Evidence for a Functional HipBA Toxin–Antitoxin System in Acidovorax citrulli
Abstract
1. Introduction
2. Results
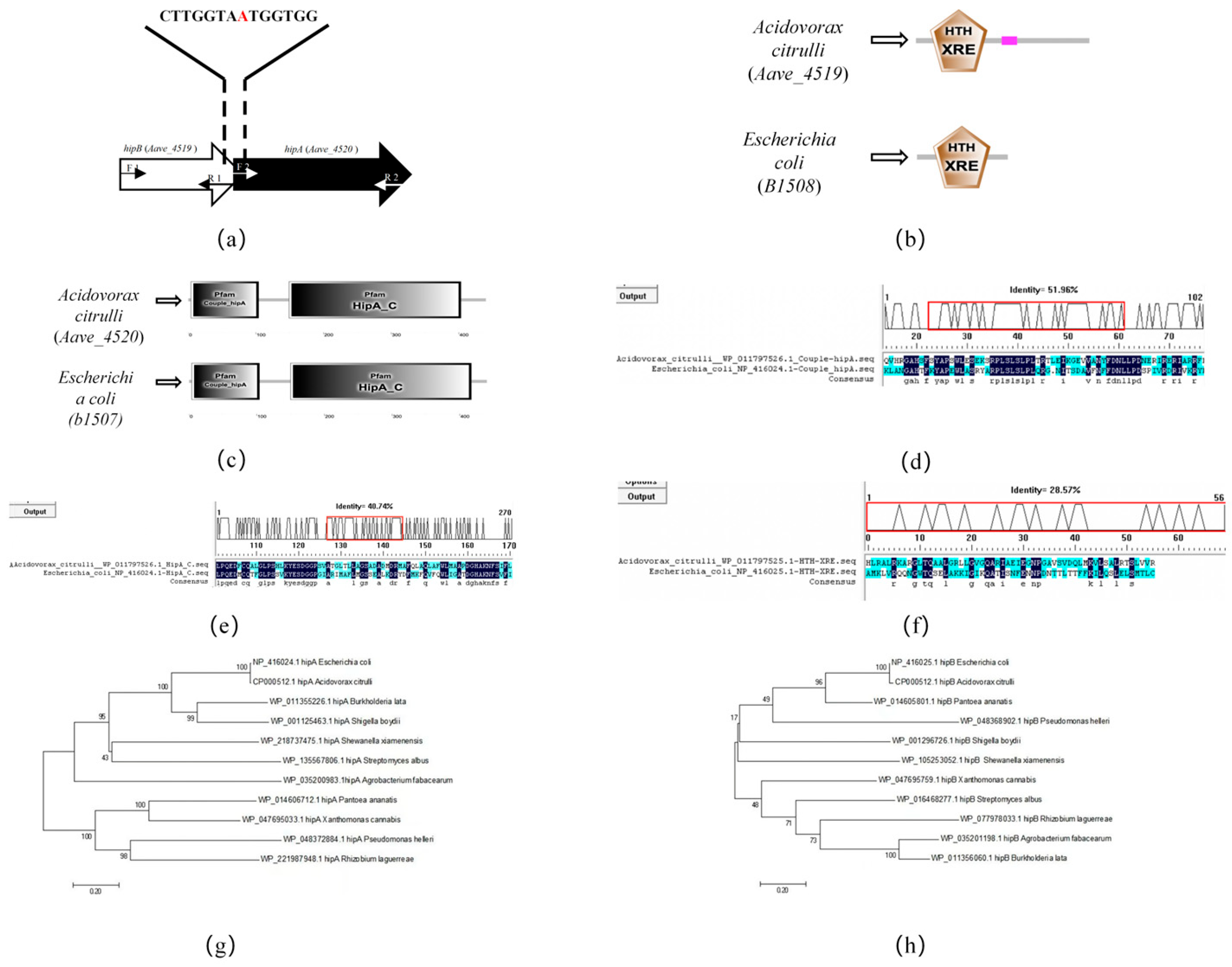
2.1. Bioinformatic Analysis of hipA and hipB in Acidovorax citrulli
2.2. Co-Transcription Test
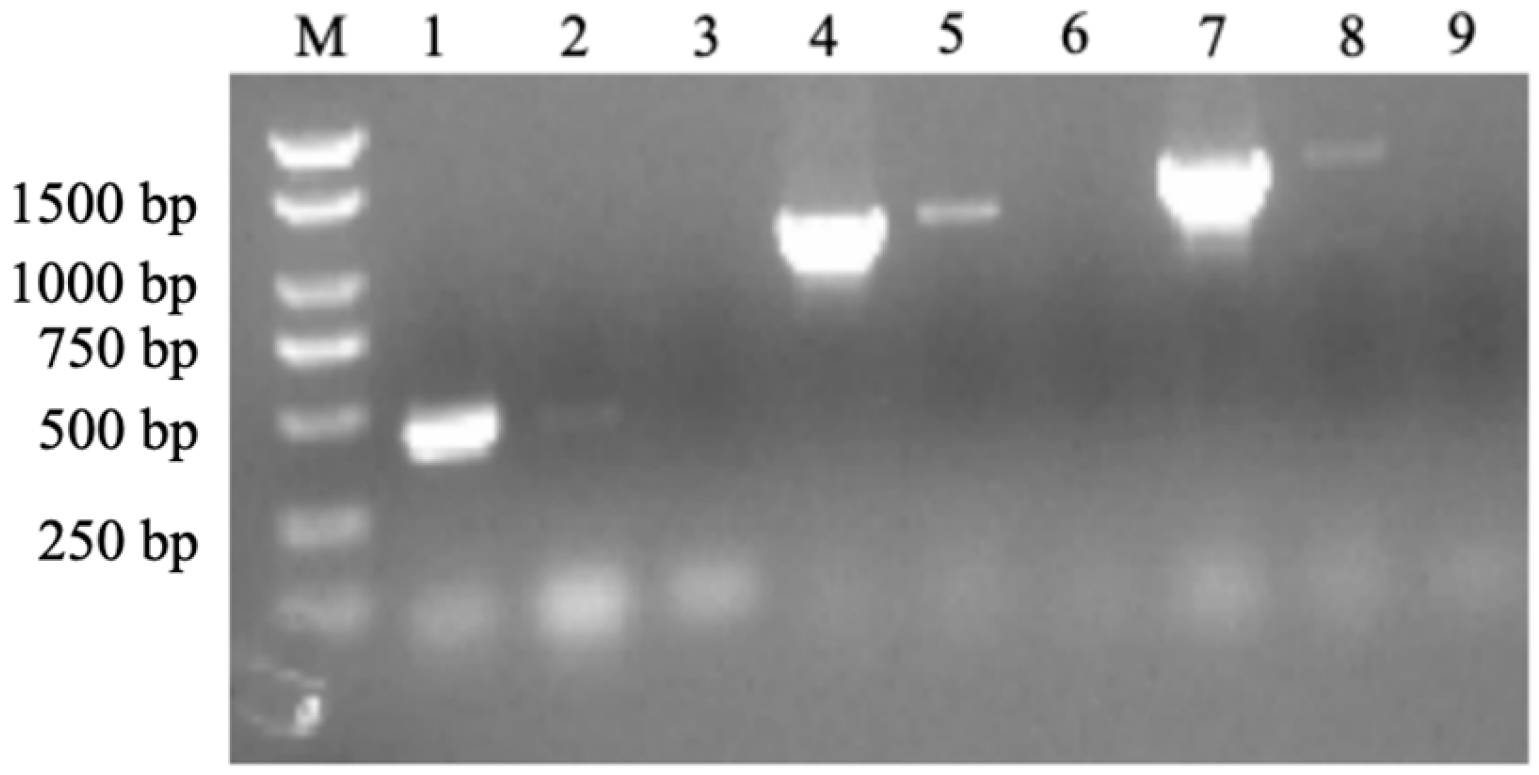
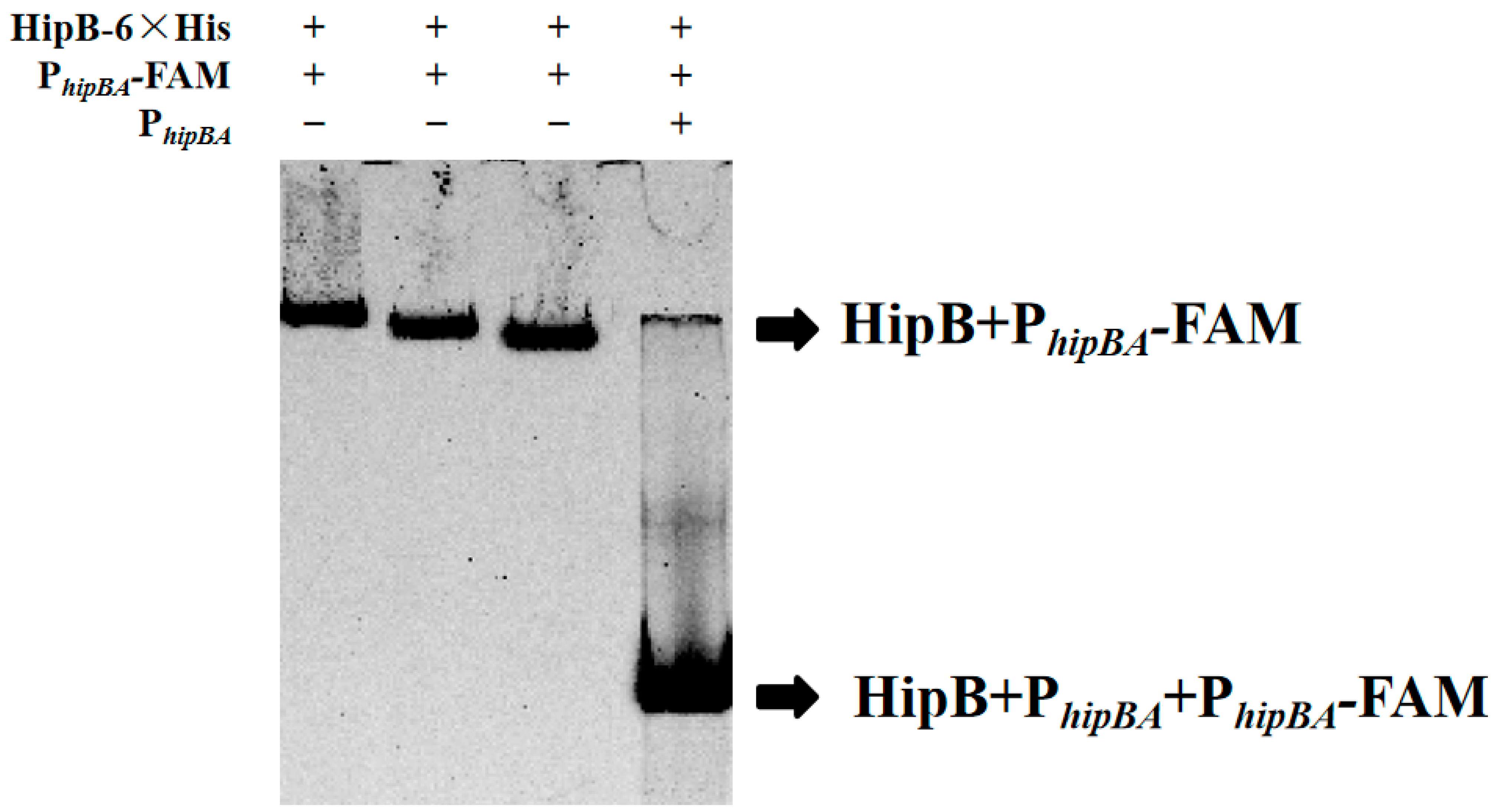
2.3. HipB Binds to the Promoter of the hipBA Operon Region in Aac5
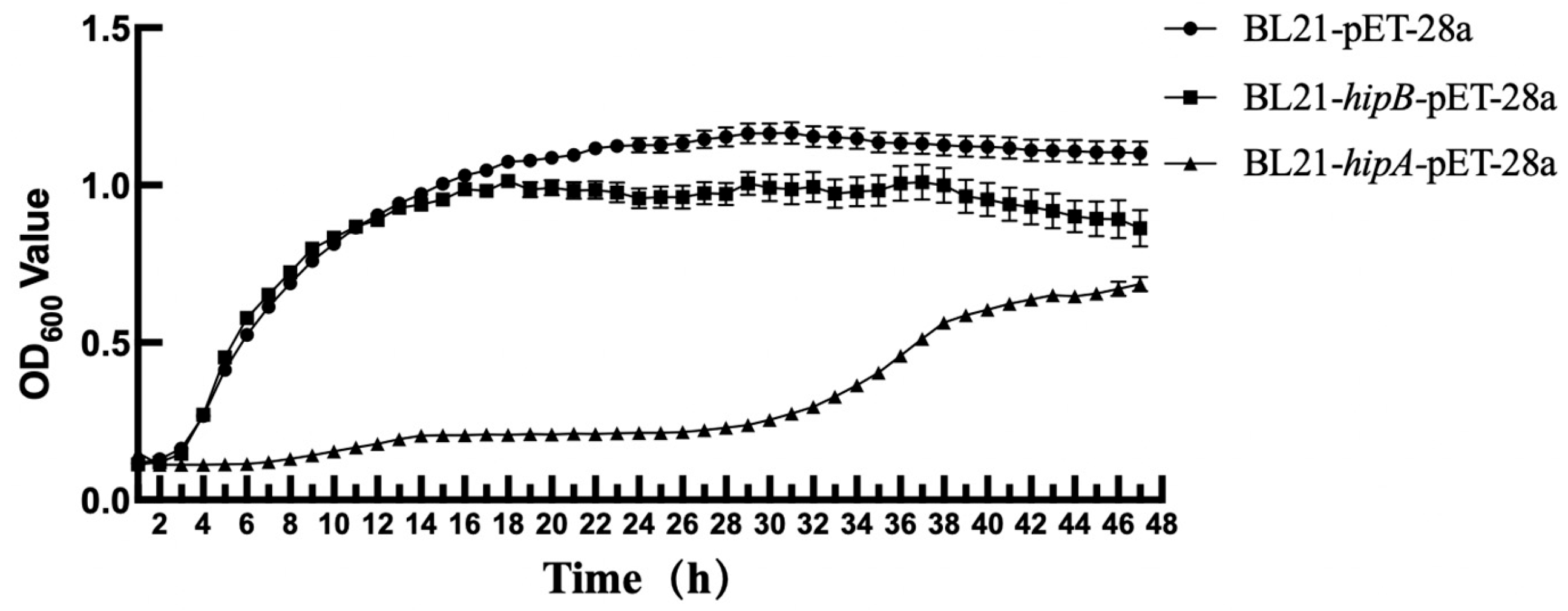
2.4. Effect of Heterologous Expression of Putative HipA and HipB on E. coli Growth
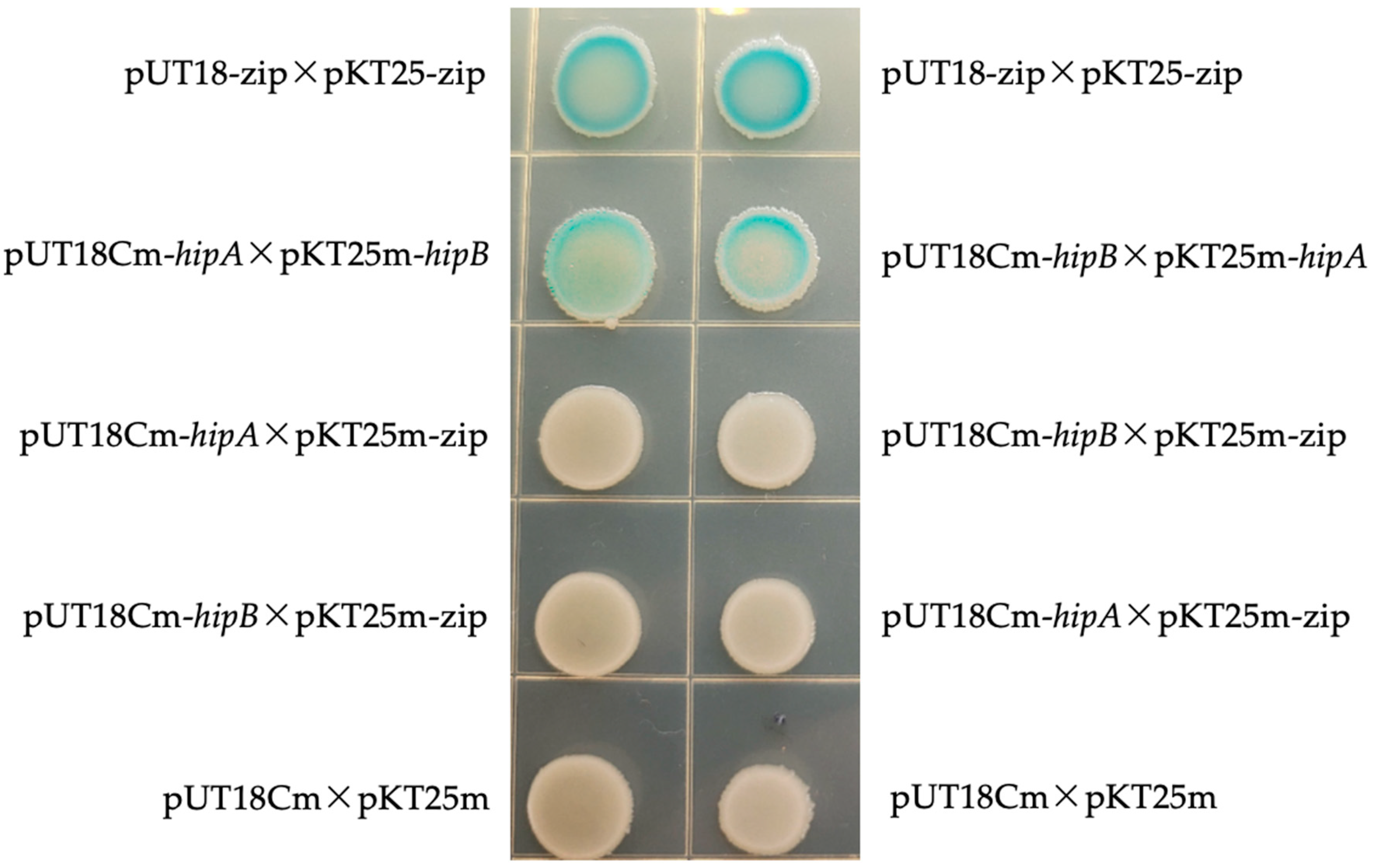
2.5. Bacterial Two-Hybrid Experiment
2.6. Quantitative Real-Time PCR Analyses of hipA and hipB mRNA Levels in A. citrulli Aac5 Under Various Environmental Stress Conditions
2.6.1. Analysis of hipA and hipB Transcription Levels in Aac5 Under Chloramphenicol Stress
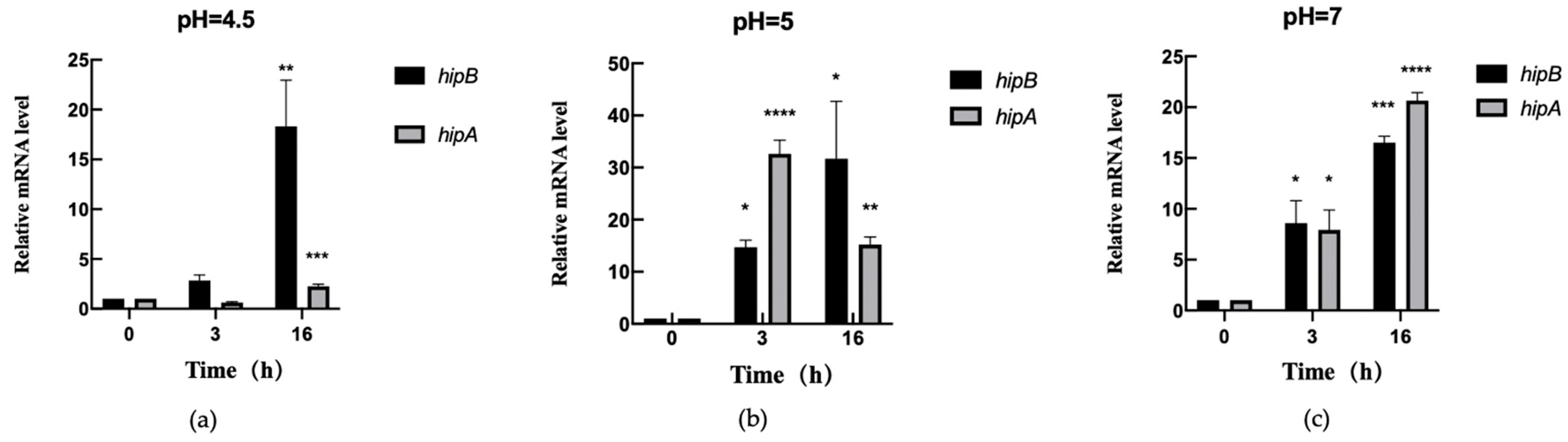
2.6.2. Transcription Level Analysis of hipA and hipB in Aac5 Under pH Stress
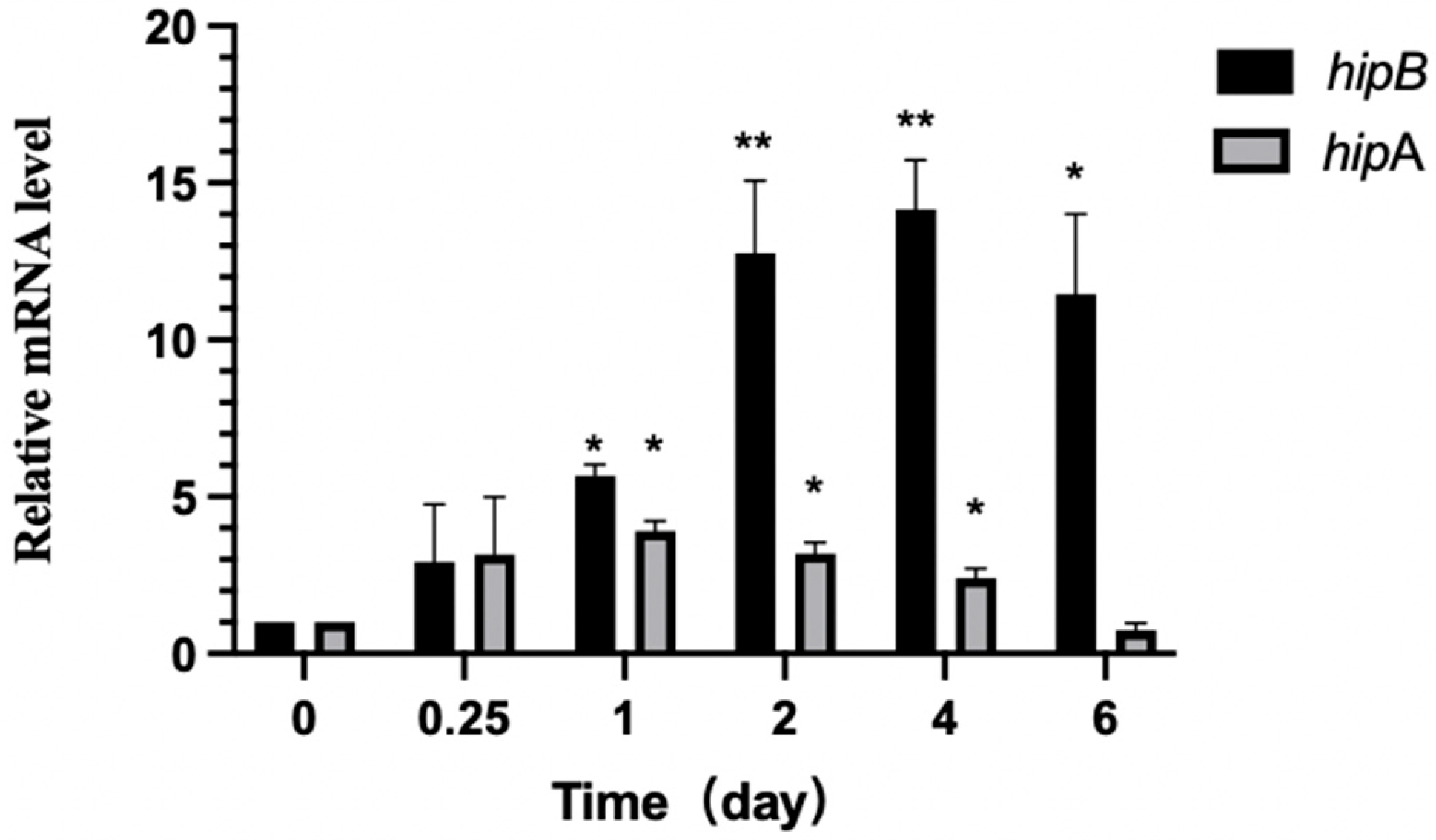
2.6.3. Differential Expression of hipA and hipB in Aac5 During Pathogen–Host Interaction
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Plasmids, Plant Materials, and Growth Conditions
4.2. Bioinformatics Analysis
4.3. Co-Transcription Test
4.4. Effect of Heterologous Expression of Predictable Proteins HipA on E. coli Growth
4.5. Bacterial Two-Hybrid Experiment
4.6. Induced Expression and Purification of Target Proteins
4.7. Western Blot
4.8. EMSA
4.9. Quantitative Real-Time (qRT) PCR Analyses of hipA and hipB in mRNA Expression Levels in A. citrulli Aac5
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Strains or Plasmids | Description | Reference or Source |
|---|---|---|
| Strains | ||
| Acidovorax citrulli Aac5 | Wild-type, AmpR | (Yan et al. [8]) |
