Lactylation in Glioblastoma: A Novel Epigenetic Modifier Bridging Epigenetic Plasticity and Metabolic Reprogramming
Abstract
1. Introduction
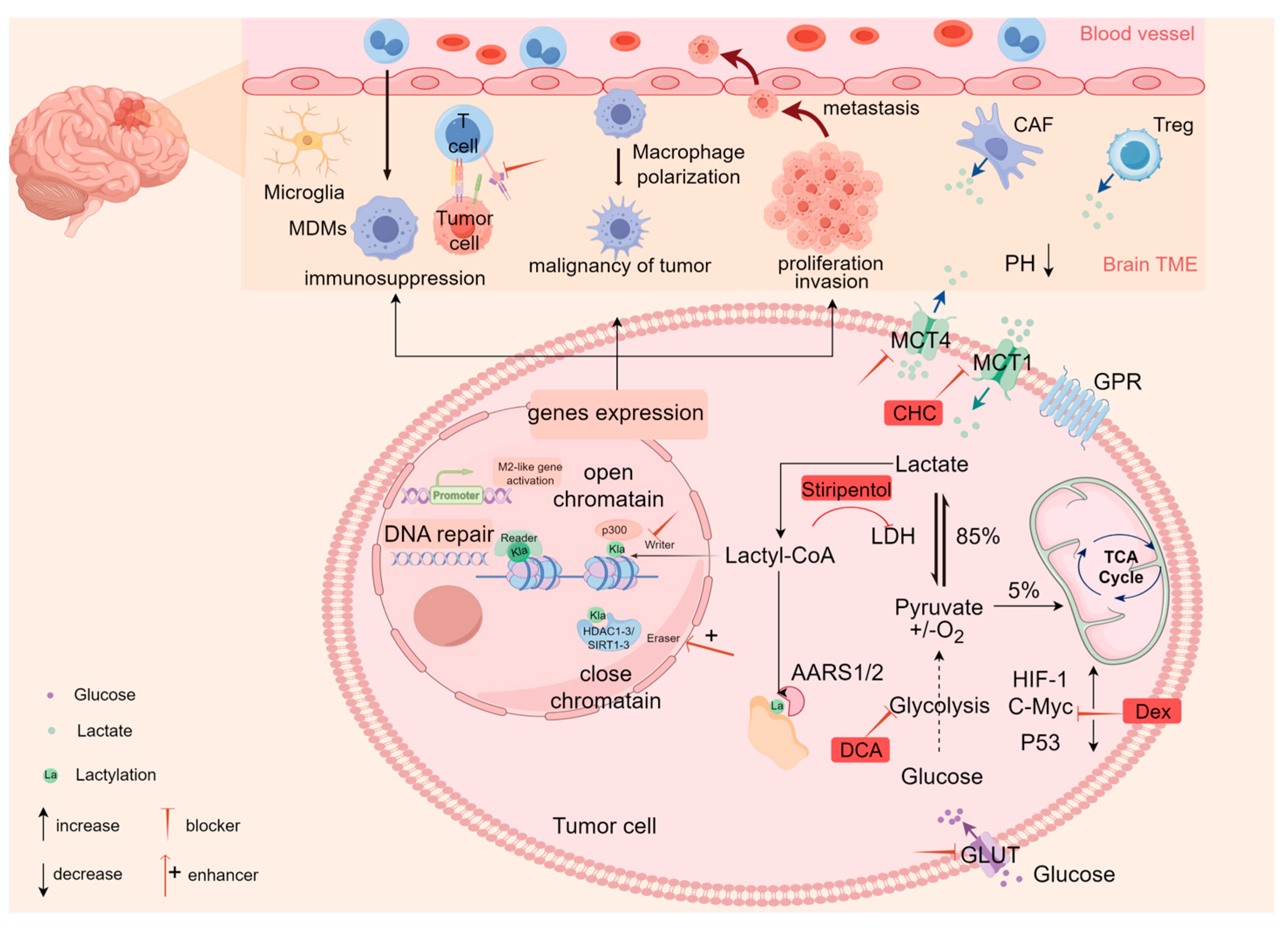
2. Glycolytic Metabolic Reprogramming in Glioblastoma
2.1. Lactate Accumulation
2.2. Lactate Transport
2.3. The Biological Role of Lactate in GBM
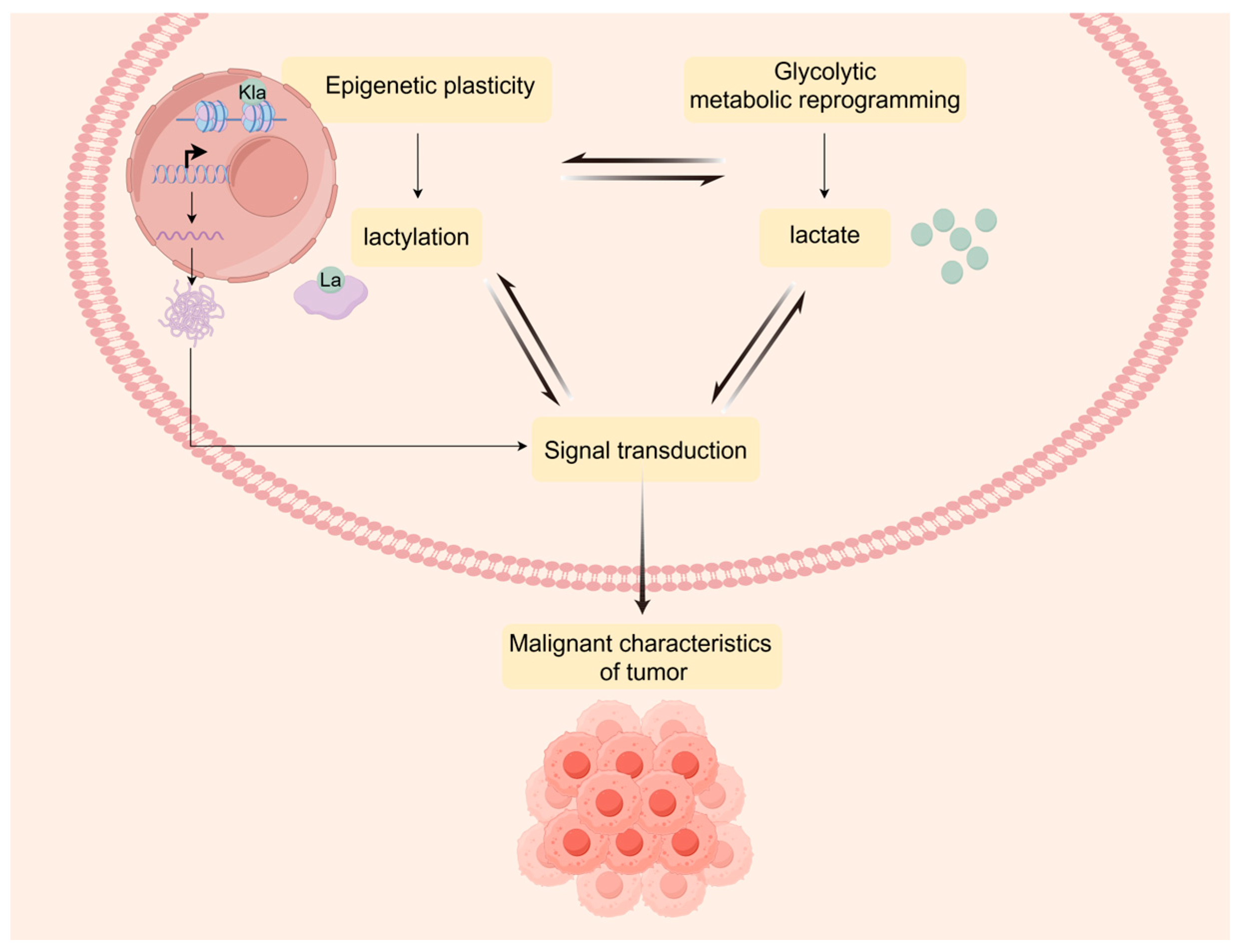
3. Lactylation Landscape Bridges Metabolic Reprogramming and Epigenetic Plasticity
3.1. The Regulatory Mechanisms of Lactylation
3.2. Detection Methods for Lactylation
3.3. Metabolic Interaction of Lactylation in Glioblastoma
3.3.1. Lactylation Promotes GBM Cell Proliferation
3.3.2. The Effect of Lactylation on the TME of GBM
4. The Clinical Application Prospect of Lactylation in GBM
4.1. Chemoradiotherapy Resistance and Prognostic Biomarkers
4.2. Unraveling Therapeutic Targets and Strategies
4.2.1. Targeting Lactate Metabolism
4.2.2. Targeting Lactate Transport
4.2.3. Targeting Lactylation
5. Decoding the Glioblastoma Lactylation Landscape: Challenges and Emerging Avenues
6. Outlook for the Future Research Direction
Author Contributions
Funding
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| GBM | glioblastoma |
| PTM | post-translational modification |
| HIF-1α | hypoxia-inducible factor 1 alpha |
| GLUT1 | glucose transporter 1 |
| TIGAR | the glycolytic and apoptotic regulator |
| mTOR | mammalian targets of rapamycin |
| TME | tumor microenvironment |
| MCTs | monocarboxylate transporters |
| GPCRs | G-protein-coupled receptors |
| HCAR1 | hydroxycarboxylic acid receptor 1 |
| CAFs | cancer-associated fibroblasts |
| TAMs | tumor-associated macrophages |
| Tregs | regulatory T cells |
| VEGF | vascular endothelial growth factor |
| Arg1 | arginase 1 |
| LDHA | lactate dehydrogenase A |
| APC/C | anaphase-promoting complex |
| LC-MS/MS | liquid chromatography–tandem mass spectrometry |
| LGSH | lactoyl-glutathione |
| EMT | epithelial–mesenchymal transition |
| cSCC | cutaneous squamous cell carcinoma |
| MMR | mismatch repair |
| DCA | dichloroacetate |
| Dex | dexmedetomidine |
| lactyl-CoA | lactyl-coenzyme A |
| HDAC | histone deacetylase |
| MDMs | monocyte-derived macrophages |
| CHC | α-cyano-4-hydroxycinnamate |
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; Price, M.; Neff, C.; Cioffi, G.; Waite, K.A.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2016–2020. Neuro-Oncology 2023, 25, iv1. [Google Scholar]
- Schaff, L.R.; Mellinghoff, I.K. Glioblastoma and Other Primary Brain Malignancies in Adults: A Review. JAMA 2023, 329, 574–587. [Google Scholar] [CrossRef] [PubMed]
- Bent, M.J.v.D.; Geurts, M.; French, P.J.; Smits, M.; Capper, D.; Bromberg, J.E.C.; Chang, S.M. Primary brain tumours in adults. Lancet 2023, 402, 1564–1579. [Google Scholar] [CrossRef] [PubMed]
- McKinnon, C.; Nandhabalan, M.; Murray, S.A.; Plaha, P. Glioblastoma: Clinical presentation, diagnosis, and management. BMJ-Br. Med. J. 2021, 374, n1560. [Google Scholar]
- Bent, M.J.v.D.; Smits, M.; Kros, J.M.; Chang, S.M. Diffuse Infiltrating Oligodendroglioma and Astrocytoma. J. Clin. Oncol. 2017, 35, 2394–2401. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro-Oncology 2021, 23, 1231–1251. [Google Scholar] [CrossRef]
- Weller, M.; Butowski, N.; Tran, D.D.; Recht, L.D.; Lim, M.; Hirte, H.; Ashby, L.; Mechtler, L.; Goldlust, S.A.; Iwamoto, F.; et al. Rindopepimut with temozolomide for patients with newly diagnosed, EGFRvIII-expressing glioblastoma (ACT IV): A randomised, double-blind, international phase 3 trial. Lancet Oncol. 2017, 18, 1373–1385. [Google Scholar] [CrossRef]
- Tan, A.C.; Ashley, D.M.; López, G.Y.; Malinzak, M.; Friedman, H.S.; Khasraw, M. Management of glioblastoma: State of the art and future directions. CA Cancer J. Clin. 2020, 70, 299–312. [Google Scholar] [CrossRef]
- Li, X.; Yang, Y.; Zhang, B.; Lin, X.; Fu, X.; An, Y.; Zou, Y.; Wang, J.-X.; Wang, Z.; Yu, T. Lactate metabolism in human health and disease. Signal Transduct. Target. Ther. 2022, 7, 305. [Google Scholar] [CrossRef]
- Zhao, S.; Allis, C.D.; Wang, G.G. The language of chromatin modification in human cancers. Nat. Rev. Cancer 2021, 21, 413–430. [Google Scholar] [CrossRef] [PubMed]
- Feng, F.; Wu, J.; Chi, Q.; Wang, S.; Liu, W.; Yang, L.; Song, G.; Pan, L.; Xu, K.; Wang, C. Lactylome Analysis Unveils Lactylation-Dependent Mechanisms of Stemness Remodeling in the Liver Cancer Stem Cells. Adv. Sci. 2024, 11, e2405975. [Google Scholar] [CrossRef]
