzan, E.; Seyedpour, S.; Rohlfs, M.; Klein, C.; Babaie, D.; Rezaei, N. DNAH11 and a Novel Genetic Variant Associated with Situs Inversus: A Case Report and Review of the Literature. Case Rep. Med. 2023, 2023, 8436715. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Schultz, R.; Elenius, V.; Lukkarinen, H.; Saarela, T. Two novel mutations in the DNAH11 gene in primary ciliary dyskinesia (CILD7) with considerable variety in the clinical and beating cilia phenotype. BMC Med. Genet. 2020, 2, 237. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Wu, Y.; Yu, J.; Qiu, Y.; Yi, T.; Luo, C.; Zhang, J.; Lu, G.; Li, X.; Xiong, F.; et al. Impact of genomic and epigenomic alterations of multigene on a multicancer pedigree. Cancer Med. 2024, 13, e7394. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Verma, S.; Bakshi, D.; Sharma, V.; Sharma, I.; Shah, R.; Bhat, A.; Bhat, G.R.; Sharma, B.; Wakhloo, A.; Kaul, S.; et al. Genetic variants of DNAH11 and LRFN2 genes and their association with ovarian and breast cancer. Int. J. Gynaecol. Obstet. 2020, 148, 118–122. [Google Scholar] [CrossRef] [PubMed]
- Taddesse-Heath, L.; Meloni-Ehrig, A.; Scheerle, J.; Kelly, J.C.; Jaffe, E.S. Plasmablastic lymphoma with MYC translocation: Evidence for a common pathway in the generation of plasmablastic features. Mod. Pathol. 2010, 23, 991–999. [Google Scholar] [CrossRef]
- Luo, S.; Zheng, N.; Lang, B. ULK4 in Neurodevelopmental and Neuropsychiatric Disorders. Front. Cell Dev. Biol. 2022, 10, 873706. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mecklenburg, N.; Kowalczyk, I.; Witte, F.; Görne, J.; Laier, A.; Mamo, T.M.; Gonschior, H.; Lehmann, M.; Richter, M.; Sporbert, A.; et al. Identification of disease-relevant modulators of the SHH pathway in the developing brain. Development 2021, 148, dev199307. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, A.J.; Lee, A.M.; McDonnell, S.K.; Cerhan, J.R.; Liebow, M.; Larson, D.R.; Colby, C.L.; Norman, A.D.; Kyle, R.A.; Kumar, S.; et al. Single-nucleotide polymorphism rs1052501 associated with monoclonal gammopathy of undetermined significance and multiple myeloma. Leukemia 2013, 27, 515–516. [Google Scholar] [CrossRef]
- Went, M.; Kinnersley, B.; Sud, A.; Johnson, D.C.; Weinhold, N.; Försti, A.; van Duin, M.; Orlando, G.; Mitchell, J.S.; Kuiper, R.; et al. Transcriptome-wide association study of multiple myeloma identifies candidate susceptibility genes. Hum. Genom. 2019, 13, 37. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Blake, D.J.; Nawrotzki, R.; Loh, N.Y.; Górecki, D.C.; Davies, K.E. beta-dystrobrevin, a member of the dystrophin-related protein family. Proc. Natl. Acad. Sci. USA 1998, 95, 241–246. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Quaranta, M.T.; Spinello, I.; Paolillo, R.; Macchia, G.; Boe, A.; Ceccarini, M.; Labbaye, C.; Macioce, P. Identification of β-Dystrobrevin as a Direct Target of miR-143: Involvement in Early Stages of Neural Differentiation. PLoS ONE 2016, 11, e0156325. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sun, J.; Li, Y.; Bie, B.; Tian, H.; Li, J.; Yang, L.; Zhou, Z.; Mu, Y.; Li, Z. Dystrobrevin beta is a promising prognostic biomarker and therapeutic target for hepatocellular carcinoma. Am. J. Transl. Res. 2024, 16, 6072–6096. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lyu, C.; Yin, X.; Li, Z.; Wang, T.; Xu, R. Vitamin D receptor gene polymorphisms and multiple myeloma: A meta-analysis. Clin. Exp. Med. 2024, 24, 118. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bouillon, R.; Carmeliet, G.; Verlinden, L.; van Etten, E.; Verstuyf, A.; Luderer, H.F.; Lieben, L.; Mathieu, C.; Demay, M. Vitamin D and human health: Lessons from vitamin D receptor null mice. Endocr. Rev. 2008, 29, 726–776. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ismail, N.H.; Mussa, A.; Al-Khreisat, M.J.; Mohamed Yusoff, S.; Husin, A.; Johan, M.F.; Islam, M.A. The Global Prevalence of Vitamin D Deficiency and Insufficiency in Patients with Multiple Myeloma: A Systematic Review and Meta-Analysis. Nutrients 2023, 15, 3227. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]


| Authors, Year | Country | Participants (Male/Female) | Mean Age | Mean Age at Diagnosis (Years) | Genotyping Method | MM Histologically Confirmed | Ethnicity | Main Results |