| BL21(DE3) | F-ompT hsdSB(rB-mB-)galdcm(DE3) | Biomed Company, Beijing |
| BTH101 | F⁻ cya-99 araD139 galE15 galK16 rpsL1 hsdR2 mcrA1 mcrB1 | This study |
| Plasmids | ||
| pET-28a | Prokaryotic expression vector, KanR | Zhao Lab collection |
| hipA-pET-28a | Prokaryotic expression vector carrying hipA gene, KanR | This study |
| DH5ɑ-pRK600 | Helper strain in tri-parental mating, CmR | Zhao Lab collection |
| hipB-pET-28a | pET-28a carrying hipB gene, KanR | This study |
| His-hipA-pET-28a | pET-28a carrying hipA gene with a His tag, KanR | This study |
| His-hipB-pET-28a | pET-28a carrying hipB gene with a His tag, KanR | This study |
| pUT18Cm-hipA | pUT18 carrying hipA gene, AmpR | This study |
| pUT18Cm-hipB | pUT18 carrying hipB gene, AmpR | This study |
| pKT25m-hipB | pKT25 carrying hipB gene, KanR | This study |
| pKT25m-hipA | pKT25 carrying hipA gene, KanR | This study |
| Primers | Primer Sequence (5′ > 3′) | Product Length/bp |
|---|---|---|
| hipA-F | ATGGTGGCGCTCGACGTGA | 1323 |
| hipA-R | TCAAGGGAGGCTGGCG | |
| hipB-F | ATGGACTATCCCATTCACCTGA | 507 |
| hipB-R | TTACCAAGAGCCCTTCTTGGG | |
| GZ4519F | GCCAACTCCGTGAGCACC | 428 |
| GZ4519R | TGACATCGGTGTTCGGGT | |
| GZ4520F | GCGTGGATGAATGGCGAA | 1100 |
| GZ4520R | GGCCCGGATGGTGTGTAT | |
| gz1920F | CTGGTGGTGAGAGATGACGC | 1328 |
| gz1920R | CGTAGGGCCAGACGGAAA | |
| hipB-pKT25-F | AGGGACTCTAGAGGATCCATGGACTATCCCATTCACCTGA | 507 |
| hipB-pKT25-R | TCTTAGTTACTTAGGTACTTACCAAGAGCCCTTCTTGGG | |
| hipA-pKT25-F | AGGGACTCTAGAGGATCCATGGTGGCGCTCGACGTGA | 1323 |
| hipA-pKT25-R | TCTTAGTTACTTAGGTACTCAAGGGAGGCTGGCG2 | |
| hipB-pUT18-F | GTCGACTCTAGAGGATCCATGGACTATCCCATTCACC | 507 |
| hipA-pUT18-R | GAATTCGAGCTCGGTACCGGGAGGCTGGCG | |
| rpoB-F | GCGACAGCGTGCTCAAAGTG | 104 |
| rpoB-R | GCCTTCGTTGGTGCGTTTCT | |