- Li, F.; Si, W.; Xia, L.; Yin, D.; Wei, T.; Tao, M.; Cui, X.; Yang, J.; Hong, T.; Wei, R. Positive feedback regulation between glycolysis and histone lactylation drives oncogenesis in pancreatic ductal adenocarcinoma. Mol. Cancer 2024, 23, 90. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Chai, P.; Xie, M.; Ge, S.; Ruan, J.; Fan, X.; Jia, R. Histone lactylation drives oncogenesis by facilitating m6A reader protein YTHDF2 expression in ocular melanoma. Genome Biol. 2021, 22, 85. [Google Scholar] [CrossRef] [PubMed]
- Stine, Z.E.; Schug, Z.T.; Salvino, J.M.; Dang, C.V. Targeting cancer metabolism in the era of precision oncology. Nat. Rev. Drug Discov. 2021, 21, 141–162. [Google Scholar] [CrossRef]
- Agnihotri, S.; Zadeh, G. Metabolic reprogramming in glioblastoma: The influence of cancer metabolism on epigenetics and unan-swered questions. Neuro-Oncology 2016, 18, 160–172. [Google Scholar]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Skvortsova, K.; Iovino, N.; Bogdanović, O. Functions and mechanisms of epigenetic inheritance in animals. Nat. Rev. Mol. Cell Biol. 2018, 19, 774–790. [Google Scholar] [CrossRef]
- Sun, L.; Zhang, H.; Gao, P. Metabolic reprogramming and epigenetic modifications on the path to cancer. Protein Cell 2022, 13, 877–919. [Google Scholar] [CrossRef]
- Reid, M.A.; Dai, Z.; Locasale, J.W. The impact of cellular metabolism on chromatin dynamics and epigenetics. Nat. Cell Biol. 2017, 19, 1298–1306. [Google Scholar] [CrossRef]
- Kaelin, W.G.; McKnight, S.L. Influence of Metabolism on Epigenetics and Disease. Cell 2013, 153, 56–69. [Google Scholar] [CrossRef] [PubMed]
- Guccione, E.; Richard, S. The regulation, functions and clinical relevance of arginine methylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 642–657. [Google Scholar] [CrossRef] [PubMed]
- Warburg, O. On respiratory impairment in cancer cells. Science 1956, 124, 269–270. [Google Scholar] [CrossRef]
- Hsu, P.P.; Sabatini, D.M. Cancer Cell Metabolism: Warburg and Beyond. Cell 2008, 134, 703–707. [Google Scholar] [CrossRef]
- DeBerardinis, R.J.; Mancuso, A.; Daikhin, E.; Nissim, I.; Yudkoff, M.; Wehrli, S.; Thompson, C.B. Beyond aerobic glycolysis: Transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc. Natl. Acad. Sci. USA 2007, 104, 19345–19350. [Google Scholar] [CrossRef] [PubMed]
- Pavlova, N.N.; Thompson, C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef]
- Heiden, M.G.V.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg Effect: The Metabolic Requirements of Cell Prolif-eration. Science 2009, 324, 1029–1033. [Google Scholar]
- Mossmann, D.; Park, S.; Hall, M.N. mTOR signalling and cellular metabolism are mutual determinants in cancer. Nat. Rev. Cancer 2018, 18, 744–757. [Google Scholar] [CrossRef]
- Bensaad, K.; Tsuruta, A.; Selak, M.A.; Vidal, M.N.C.; Nakano, K.; Bartrons, R.; Gottlieb, E.; Vousden, K.H. TIGAR, a p53-Inducible Regulator of Glycolysis and Apoptosis. Cell 2006, 126, 107–120. [Google Scholar] [CrossRef]
- Rodríguez-García, A.; Samsó, P.; Fontova, P.; Simon, H.; Manzano, A.; Castaño, E.; Rosa, J.L.; Martinez-Outshoorn, U.; Ventura, F.; Navarro-Sabaté, À.; et al. TGF-β1 targets Smad, p38 MAPK, and PI3K/Akt signaling pathways to induce PFKFB3 gene expression and glycolysis in glioblastoma cells. FEBS J. 2017, 284, 3437–3454. [Google Scholar] [CrossRef]
- Masui, K.; Tanaka, K.; Akhavan, D.; Babic, I.; Gini, B.; Matsutani, T.; Iwanami, A.; Liu, F.; Villa, G.R.; Gu, Y.; et al. mTOR Complex 2 Controls Glycolytic Metabolism in Glioblastoma through FoxO Acetylation and Upregulation of c-Myc. Cell Metab. 2013, 18, 726–739. [Google Scholar] [CrossRef] [PubMed]
- Hirayama, A.; Kami, K.; Sugimoto, M.; Sugawara, M.; Toki, N.; Onozuka, H.; Kinoshita, T.; Saito, N.; Ochiai, A.; Tomita, M.; et al. Quantitative Metabolome Profiling of Colon and Stomach Cancer Microenvironment by Capillary Electrophoresis Time-of-Flight Mass Spectrometry. Cancer Res. 2009, 69, 4918–4925. [Google Scholar] [CrossRef] [PubMed]
- Poff, A.; Koutnik, A.P.; Egan, K.M.; Sahebjam, S.; D’agostino, D.; Kumar, N.B. Targeting the Warburg effect for cancer treatment: Ketogenic diets for management of glioma. Semin. Cancer Biol. 2019, 56, 135–148. [Google Scholar] [CrossRef] [PubMed]