|---|---|---|---|---|---|---|---|---|
| Broderick et al., 2011 [13] | United Kingdom Germany TOTAL | 1371 (819/552) 384 (229/155) 1675 (1048/627) | NR NR NR | 64.1 ± 10.3 54.5 ± 8.0 NR | Microarray | Yes | European | SNP rs1052501within gene ULK4 and SNP rs4487645 were associated with higher risk of MM |
| Martino et al., 2012 [16] | Germany | 1139 | NR | NR | PCR | Yes | European | SNPs rs4487645 and rs6746082 were linked to higher risk of MM |
| Erickson et al., 2014 [18] | United States of America | 1202 | 57.7 | 61.0 | PCR | NR | European/North American | The SNPs rs12614346 and rs73486634 were linked to an increased risk of MM. |
| Hosgood III et al., 2008 [19] | United States of America | 128 (0/128) | NR | NR | qPCR | Yes | North American | CASP3 and CASP9 polymorphisms were associated with decreased risk of MM. |
| Karabon et al., 2012 [20] | Poland | 200 (94/106) | 67 ± 10.9 | 63.5 ± 11.2 | PCR-RFLP | Yes | Danish | Three CTLA variations were identified more frequently in MM patients compared to the control group. Five CTLA polymorphisms were associated with higher risk of MM. |
| Juan Du et al., 2011 [21] | China | 252 (161/91) | 58 | NR | RT-PCR | NR | Chinese | TRAF3 rs12147254 variant is linked to a reduced risk of developing multiple myeloma, while the rs11160707 genotype has been correlated with improved progression-free survival. |
| Campa et al., 2011 [22] | German | 1188 (643/545) | 58.62 ± 10.15 | NR | castPCR | Yes | European | Rs2456449 was correlated with the risk of multiple myeloma. |
| Chubb et al., 2013 [23] | German/United Kingdom | UK-replication-1—812 (412/400) UK-replication-2—396 (181/215) German-replication—1149 (676/473) | NR NR NR | NR 66.0 57.6 | PCR KASPar | Yes | European | The variants rs10936599, rs2285803, rs4273077, and rs877529 were correlated with the risk of multiple myeloma. |
| Martino et al., 2014 [24] | Italy/Poland/Spain/France/Portugal/Hungary/Denmark | 1498 (756/742) | 60.9 ± 10.6 | NR | castPCR | Yes | European | A possible association between the SNP rs2227667 (SERPINE1) and the risk of multiple myeloma in women has been identified. |
| Faber et al., 2011 [25] | Brazil | 150 (81/69) | 54 | NR | mPCR/ RT-PCR | NR | White people/African-Brazilians | An increased risk of MM was observed in individuals with the VEGF CC genotype combined with GSTM1 undeleted and GSTT1 null genotypes. |
| Vangsted et al., 2012 [26] | Denmark | 348 | NR | NR | ABI 7500 or HT7900 systems | NR | Danish | Association between IL1B expression and risk of MM. |
| Scionti et al., 2022 [27] | Italy | 64 | NR | NR | DMET Console software version 1.1 | NR | Italian | Polymorphism in ADME genes were associated with susceptibility for MM. |
| Wang et al., 2024 [28] | China | 94 (49/45) | 59.25 ± 9.36 | NR | UE Blood genomic DNA preparation kit | NR | Chinese | Polymorphisms in BNIP3L were associated with susceptibility and prognosis of MM in chinese population. |
| Hongbing Rui et al., 2019 [14] | China | 40 (23/17) | 59.5 | NR | PCR | NR | Chinese | Association between polymorphisms in VDR gene with increased risk of MM. |
| Niebudek et al., 2018 [29] | Poland | 91 (41/50) | 63 | NR | PCR-RFLP | NR | Polish | Polymorphism T-129C in ABCB1 gene was not associated with the increased risk of MM development in the polish population. |
| Kumar et al., 2020 [15] | India | 75 (54/21) | 57 | NR | locus-specific PCR | NR | Indian | FokI, ApaI, and BsmI genotypes were associated with risk of MM in the Indian population. |
| Peng et al., 2017 [30] | China | 827 (473/354) | 59.35 ± 9.95 | over 60 years | locus-specific PCR | yes | Chinese | rs79480871 were associated with susceptibility for MM. |
| Li et al., 2019 [31] | China | 739 (415/324) | 59.27 ± 10.11 | older than 65 years | locus-specific PCR | Yes | Chinese | Association for susceptibility in MM by rs61070260 in LRP1B. |
| Szemraj-Rogucka, 2013 [32] | Poland | 174 (93/81) | 61 | 68 | PCR-RFLP | NR | Caucasian | Predisposition of MM were associated for rs6449182 of CD38. |
| Brito et al., 2014 [33] | Brazil | 192 (99/93) | 62 | over 60 years | RT-PCR | NR | Caucasian and Non-Caucasian | Aggressiveness and risk of MM. were associated with VEGF and VEGFR2. |