| PhipBA-FAM-F | TCACCACGCTCTACAGGGACTT | 396 |
| PhipBA-FAM-R | CATCTATATCGGTTCGTGCCAA | |
| PhipBA-F | TCACCACGCTCTACAGGGACTT | 396 |
| PhipBA-R | TCACCACGCTCTACAGGGACTT |
| System 1 | System 2 | System 3 | System 4 | |
|---|---|---|---|---|
| HipB (μL) | 3 | 2 | 1 | 1 |
| FAM + DNA (μL) | 2.2 | 2.2 | 2.2 | 2.2 |
| Buffer (μL) | 2 | 2 | 2 | 2 |
| Non-specific Competitor DNA (μL) | 0 | 0 | 0 | 4.8 |
| ddH2O (μL) | 2.8 | 3.8 | 4.8 | 0 |
References
- Burdman, S.; Kots, N.; Kritzman, G.; Kopelowitz, J. Molecular, physiological, and host-range characterization of Acidovorax avenae subsp. citrulli isolates from watermelon and melon in Israel. Plant Dis. 2005, 89, 1339–1347. [Google Scholar] [CrossRef] [PubMed]
- Crall, J.M.; Schenck, N.C. Bacterial fruit rot of watermelon in Florida. Plant Dis. 1969, 53, 74–75. [Google Scholar] [CrossRef]
- Martin, H.L.; Horlock, C.M. First report of Acidovorax avenae subsp. citrulli as a pathogen of gramma in Australia. Plant Dis. 2002, 86, 1406. [Google Scholar] [CrossRef] [PubMed]
- Palkovics, L.; Petroczy, M.; Kertesz, B.; Nemeth, J.; Barsony, C.; Mike, Z.; Hevesi, M. First report of bacterial fruit blotch of watermelon caused by Acidovorax avenae subsp. citrulli in Hungary. Plant Dis. 2008, 92, 834. [Google Scholar] [CrossRef]
- Silva, G.M.; Souza, R.M.; Yan, L.; Júnior, R.S.; Medeiros, F.H.; Walcott, R.R. Strains of the group I lineage of Acidovorax citrulli, the causal agent of bacterial fruit blotch of cucurbitaceous crops, are predominant in Brazil. Phytopathology 2016, 106, 1486–1494. [Google Scholar] [CrossRef]
- Fujiwara, S.; Toshio, M.; Nakayama, E.; Tanaka, N.; Tabuchi, M. Host-specific activation of a pathogen effector Aave_4606 from Acidovorax citrulli, the causal agent for bacterial fruit blotch. Biochem. Biophys. Res. Commun. 2022, 616, 41–48. [Google Scholar] [CrossRef]