- Brooks, G.A. The Science and Translation of Lactate Shuttle Theory. Cell Metab. 2018, 27, 757–785. [Google Scholar] [CrossRef]
- Hadzic, A.; Nguyen, T.D.; Hosoyamada, M.; Tomioka, N.H.; Bergersen, L.H.; Storm-Mathisen, J.; Morland, C. The Lactate Receptor HCA1 Is Present in the Choroid Plexus, the Tela Choroidea, and the Neuroepithelial Lining of the Dorsal Part of the Third Ventricle. Int. J. Mol. Sci. 2020, 21, 6457. [Google Scholar] [CrossRef]
- Torrini, C.; Nguyen, T.T.T.; Shu, C.; Mela, A.; Humala, N.; Mahajan, A.; Seeley, E.H.; Zhang, G.; Westhoff, M.-A.; Karpel-Massler, G.; et al. Lactate is an epigenetic metabolite that drives survival in model systems of glioblastoma. Mol. Cell 2022, 82, 3061–3076.e6. [Google Scholar] [CrossRef]
- Mächler, P.; Wyss, M.T.; Elsayed, M.; Stobart, J.; Gutierrez, R.; Von Faber-Castell, A.; Kaelin, V.; Zuend, M.; San Martín, A.; Romero-Gómez, I.; et al. In Vivo Evidence for a Lactate Gradient from Astrocytes to Neurons. Cell Metab. 2016, 23, 94–102. [Google Scholar] [CrossRef]
- Xie, Q.; Zhu, Z.; He, Y.; Zhang, Z.; Zhang, Y.; Wang, Y.; Luo, J.; Peng, T.; Cheng, F.; Gao, J.; et al. A lactate-induced Snail/STAT3 pathway drives GPR81 expression in lung cancer cells. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2020, 1866, 165576. [Google Scholar] [CrossRef]
- Miranda-Gonçalves, V.; Bezerra, F.; Costa-Almeida, R.; Freitas-Cunha, M.; Soares, R.; Martinho, O.; Reis, R.M.; Pinheiro, C.; Baltazar, F. Monocarboxylate transporter 1 is a key player in glioma-endothelial cell crosstalk. Mol. Carcinog. 2017, 56, 2630–2642. [Google Scholar] [CrossRef]
- Longhitano, L.; Vicario, N.; Tibullo, D.; Giallongo, C.; Broggi, G.; Caltabiano, R.; Barbagallo, G.M.V.; Altieri, R.; Baghini, M.; Di Rosa, M.; et al. Lactate Induces the Expressions of MCT1 and HCAR1 to Promote Tumor Growth and Progression in Glioblastoma. Front. Oncol. 2022, 12, 871798. [Google Scholar] [CrossRef]
- Fischer, K.; Hoffmann, P.; Voelkl, S.; Meidenbauer, N.; Ammer, J.; Edinger, M.; Gottfried, E.; Schwarz, S.; Rothe, G.; Hoves, S.; et al. Inhibitory effect of tumor cell–derived lactic acid on human T cells. Blood 2007, 109, 3812–3819. [Google Scholar] [CrossRef] [PubMed]
- Payen, V.L.; Mina, E.; Van Hee, V.F.; Porporato, P.E.; Sonveaux, P. Monocarboxylate transporters in cancer. Mol. Metab. 2020, 33, 48–66. [Google Scholar]
- Watson, M.J.; Vignali, P.D.A.; Mullett, S.J.; Overacre-Delgoffe, A.E.; Peralta, R.M.; Grebinoski, S.; Menk, A.V.; Rittenhouse, N.L.; DePeaux, K.; Whetstone, R.D.; et al. Metabolic support of tumour-infiltrating regulatory T cells by lactic acid. Nature 2021, 591, 645–651. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Outschoorn, U.E.; Sotgia, F.; Lisanti, M.P. Power surge: Supporting cells “fuel” cancer cell mitochondria. Cell Metab. 2012, 15, 4–5. [Google Scholar]
- Chen, J.; Zhu, Y.; Wu, C.; Shi, J. Engineering lactate-modulating nanomedicines for cancer therapy. Chem. Soc. Rev. 2023, 52, 973–1000. [Google Scholar] [CrossRef]
- Ippolito, L.; Morandi, A.; Giannoni, E.; Chiarugi, P. Lactate: A Metabolic Driver in the Tumour Landscape. Trends Biochem. Sci. 2019, 44, 153–166. [Google Scholar] [CrossRef]
- Hirschhaeuser, F.; Sattler, U.G.A.; Mueller-Klieser, W. Lactate: A Metabolic Key Player in Cancer. Cancer Res. 2011, 71, 6921–6925. [Google Scholar] [CrossRef]
- Hui, S.; Ghergurovich, J.M.; Morscher, R.J.; Jang, C.; Teng, X.; Lu, W.; Esparza, L.A.; Reya, T.; Zhan, L.; Guo, J.Y.; et al. Glucose feeds the TCA cycle via circulating lactate. Nature 2017, 551, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Certo, M.; Tsai, C.-H.; Pucino, V.; Ho, P.-C.; Mauro, C. Lactate modulation of immune responses in inflammatory versus tumour microenvironments. Nat. Rev. Immunol. 2020, 21, 151–161. [Google Scholar] [CrossRef]
- Colegio, O.R.; Chu, N.-Q.; Szabo, A.L.; Chu, T.; Rhebergen, A.M.; Jairam, V.; Cyrus, N.; Brokowski, C.E.; Eisenbarth, S.C.; Phillips, G.M.; et al. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature 2014, 513, 559–563. [Google Scholar] [CrossRef]
- Liu, W.; Wang, Y.; Bozi, L.H.M.; Fischer, P.D.; Jedrychowski, M.P.; Xiao, H.; Wu, T.; Darabedian, N.; He, X.; Mills, E.L.; et al. Lactate regulates cell cycle by remodelling the anaphase promoting complex. Nature 2023, 616, 790–797. [Google Scholar] [CrossRef] [PubMed]