| Weinhold et al., 2014 [17] | United Kingdom/Germany | 492 (234/258) | 67.5 | NR | Kompetitive Allele Specific Polymerase (KASP) chain reaction | Yes | European | rs1052501, rs2285803, rs4487645, and rs4273077 increased MM risk. |
| SNPs | Gene | Location | Alleles | Ancestral | Functional Consequence | Clinical Significance |
|---|---|---|---|---|---|---|
| rs1544410 | VDR | Chromosome 12:47846052 | C/A/G/T | C | Intron variant | Benign, likely risk allele |
| rs6746082 | DTNB | Chromosome 2:25436375 | A/C/G/T | A | Intron variant | NR |
| rs1052501 | ULK4 | Chromosome 3:41883906 | C/G/T | T | Missense variant | Benign |
| rs4487645 | DNAH11 | Chromosome 7:21898622 | C/A/T | C | Intron variant | NR |
| Studies | Selection | Item 2 | Item 3 | Item 4 | Comparability | Item 1b | Exposure | Item 2 | Item 3 | Total Score |
|---|---|---|---|---|---|---|---|---|---|---|
| Item 1 | Item 1a | Item 1 | ||||||||
| Broderick et al., 2011 [13] | * | * | * | * | * | * | * | 7 | ||
| Hongbing Rui et al., 2019 [14] | * | * | * | * | * | * | * | 7 | ||
| Kumar et al., 2020 [15] | * | * | * | * | * | * | * | 7 | ||
| Martino et al., 2012 [16] | * | * | * | * | * | * | * | 7 | ||
| Weinhold et al., 2014 [17] | * | * | * | * | * | * | * | 7 |
| Studies | Description of Genotyping Methods and Errors | Description of Modeling Population Stratification | Description of Modeling Haplotype Variation | Hardy–Weinberg Equilibrium Was Considered | Statement of Whether the Study Is the First Report of a Genetic Association, a Replication Effort, or Both | Score | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Genotyping Methods and Platforms | Error Rates and Call Rates | Genotyping in Batches | Laboratory/Center Where Genotyping Was Performed | The Number of Individuals Successful in Genotyping | ||||||
| Broderick et al., 2011 [13] | Yes | Yes | Yes | Yes | Yes | Yes | No | Yes | No | 7 |
| Hongbing Rui et al., 2019 [14] | Yes | Yes | No | Yes | Yes | Yes | Yes | Yes | No | 7 |
| Kumar et al., 2020 [15] | Yes | Yes | Yes | Yes | Yes | Yes | No | Yes | No | 7 |
| Martino et al., 2012 [16] | Yes | Yes | Yes | Yes | Yes | Yes | No | Yes | Yes | 8 |
| Weinhold et al., 2014 [17] | Yes | Yes | Yes | Yes | Yes | Yes | No | Yes | No | 7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gilioli da Costa Nunes, G.; Cezar Aquino de Moraes, F.; Carvalho de Almeida, A.B.; Goes Costa, F.; Duarte de Andrade Junior, L.F.; Sabino Hupp, M.V.; Rotondano Assunção, R.; Rodrigues Fernandes, M.; Emanuel Batista dos Santos, S.; Pereira Carneiro dos Santos, N. Single-Nucleotide Polymorphisms Related to Multiple Myeloma Risk: A Systematic Review and Meta-Analysis. Int. J. Mol. Sci. 2025, 26, 3369. https://doi.org/10.3390/ijms26073369
Gilioli da Costa Nunes G, Cezar Aquino de Moraes F, Carvalho de Almeida AB, Goes Costa F, Duarte de Andrade Junior LF, Sabino Hupp MV, Rotondano Assunção R, Rodrigues Fernandes M, Emanuel Batista dos Santos S, Pereira Carneiro dos Santos N. Single-Nucleotide Polymorphisms Related to Multiple Myeloma Risk: A Systematic Review and Meta-Analysis. International Journal of Molecular Sciences. 2025; 26(7):3369. https://doi.org/10.3390/ijms26073369
Chicago/Turabian StyleGilioli da Costa Nunes, Giovanna, Francisco Cezar Aquino de Moraes, Aline Beatriz Carvalho de Almeida, Felipe Goes Costa, Luiz Fernando Duarte de Andrade Junior, Maria Vitória Sabino Hupp, Ruan Rotondano Assunção, Marianne Rodrigues Fernandes, Sidney Emanuel Batista dos Santos, and Ney Pereira Carneiro dos Santos. 2025. "Single-Nucleotide Polymorphisms Related to Multiple Myeloma Risk: A Systematic Review and Meta-Analysis" International Journal of Molecular Sciences 26, no. 7: 3369. https://doi.org/10.3390/ijms26073369
APA StyleGilioli da Costa Nunes, G., Cezar Aquino de Moraes, F., Carvalho de Almeida, A. B., Goes Costa, F., Duarte de Andrade Junior, L. F., Sabino Hupp, M. V., Rotondano Assunção, R., Rodrigues Fernandes, M., Emanuel Batista dos Santos, S., & Pereira Carneiro dos Santos, N. (2025). Single-Nucleotide Polymorphisms Related to Multiple Myeloma Risk: A Systematic Review and Meta-Analysis. International Journal of Molecular Sciences, 26(7), 3369. https://doi.org/10.3390/ijms26073369