- Charlermroj, R.; Makornwattana, M.; Himananto, O.; Seepiban, C.; Phuengwas, S.; Warin, N.; Gajanandana, O.; Karoonuthaisiri, N. An accurate, specific, sensitive, high-throughput method based on a microsphere immunoassay for multiplex detection of three viruses and bacterial fruit blotch bacterium in cucurbits. J. Virol. Methods 2017, 247, 6–14. [Google Scholar] [CrossRef]
- Yan, S.; Yang, Y.; Wang, T. Genetic diversity analysis of Acidovorax citrulli in China. Eur. J. Plant Pathol. 2013, 136, 171–181. [Google Scholar] [CrossRef]
- Fan, H.Y.; Zhang, Z.W.; Li, Y.; Zhang, X.; Duan, Y.; Wang, Q. Biocontrol of bacterial fruit blotch by Bacillus subtilis 9407 via surfactin-mediated antibacterial activity and colonization. Front. Microbiol. 2017, 8, 1973. [Google Scholar] [CrossRef]
- Jiang, C.H.; Wu, F.; Yu, Z.Y.; Xie, P.; Ke, H.J.; Li, H.W.; Yu, Y.Y.; Guo, J.H. Study on screening and antagonistic mechanisms of Bacillus amyloliquefaciens 54 against bacterial fruit blotch (BFB) caused by Acidovorax avenae subsp. citrulli. Microbiol. Res. 2015, 170, 95–104. [Google Scholar] [CrossRef]
- Xie, Y.; Wei, Y.; Shen, Y.; Li, X.; Zhou, H.; Tai, C.; Deng, Z.; Ou, H.Y. TADB 2.0: An updated database of bacterial type II toxin-antitoxin loci. Nucleic. Acids. Res. 2018, 46, D749–D753. [Google Scholar] [CrossRef] [PubMed]
- Aakre, C.D.; Phung, T.N.; Huang, D.; Laub, M.T. A bacterial toxin inhibits DNA replication elongation through a direct interaction with the beta sliding clamp. Mol. Cell. 2013, 52, 617–628. [Google Scholar] [CrossRef] [PubMed]