- Torrini, C.; Nguyen, T.; Shu, C.; Mela, A.; Humala, N.; Mahajan, A.; Karpel-Massler, G.; Bruce, J.; Canoll, P.; Siegelin, M. EPCO-16. Lactic Acid Is an Epigenetic Metabolite That Drives Glioblastoma Survival and Growth. Neuro-Oncology 2020, 22, ii72. [Google Scholar] [CrossRef]
- Lee, D.C.; Sohn, H.A.; Park, Z.-Y.; Oh, S.; Kang, Y.K.; Lee, K.-M.; Kang, M.; Jang, Y.J.; Yang, S.-J.; Hong, Y.K.; et al. A Lactate-Induced Response to Hypoxia. Cell 2015, 161, 595–609. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Rodado, V.; Malta, T.M.; Seki, T.; Lita, A.; Dowdy, T.; Celiku, O.; Cavazos-Saldana, A.; Li, A.; Liu, Y.; Han, S.; et al. Metabolic reprogramming associated with aggressiveness occurs in the G-CIMP-high molecular subtypes of IDH1mut lower grade gliomas. Neuro-Oncology 2019, 22, 480–492. [Google Scholar] [CrossRef]
- Nikoobakht, M.; Shamshiripour, P.; Nekoo, Z.A.; Haghmohammadi, S.F. Elevated Lactate and Total Protein Levels in Stereotactic Brain Biopsy Specimen; Potential Biomarkers of Malignancy and Poor Prognosis. Arch. Iran. Med. 2019, 22, 125–131. [Google Scholar]
- Saraswathy, S.; Crawford, F.W.; Lamborn, K.R.; Pirzkall, A.; Chang, S.; Cha, S.; Nelson, S.J. Evaluation of MR markers that predict survival in patients with newly diagnosed GBM prior to adjuvant therapy. J. Neuro-Oncol. 2008, 91, 69–81. [Google Scholar] [CrossRef]
- Zhang, D.; Tang, Z.; Huang, H.; Zhou, G.; Cui, C.; Weng, Y.; Liu, W.; Kim, S.; Lee, S.; Perez-Neut, M.; et al. Metabolic regulation of gene expression by histone lactylation. Nature 2019, 574, 575–580. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhao, F.; Qu, Y. Lactylation: A Novel Post-Translational Modification with Clinical Implications in CNS Diseases. Biomolecules 2024, 14, 1175. [Google Scholar] [CrossRef]
- Wang, J.-H.; Mao, L.; Zhang, X.; Wu, M.; Wen, Q.; Yu, S.-C. Beyond metabolic waste: Lysine lactylation and its potential roles in cancer progression and cell fate determination. Cell. Oncol. 2023, 46, 465–480. [Google Scholar] [CrossRef]
- Liberti, M.V.; Locasale, J.W. Histone Lactylation: A New Role for Glucose Metabolism. Trends Biochem. Sci. 2020, 45, 179–182. [Google Scholar] [CrossRef]
- Moreno-Yruela, C.; Zhang, D.; Wei, W.; Bæk, M.; Liu, W.; Gao, J.; Danková, D.; Nielsen, A.L.; Bolding, J.E.; Yang, L.; et al. Class I histone deacetylases (HDAC1–3) are histone lysine delactylases. Sci. Adv. 2022, 8, eabi6696. [Google Scholar] [CrossRef]
- Mao, Y.; Zhang, J.; Zhou, Q.; He, X.; Zheng, Z.; Wei, Y.; Zhou, K.; Lin, Y.; Yu, H.; Zhang, H.; et al. Hypoxia induces mitochondrial protein lactylation to limit oxidative phosphorylation. Cell Res. 2024, 34, 13–30. [Google Scholar] [CrossRef]
- Zu, H.; Li, C.; Dai, C.; Pan, Y.; Ding, C.; Sun, H.; Zhang, X.; Yao, X.; Zang, J.; Mo, X. SIRT2 functions as a histone delactylase and inhibits the proliferation and migration of neuroblastoma cells. Cell Discov. 2022, 8, 54. [Google Scholar] [CrossRef] [PubMed]
- Kunadis, E.; Piperi, C. Exploring the Multi-Faceted Role of Sirtuins in Glioblastoma Pathogenesis and Targeting Options. Int. J. Mol. Sci. 2022, 23, 12889. [Google Scholar] [CrossRef] [PubMed]
- Park, H.-K.; Hong, J.-H.; Oh, Y.T.; Kim, S.S.; Yin, J.; Lee, A.-J.; Chae, Y.C.; Kim, J.H.; Park, S.-H.; Park, C.-K.; et al. Interplay between TRAP1 and Sirtuin-3 Modulates Mitochondrial Respiration and Oxidative Stress to Maintain Stemness of Glioma Stem Cells. Cancer Res. 2019, 79, 1369–1382. [Google Scholar] [CrossRef]
- Gao, M.; Zhang, N.; Liang, W. Systematic Analysis of Lysine Lactylation in the Plant Fungal Pathogen Botrytis cinerea. Front. Microbiol. 2020, 11, 594743. [Google Scholar] [CrossRef]
- Gaffney, D.O.; Jennings, E.Q.; Anderson, C.C.; Marentette, J.O.; Shi, T.; Oxvig, A.-M.S.; Streeter, M.D.; Johannsen, M.; Spiegel, D.A.; Chapman, E.; et al. Non-enzymatic Lysine Lactoylation of Glycolytic Enzymes. Cell Chem. Biol. 2019, 27, 206–213.e6. [Google Scholar] [CrossRef]
- Zong, Z.; Xie, F.; Wang, S.; Wu, X.; Zhang, Z.; Yang, B.; Zhou, F. Alanyl-tRNA synthetase, AARS1, is a lactate sensor and lactyltransferase that lactylates p53 and contributes to tu-morigenesis. Cell 2024, 187, 2375–2392. [Google Scholar]
- Wan, N.; Wang, N.; Yu, S.; Zhang, H.; Tang, S.; Wang, D.; Lu, W.; Li, H.; Delafield, D.G.; Kong, Y.; et al. Cyclic immonium ion of lactyllysine reveals widespread lactylation in the human proteome. Nat. Methods 2022, 19, 854–864. [Google Scholar] [CrossRef]