- Aizenman, E.; Engelberg-Kulka, H.; Glaser, G. An Escherichia coli chromosomal addiction module regulated by guanosine 3′,5′-bispyrophosphate—A model for programmed bacterial cell death. Proc. Natl. Acad. Sci. USA 1996, 93, 6059–6063. [Google Scholar] [CrossRef] [PubMed]
- Harms, A.; Brodersen, D.E.; Mitarai, N.; Gerdes, K. Toxins, targets, and triggers: An overview of toxin-antitoxin biology. Mol. Cell. 2018, 70, 768–784. [Google Scholar] [CrossRef]
- Ogura, T.; Hiraga, S. Mini-F plasmid genes that couple host cell division to plasmid proliferation. Proc. Natl. Acad. Sci. USA 1983, 80, 4784–4788. [Google Scholar] [CrossRef]
- Gerdes, K.; Rasmussen, P.B.; Molin, S. Unique type of plasmid maintenance function: Postsegregational killing of plasmid-free cells. Proc. Natl. Acad. Sci. USA 1986, 83, 3116–3120. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Park, J.H.; Inouye, M. Toxin-antitoxin systems in bacteria and archaea. Annu. Rev. Genet. 2011, 45, 61–79. [Google Scholar]
- Guo, Y.; Yao, J.; Sun, C.; Wen, Z.; Wang, X. Characterization of the deep-sea Streptomyces sp. SCSIO 02999 derived VapC/VapB Toxin-antitoxin system in Escherichia coli. Toxins 2016, 8, 195. [Google Scholar] [CrossRef]
- Sevin, E.W.; Barloy-Hubler, F. RASTA-Bacteria: A web-based tool for identifying toxin-antitoxin loci in prokaryotes. Genome Bio. 2007, 8, R155. [Google Scholar] [CrossRef]
- Wang, X.; Kan, Y.; Bai, K.; Xu, X.; Chen, X.; Yu, C.; Shi, J.; Jiang, N.; Li, J.; Luo, L. A novel double-ribonuclease toxin-antitoxin system linked to the stress response and survival of Acidovorax citrulli. Microbiol. Spectr. 2023, 11, e0216923. [Google Scholar] [CrossRef]
- Moyed, H.S.; Bertrand, K.P. hipA a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J. Bacteriol. 1983, 155, 768–775. [Google Scholar] [CrossRef] [PubMed]