- Kim, M.M.; Parolia, A.; Dunphy, M.P.; Venneti, S. Non-invasive metabolic imaging of brain tumours in the era of precision medicine. Nat. Rev. Clin. Oncol. 2016, 13, 725–739. [Google Scholar] [CrossRef]
- Viswanath, P.; Batsios, G.; Mukherjee, J.; Gillespie, A.M.; Larson, P.E.Z.; Luchman, H.A.; Phillips, J.J.; Costello, J.F.; Pieper, R.O.; Ronen, S.M. Non-invasive assessment of telomere maintenance mechanisms in brain tumors. Nat. Commun. 2021, 12, 92. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, V.; Daoud, E.V.; Hatanpaa, K.J.; Gao, A.; Zhang, S.; An, Z.; Ganji, S.K.; Raisanen, J.M.; Lewis, C.M.; Askari, P.; et al. Glycine by MR spectroscopy is an imaging biomarker of glioma aggressiveness. Neuro-Oncology 2020, 22, 1018–1029. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Zhou, Y.; Liao, H.; Rowland, B.C.; Kong, X.; Arvold, N.D.; A Reardon, D.; Wen, P.Y.; Lin, A.P.; Huang, R.Y. Diagnostic accuracy of 2-hydroxyglutarate magnetic resonance spectroscopy in newly diagnosed brain mass and suspected recurrent gliomas. Neuro-Oncology 2018, 20, 1262–1271. [Google Scholar] [CrossRef] [PubMed]
- Hua, W.; Zhang, W.; Brown, H.; Wu, J.; Fang, X.; Shahi, M.; Chen, R.; Zhang, H.; Jiao, B.; Wang, N.; et al. Rapid detection of IDH mutations in gliomas by intraoperative mass spectrometry. Proc. Natl. Acad. Sci. USA 2024, 121, e2318843121. [Google Scholar] [CrossRef]
- Yue, Q.; Wang, Z.; Shen, Y.; Lan, Y.; Zhong, X.; Luo, X.; Yang, T.; Zhang, M.; Zuo, B.; Zeng, T.; et al. Histone H3K9 Lactylation Confers Temozolomide Resistance in Glioblastoma via LUC7L2-Mediated MLH1 Intron Retention. Adv. Sci. 2024, 11, 2309290. [Google Scholar] [CrossRef]
- Li, G.; Wang, D.; Zhai, Y.; Pan, C.; Zhang, J.; Wang, C.; Huang, R.; Yu, M.; Li, Y.; Liu, X.; et al. Glycometabolic reprogramming-induced XRCC1 lactylation confers therapeutic resistance in ALDH1A3-overexpressing glioblastoma. Cell Metab. 2024, 36, 1696–1710.e10. [Google Scholar] [CrossRef]
- Li, F.; Zhang, H.; Huang, Y.; Li, D.; Zheng, Z.; Xie, K.; Cao, C.; Wang, Q.; Zhao, X.; Huang, Z.; et al. Single-cell transcriptome analysis reveals the association between histone lactylation and cisplatin resistance in bladder cancer. Drug Resist. Updat. 2024, 73, 101059. [Google Scholar] [CrossRef]
- Jiang, P.; Ning, W.; Shi, Y.; Liu, C.; Mo, S.; Zhou, H.; Liu, K.; Guo, Y. FSL-Kla: A few-shot learning-based multi-feature hybrid system for lactylation site prediction. Comput. Struct. Biotechnol. J. 2021, 19, 4497–4509. [Google Scholar] [CrossRef]
- Li, L.; Chen, K.; Wang, T.; Wu, Y.; Xing, G.; Chen, M.; Hao, Z.; Zhang, C.; Zhang, J.; Ma, B.; et al. Glis1 facilitates induction of pluripotency via an epigenome–metabolome–epigenome signalling cascade. Nat. Metab. 2020, 2, 882–892. [Google Scholar] [CrossRef]
- Izzo, L.T.; Wellen, K.E. Histone lactylation links metabolism and gene regulation. Nature 2019, 574, 492–493. [Google Scholar] [CrossRef]
- Chen, A.-N.; Luo, Y.; Yang, Y.-H.; Fu, J.-T.; Geng, X.-M.; Shi, J.-P.; Yang, J. Lactylation, a Novel Metabolic Reprogramming Code: Current Status and Prospects. Front. Immunol. 2021, 12, 688910. [Google Scholar] [CrossRef]
- Li, L.; Li, Z.; Meng, X.; Wang, X.; Song, D.; Liu, Y.; Xu, T.; Qin, J.; Sun, N.; Tian, K.; et al. Histone lactylation-derived LINC01127 promotes the self-renewal of glioblastoma stem cells via the cis-regulating the MAP4K4 to activate JNK pathway. Cancer Lett. 2023, 579, 216467. [Google Scholar] [CrossRef]
- Gu, X.; Zhuang, A.; Yu, J.; Yang, L.; Ge, S.; Ruan, J.; Jia, R.; Fan, X.; Chai, P. Histone lactylation-boosted ALKBH3 potentiates tumor progression and diminished promyelocytic leukemia protein nuclear condensates by m1A demethylation of SP100A. Nucleic Acids Res. 2023, 52, 2273–2289. [Google Scholar] [CrossRef]
- Li, M.; Sun, P.; Tu, B.; Deng, G.; Li, D.; He, W. Hypoxia conduces the glioma progression by inducing M2 macrophage polarization via elevating TNFSF9 level in a histone-lactylation-dependent manner. Am. J. Physiol. Physiol. 2024, 327, C487–C504. [Google Scholar] [CrossRef]
- Wang, Y.; Li, H.; Jiang, S.; Fu, D.; Lu, X.; Lu, M.; Li, Y.; Luo, D.; Wu, K.; Xu, Y.; et al. The glycolytic enzyme PFKFB3 drives kidney fibrosis through promoting histone lactylation-mediated NF-κB family activation. Kidney Int. 2024, 106, 226–240. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, J.; Lin, R.; Huang, Y.; Wang, Z.; Xu, S.; Wang, L.; Chen, F.; Zhang, J.; Pan, K.; et al. Lactate drives epithelial-mesenchymal transition in diabetic kidney disease via the H3K14la/KLF5 pathway. Redox Biol. 2024, 75, 103246. [Google Scholar] [CrossRef]
- Noe, J.T.; Rendon, B.E.; Geller, A.E.; Conroy, L.R.; Morrissey, S.M.; Young, L.E.; Bruntz, R.C.; Kim, E.J.; Wise-Mitchell, A.; Rizzo, M.B.d.S.; et al. Lactate supports a metabolic-epigenetic link in macrophage polarization. Sci. Adv. 2021, 7, eabi8602. [Google Scholar] [CrossRef]
- De Leo, A.; Ugolini, A.; Yu, X.; Scirocchi, F.; Scocozza, D.; Peixoto, B.; Pace, A.; D’Angelo, L.; Liu, J.K.; Etame, A.B.; et al. Glucose-driven histone lactylation promotes the immunosuppressive activity of monocyte-derived macrophages in glioblastoma. Immunity 2024, 57, 1105–1123.e8. [Google Scholar]
- Li, H.; Liu, C.; Li, R.; Zhou, L.; Ran, Y.; Yang, Q.; Huang, H.; Lu, H.; Song, H.; Yang, B.; et al. AARS1 and AARS2 sense l-lactate to regulate cGAS as global lysine lactyltransferases. Nature 2024, 634, 1229–1237. [Google Scholar] [CrossRef]
- Raychaudhuri, D.; Singh, P.; Chakraborty, B.; Hennessey, M.; Tannir, A.J.; Byregowda, S.; Natarajan, S.M.; Trujillo-Ocampo, A.; Im, J.S.; Goswami, S. Histone lactylation drives CD8+ T cell metabolism and function. Nat. Immunol. 2024, 25, 2140–2151. [Google Scholar] [CrossRef]
- Krol, A.L.; Nehring, H.P.; Krause, F.F.; Wempe, A.; Raifer, H.; Nist, A.; Stiewe, T.; Bertrams, W.; Schmeck, B.; Luu, M.; et al. Lactate induces metabolic and epigenetic reprogramming of pro-inflammatory Th17 cells. Embo Rep. 2022, 23, e54685. [Google Scholar] [CrossRef]
- Gu, J.; Zhou, J.; Chen, Q.; Xu, X.; Gao, J.; Li, X.; Shao, Q.; Zhou, B.; Zhou, H.; Wei, S.; et al. Tumor metabolite lactate promotes tumorigenesis by modulating MOESIN lactylation and enhancing TGF-β signaling in regulatory T cells. Cell Rep. 2022, 39, 110986. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Outschoorn, U.E.; Peiris-Pagés, M.; Pestell, R.G.; Sotgia, F.; Lisanti, M.P. Cancer metabolism: A therapeutic perspective. Nat. Rev. Clin. Oncol. 2016, 14, 11–31. [Google Scholar] [CrossRef]
- Lin, J.; Liu, G.; Chen, L.; Kwok, H.F.; Lin, Y. Targeting lactate-related cell cycle activities for cancer therapy. Semin. Cancer Biol. 2022, 86, 1231–1243. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Li, Y.; Li, H.; Chen, X.; Fu, H.; Mao, D.; Chen, W.; Lan, L.; Wang, C.; Hu, K.; et al. NBS1 lactylation is required for efficient DNA repair and chemotherapy resistance. Nature 2024, 631, 663–669. [Google Scholar] [CrossRef]
- Chen, Y.; Wu, J.; Zhai, L.; Zhang, T.; Yin, H.; Gao, H.; Zhao, F.; Wang, Z.; Yang, X.; Jin, M.; et al. Metabolic regulation of homologous recombination repair by MRE11 lactylation. Cell 2023, 187, 294–311.e21. [Google Scholar] [CrossRef]
- Liu, Z.; Huang, Y.; Liu, X. Lactylation regulated DNA damage repair and cancer cell chemosensitivity. Sci. Bull. 2024, 69, 1185–1187. [Google Scholar] [CrossRef]
- Sun, P.; Ma, L.; Lu, Z. Lactylation: Linking the Warburg effect to DNA damage repair. Cell Metab. 2024, 36, 1637–1639. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Ren, X.; Park, Y.E.; Feng, H.; Sheng, X.; Song, X.; AminiTabrizi, R.; Shah, H.; Li, L.; Zhang, Y.; et al. Nuclear GTPSCS functions as a lactyl-CoA synthetase to promote histone lactylation and gliomagenesis. Cell Metab. 2024, 37, 377–394.e9. [Google Scholar] [CrossRef]
- Marko, N.F.; Weil, R.J.; Schroeder, J.L.; Lang, F.F.; Suki, D.; Sawaya, R.E. Extent of Resection of Glioblastoma Revisited: Personalized Survival Modeling Facilitates More Accurate Survival Prediction and Supports a Maximum-Safe-Resection Approach to Surgery. J. Clin. Oncol. 2014, 32, 774–782. [Google Scholar] [CrossRef]
- Janjua, T.I.; Rewatkar, P.; Ahmed-Cox, A.; Saeed, I.; Mansfeld, F.M.; Kulshreshtha, R.; Kumeria, T.; Ziegler, D.S.; Kavallaris, M.; Mazzieri, R.; et al. Frontiers in the treatment of glioblastoma: Past, present and emerging. Adv. Drug Deliv. Rev. 2021, 171, 108–138. [Google Scholar] [CrossRef] [PubMed]
- Weller, M.; Wen, P.Y.; Chang, S.M.; Dirven, L.; Lim, M.; Monje, M.; Reifenberger, G. Glioma. Nat. Rev. Dis. Primers 2024, 10, 33. [Google Scholar] [CrossRef] [PubMed]