- Shavit, R.; Lebendiker, M.; Pasternak, Z.; Burdman, S.; Helman, Y. The vapB-vapC operon of Acidovorax citrulli functions as a bona-fide toxin-antitoxin module. Front. Microbiol. 2016, 6, 1499. [Google Scholar] [CrossRef]
- Cheverton, A.M.; Gollan, B.; Przydacz, M.; Wong, C.T.; Mylona, A.; Hare, S.A.; Helaine, S. A Salmonella toxin promotes persister formation through acetylation of tRNA. Mol. Cell. 2016, 63, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Kessler, D.A.; Ben-Jacob, E.; Levine, H. Growth feedback as a basis for persister bistability. Proc. Natl. Acad. Sci. USA 2014, 7, 544. [Google Scholar] [CrossRef]
- Schumacher, M.A.; Balani, P.; Min, J.; Chinnam, N.B.; Hansen, S.; Vulić, M.; Lewis, K.; Brennan, R.G. HipBA-promoter structures reveal the basis of heritable multidrug tolerance. Nature 2015, 524, 59–64. [Google Scholar] [CrossRef]
- Wood, T.K.; Knabel, S.J.; Kwan, B.W. Bacterial persister cell formation and dormancy. Appl. Environ. Microbiol. 2013, 79, 7116–7121. [Google Scholar] [CrossRef]
- Korch, S.B.; Henderson, T.A.; Hill, T.M. Characterization of the hipA7 allele of Escherichia coli and evidence that high persistence is governed by (p) ppGpp synthesis. Mol. Microbiol. 2003, 50, 1199–1213. [Google Scholar] [CrossRef]
- Qiao, P.; Zhao, M.; Guan, W.; Walcott, R.; Ye, Y.F.; Yang, Y.W.; Zhao, T.C. A putative multi-sensor hybrid histidine kinase, BarAAc, inhibits the expression of the type III secretion system regulator HrpG in Acidovorax citrulli. Front. Microbiol. 2022, 13, 1064577. [Google Scholar] [CrossRef]
- Wang, Y.; Zhou, Q.; Li, B.; Wu, G.; Ibrahim, M.; Xie, G.; Li, H.; Sun, G. Differentiation in MALDI-TOF MS and FTIR spectra between two closely related species Acidovorax oryzae and Acidovorax citrulli. BMC Microbiol. 2012, 12, 182. [Google Scholar] [CrossRef]
- Huang, Z.; Ji, Z.; Wang, J.; Li, Z.; Jiang, Z.; Ni, W.; Chen, H.; Wei, T.; Zhang, X. Rice yellow stunt virus p3 protein enters the nucleus of leafhopper cell and localizes to viroplasm through interaction with N protein via importin α3-mediated pathway. Phytopathol. Res. 2023, 5, 48. [Google Scholar]
- Tandon, H.; Sharma, A.; Wadhwa, S.; Varadarajan, R.; Singh, R.; Srinivasan, N.; Sandhya, S. Bioinformatic and mutational studies of related toxin-antitoxin pairs in Mycobacterium tuberculosis predict and identify key functional residues. J. Biol. Chem. 2019, 294, 9048–9063. [Google Scholar] [CrossRef] [PubMed]
- Kędzierska, B.; Hayes, F. Emerging roles of toxin-antitoxin modules in bacterial pathogenesis. Molecules 2016, 21, 790. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Guo, Y.; Tang, K.; Wen, Z.; Li, B.; Zeng, Z.; Wang, X. MqsR/MqsA toxin/antitoxin system regulates persistence and biofilm formation in Pseudomonas putida KT2440. Front. Microbiol. 2017, 8, 840. [Google Scholar] [CrossRef]
- Fraikin, N.; Goormaghtigh, F.; Vanmelderen, L. Type II toxin-antitoxin systems: Evolution and revolutions. J. Bacteriol. 2020, 202, e00763-19. [Google Scholar] [CrossRef]
- Afif, H.; Allali, N.; Couturier, M. The ratio between CcdA and CcdB modulates the transcriptional repression of the ccd poison-antidote system. Mol. Microbiol. 2001, 41, 73–82. [Google Scholar] [CrossRef]
- Christensen, S.K.; Mikkelsen, M.; Pedersen, K. RelE, a global inhibitor of translation, is activated during nutritional stress. Proc. Natl. Acad. Sci. USA 2001, 98, 14328–14333. [Google Scholar] [CrossRef]
- Bodogai, M.; Ferenczi, S.; Bashtovyy, D.; Miclea, P.; Papp, P.; Dusha, I. The ntrPR operon of Sinorhizobium melilotiis organized and functions as a toxin-antitoxin module. Mol. Plant Microbe Interact. 2006, 19, 811–822. [Google Scholar] [CrossRef]