- Cuddapah, V.A.; Robel, S.; Watkins, S.; Sontheimer, H. A neurocentric perspective on glioma invasion. Nat. Rev. Neurosci. 2014, 15, 455–465. [Google Scholar] [CrossRef] [PubMed]
- Oldrini, B.; Vaquero-Siguero, N.; Mu, Q.; Kroon, P.; Zhang, Y.; Galán-Ganga, M.; Bao, Z.; Wang, Z.; Liu, H.; Sa, J.K.; et al. MGMT genomic rearrangements contribute to chemotherapy resistance in gliomas. Nat. Commun. 2020, 11, 3883. [Google Scholar] [CrossRef]
- Dai, X.; Lv, X.; Thompson, E.W.; Ostrikov, K. Histone lactylation: Epigenetic mark of glycolytic switch. Trends Genet. 2022, 38, 124–127. [Google Scholar] [CrossRef]
- Zhou, L.; Wang, Y.; Zhou, M.; Zhang, Y.; Wang, P.; Li, X.; Yang, J.; Wang, H.; Ding, Z. HOXA9 inhibits HIF-1α-mediated glycolysis through interacting with CRIP2 to repress cutaneous squamous cell carcinoma development. Nat. Commun. 2018, 9, 1480. [Google Scholar] [CrossRef]
- A Flavahan, W.; Wu, Q.; Hitomi, M.; Rahim, N.; Kim, Y.; E Sloan, A.; Weil, R.J.; Nakano, I.; Sarkaria, J.N.; Stringer, B.W.; et al. Brain tumor initiating cells adapt to restricted nutrition through preferential glucose uptake. Nat. Neurosci. 2013, 16, 1373–1382. [Google Scholar] [CrossRef]
- Sharma, D.; Singh, M.; Rani, R. Role of LDH in tumor glycolysis: Regulation of LDHA by small molecules for cancer therapeutics. Semin. Cancer Biol. 2022, 87, 184–195. [Google Scholar] [CrossRef]
- Di, H.; Zhang, X.; Guo, Y.; Shi, Y.; Fang, C.; Yuan, Y.; Wang, J.; Shang, C.; Guo, W.; Li, C. Silencing LDHA inhibits proliferation, induces apoptosis and increases chemosensitivity to temozolomide in glioma cells. Oncol. Lett. 2018, 15, 5131–5136. [Google Scholar] [CrossRef]
- Dunbar, E.M.; Coats, B.S.; Shroads, A.L.; Langaee, T.; Lew, A.; Forder, J.R.; Shuster, J.J.; Wagner, D.A.; Stacpoole, P.W. Phase 1 trial of dichloroacetate (DCA) in adults with recurrent malignant brain tumors. Investig. New Drugs 2013, 32, 452–464. [Google Scholar] [CrossRef]
- Michelakis, E.D.; Sutendra, G.; Dromparis, P.; Webster, L.; Haromy, A.; Niven, E.; Maguire, C.; Gammer, T.-L.; Mackey, J.R.; Fulton, D.; et al. Metabolic Modulation of Glioblastoma with Dichloroacetate. Sci. Transl. Med. 2010, 2, 31ra34. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Liao, W.; Li, C.; Zhu, L. Silencing BMAL1 promotes M1/M2 polarization through the LDHA/lactate axis to promote GBM sensitivity to bevacizumab. Int. Immunopharmacol. 2024, 134, 112187. [Google Scholar] [CrossRef]
- Mair, R.; Wright, A.J.; Ros, S.; Hu, D.E.; Booth, T.; Kreis, F.; Rao, J.; Watts, C.; Brindle, K.M. Metabolic Imaging Detects Low Levels of Glycolytic Activity That Vary with Levels of c-Myc Expression in Pa-tient-Derived Xenograft Models of Glioblastoma. Cancer Res. 2018, 78, 5408–5418. [Google Scholar]
- Nguyen, T.T.T.; Zhang, Y.; Shang, E.; Shu, C.; Torrini, C.; Zhao, J.; Bianchetti, E.; Mela, A.; Humala, N.; Mahajan, A.; et al. HDAC inhibitors elicit metabolic reprogramming by targeting super-enhancers in glioblastoma models. J. Clin. Investig. 2020, 130, 3699–3716. [Google Scholar] [CrossRef]
- Uddin, M.S.; Al Mamun, A.; Alghamdi, B.S.; Tewari, D.; Jeandet, P.; Sarwar, M.S.; Ashraf, G.M. Epigenetics of glioblastoma multiforme: From molecular mechanisms to therapeutic approaches. Semin. Cancer Biol. 2022, 83, 100–120. [Google Scholar] [PubMed]
- Vander Heiden, M.G.; DeBerardinis, R.J. Understanding the Intersections between Metabolism and Cancer Biology. Cell 2017, 168, 657–669. [Google Scholar] [CrossRef] [PubMed]
- Miranda-Gonçalves, V.; Honavar, M.; Pinheiro, C.; Martinho, O.; Pires, M.M.; Pinheiro, C.; Cordeiro, M.; Bebiano, G.; Costa, P.; Palmeirim, I.; et al. Monocarboxylate transporters (MCTs) in gliomas: Expression and exploitation as therapeutic targets. Neuro Oncol. 2012, 15, 172–188. [Google Scholar] [CrossRef]
- Marchiq, I.; Le Floch, R.; Roux, D.; Simon, M.P.; Pouyssegur, J. Genetic disruption of lactate/H+ symporters (MCTs) and their subunit CD147/BASIGIN sensitizes glycolytic tumor cells to phenformin. Cancer Res. 2015, 75, 171–180. [Google Scholar]
- Davalos, V.; Esteller, M. Cancer epigenetics in clinical practice. CA Cancer J. Clin. 2023, 73, 376–424. [Google Scholar]
- Dong, F.; Yin, H.; Zheng, Z. Hypoxia-Inducible Factor-1α Regulates BNIP3-Dependent Mitophagy and Mediates Metabolic Reprogramming Through Histone Lysine Lactylation Modification to Affect Glioma Proliferation and Invasion. J. Biochem. Mol. Toxicol. 2025, 39, e70069. [Google Scholar] [CrossRef]
- Zhu, J.; Zhang, Y. Dexmedetomidine inhibits the migration, invasion, and glycolysis of glioblastoma cells by lactylation of c-myc. Neurol. Res. 2024, 46, 1105–1112. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhou, Y.; Wang, H. The role of lactate-induced protein lactylation in gliomas: Implications for preclinical research and the development of new treatments. Front. Pharmacol. 2024, 15, 1383274. [Google Scholar] [CrossRef]
- Wang, S.; Huang, T.; Wu, Q.; Yuan, H.; Wu, X.; Yuan, F.; Duan, T.; Taori, S.; Zhao, Y.; Snyder, N.W.; et al. Lactate reprograms glioblastoma immunity through CBX3-regulated histone lactylation. J. Clin. Investig. 2024, 134, e176851. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Liu, B.; Li, Y.; Wu, J.; Cao, Y.; Yang, S.; Tan, H.; Cai, L.; Zhang, S.; Qi, X.; et al. Oxamate enhances the efficacy of CAR-T therapy against glioblastoma via suppressing ectonucleotidases and CCR8 lactylation. J. Exp. Clin. Cancer Res. 2023, 42, 523. [Google Scholar] [CrossRef]
- Yang, K.; Fan, M.; Wang, X.; Xu, J.; Wang, Y.; Tu, F.; Gill, P.S.; Ha, T.; Liu, L.; Williams, D.L.; et al. Lactate promotes macrophage HMGB1 lactylation, acetylation, and exosomal release in polymicrobial sepsis. Cell Death Differ. 2022, 29, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Zhu, L.; Qin, Z.-Y.; Guo, Y.; Wang, S.; Xue, M.; Shen, K.-Y.; Hu, B.-Y.; Wang, X.-F.; Wang, C.-Q.; et al. Modulation of cellular metabolism by protein crotonylation regulates pancreatic cancer progression. Cell Rep. 2023, 42, 112666. [Google Scholar] [CrossRef]
- Rho, H.; Terry, A.R.; Chronis, C.; Hay, N. Hexokinase 2-mediated gene expression via histone lactylation is required for hepatic stellate cell activation and liver fibrosis. Cell Metab. 2023, 35, 1406–1423.e8. [Google Scholar]
- Dai, S.-K.; Liu, P.-P.; Li, X.; Jiao, L.-F.; Teng, Z.-Q.; Liu, C.-M. Dynamic profiling and functional interpretation of histone lysine crotonylation and lactylation during neural development. Development 2022, 149, dev200049. [Google Scholar] [CrossRef]
- Jacob, F.; Salinas, R.D.; Zhang, D.Y.; Nguyen, P.T.T.; Schnoll, J.G.; Wong, S.Z.H.; Thokala, R.; Sheikh, S.; Saxena, D.; Prokop, S.; et al. A Patient-Derived Glioblastoma Organoid Model and Biobank Recapitulates Inter- and Intra-tumoral Heterogeneity. Cell 2020, 180, 188–204.e22. [Google Scholar] [CrossRef]
- Hu, J.; Wang, S.G.; Hou, Y.; Chen, Z.; Liu, L.; Li, R.; Li, N.; Zhou, L.; Yang, Y.; Wang, L.; et al. Multiomic profiling of clear cell renal cell carcinoma identifies metabolic reprogramming associated with disease progression. Nat. Genet. 2024, 56, 442–457. [Google Scholar] [CrossRef]
- Tirosh, I.; Suva, M.L. Cancer cell states: Lessons from ten years of single-cell RNA-sequencing of human tumors. Cancer Cell 2024, 42, 1497–1506. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Zhou, Z.; Qiu, P.; Xin, T. Integrated single-cell and bulk RNA-sequencing data reveal molecular subtypes based on lactylation-related genes and prognosis and therapeutic response in glioma. Heliyon 2024, 10, e30726. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qiu, Q.; Deng, H.; Song, P.; Liu, Y.; Zhang, M. Lactylation in Glioblastoma: A Novel Epigenetic Modifier Bridging Epigenetic Plasticity and Metabolic Reprogramming. Int. J. Mol. Sci. 2025, 26, 3368. https://doi.org/10.3390/ijms26073368
Qiu Q, Deng H, Song P, Liu Y, Zhang M. Lactylation in Glioblastoma: A Novel Epigenetic Modifier Bridging Epigenetic Plasticity and Metabolic Reprogramming. International Journal of Molecular Sciences. 2025; 26(7):3368. https://doi.org/10.3390/ijms26073368
Chicago/Turabian StyleQiu, Qingya, Hui Deng, Ping Song, Yushu Liu, and Mengxian Zhang. 2025. "Lactylation in Glioblastoma: A Novel Epigenetic Modifier Bridging Epigenetic Plasticity and Metabolic Reprogramming" International Journal of Molecular Sciences 26, no. 7: 3368. https://doi.org/10.3390/ijms26073368
APA StyleQiu, Q., Deng, H., Song, P., Liu, Y., & Zhang, M. (2025). Lactylation in Glioblastoma: A Novel Epigenetic Modifier Bridging Epigenetic Plasticity and Metabolic Reprogramming. International Journal of Molecular Sciences, 26(7), 3368. https://doi.org/10.3390/ijms26073368