- Robson, J.; McKenzie, J.; Cursons, L. The vapBC operon from Mycobacterium smegmatis is an autoregulated toxin-antitoxin module that controls growth via inhibition of translation. J. Mol. Biol. 2009, 390, 353–367. [Google Scholar] [CrossRef]
- Winther, K.S.; Gerdes, K. Regulation of enteric vapBC transcription: Induction by VapC toxin dimer-breaking. Nucleic Acids Res. 2012, 40, 4347–4357. [Google Scholar] [CrossRef]
- Schuster, C.F.; Bertram, R. Toxin-antitoxin systems are ubiquitous and versatile modulators of prokaryotic cell fate. FEMS Microbiol. Lett. 2013, 340, 73–85. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.; Zhao, M.; Cai, L.; Guan, W.; Yang, Y.; Walcott, R.; Zhao, W.; Zhao, T. Evidence for a Functional HipBA Toxin–Antitoxin System in Acidovorax citrulli. Int. J. Mol. Sci. 2025, 26, 3366. https://doi.org/10.3390/ijms26073366
Zhang H, Zhao M, Cai L, Guan W, Yang Y, Walcott R, Zhao W, Zhao T. Evidence for a Functional HipBA Toxin–Antitoxin System in Acidovorax citrulli. International Journal of Molecular Sciences. 2025; 26(7):3366. https://doi.org/10.3390/ijms26073366
Chicago/Turabian StyleZhang, Hao, Mei Zhao, Lulu Cai, Wei Guan, Yuwen Yang, Ron Walcott, Wenjun Zhao, and Tingchang Zhao. 2025. "Evidence for a Functional HipBA Toxin–Antitoxin System in Acidovorax citrulli" International Journal of Molecular Sciences 26, no. 7: 3366. https://doi.org/10.3390/ijms26073366
APA StyleZhang, H., Zhao, M., Cai, L., Guan, W., Yang, Y., Walcott, R., Zhao, W., & Zhao, T. (2025). Evidence for a Functional HipBA Toxin–Antitoxin System in Acidovorax citrulli. International Journal of Molecular Sciences, 26(7), 3366. https://doi.org/10.3390/ijms26073366






