Abstract
Sheep (Ovis aries), domesticated from wild Asian mouflon ~10,000 years ago, are an important livestock species adapted to various ecological environments. Recent advancements in high-throughput sequencing and global environmental databases have facilitated the exploration of genetic–environmental associations, uncovering the genetic and epigenetic mechanisms behind sheep’s adaptation to multiple environments. Studies show that HIF-1α and EPAS1 enhance high-altitude adaptation via hypoxic stress regulation; UCP1 contributes to cold adaptation through non-shivering thermogenesis; SLC4A4 and GPX3 increase drought resistance by regulating renal water reabsorption; and SOCS2 likely plays a role in metabolic and stress response regulation. Additionally, sheep adapt to temperature, drought, and environmental stress through DNA methylation, transcriptional regulation (e.g., SOD1, GPX4), heat shock proteins (e.g., HSP70), and metabolic pathways (e.g., UCP1). These findings offer valuable insights for improving sheep breeding and genetic enhancement. This review summarizes the mechanisms of adaptation to high altitude, cold, heat, drought, and comprehensive climate stress.
1. Introduction
Sheep (Ovis aries) were among the earliest domesticated livestock, with archaeological and genomic evidence tracing their domestication to ~10,000 years ago in the Fertile Crescent [1]. The Asiatic mouflon (Ovis orientalis) is widely recognized as their primary ancestor, though potential gene flow from the urial (Ovis vignei) remains uncertain [2]. By ~8000–9000 years ago, sheep had spread into Mesopotamia, Europe, and Asia, with key genetic exchanges during the Bronze and Iron Ages (~5000–3000 years ago), accelerating their global spread and adaptation [3]. Initially domesticated for meat, sheep later became a key source of wool and dairy, driving specialized breeding [4]. Beyond their economic role, they shaped early trade, cultural, and agricultural systems [5]. Today, they inhabit diverse ecosystems, exhibiting extensive genetic and phenotypic diversity shaped by natural selection and artificial breeding.
Advances in multi-omics technologies have enhanced our understanding of sheep adaptation. Genomic studies reveal key adaptations to high altitude, cold, and arid environments, with EGLN1 and HIF-1α aiding hypoxia tolerance, UCP1 in thermoregulation, and GPX3 in water metabolism [6,7]. Population genetics identifies strong selection signals and historical gene flow in different sheep populations [8]. Transcriptomics clarifies gene expression under environmental stress, while epigenomics demonstrates the role of DNA methylation and histone modifications in adaptation [9,10]. The integration of these datasets has provided new insights into the complex networks driving adaptation across diverse environments [11,12,13].
This review examines the genetic and epigenetic mechanisms underlying sheep adaptation to diverse environmental stressors. By integrating genomics, transcriptomics, epigenomics, and metabolomics, we highlight key adaptive pathways, including HIF signaling for hypoxia tolerance, thermogenic regulation for cold adaptation, and water metabolism genes in arid environments. Furthermore, we identify both conserved and lineage-specific genetic signatures through population genomics and selective sweeps. By synthesizing multi-omics data, this review not only identifies existing knowledge gaps but also underscores the potential of integrative omics and genome-assisted selection in developing climate-resilient sheep breeds, ultimately contributing to sustainable livestock production in a changing global climate.
2. Diversity, Distribution, and Adaptation of Sheep Breeds
Sheep are one of the most widely distributed livestock species and are adapted to extreme environments such as high altitude, cold, heat, and drought. These complex ecological gradients have created diverse survival pressures, driving sheep to exhibit remarkable genetic and physiological adaptations through long-term natural selection [14,15]. These pressures not only shape survival strategies but also provide valuable resources for studying biological adaptation mechanisms (Figure 1) [16].
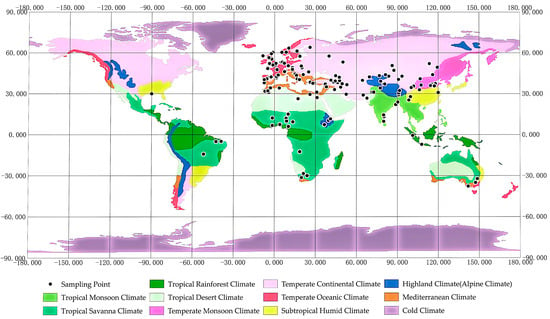
Figure 1.
Climate distribution of resequencing samples of major sheep breeds.
In high-altitude regions like the Tibetan Plateau (elevation 3500–5000 m), Tibetan sheep have adapted to low oxygen, UV radiation, and cold through the regulation of the HIF pathway and enhanced antioxidant capacity [6,7,17]. Similarly, Ethiopian Menz sheep exhibit selection in genes like PPP1R12A and RELN, which are associated with respiratory adaptation, indicating convergent evolution in high-altitude resilience [15]. In cold regions like the Mongolian Plateau and Northern China, Mongolian and Tan sheep enhance cold tolerance through fat storage and non-shivering thermogenesis, while Small-Tailed Han sheep exhibit both reproductive resilience and cold resistance in extreme climates [18,19,20]. Beyond Asia, Yakut, Baikal, Tuva, and Changthangi sheep exhibit thermogenic and metabolic adaptations, reflecting the widespread evolution of cold tolerance in sheep [21,22,23]. In hot regions like southern China and Africa, Hu sheep regulate water metabolism for thermoregulation, while fat-tailed sheep rely on tail fat reserves for energy storage and heat tolerance [24,25,26]. Globally, heat adaptation strategies vary, including heat shock protein regulation (Indian and Macheri sheep) [27,28], metabolic flexibility (Hu and Egyptian sheep) [29,30], spermatogenesis protection (Turpan black sheep) [31], and pigmentation-linked thermoregulation (Iranian sheep), highlighting genetic diversity in thermal stress resilience [32]. Drought adaptation in sheep involves diverse strategies across regions. Tan and Altay sheep enhance renal water reabsorption, while Egyptian fat-tailed sheep rely on fat metabolism for energy conservation. Taklimakan desert and Xinjiang sheep regulate osmotic balance and feed efficiency, highlighting global genetic adaptations to arid environments [33,34,35,36]. Certain breeds, such as fine-wool sheep, are both cold- and drought-tolerant, maintaining high-quality wool production across diverse ecological zones [37]. Expanding the discussion to globally distributed breeds provides a comprehensive perspective on genetic mechanisms driving environmental adaptability, contributing to sustainable breeding strategies for climate resilience.
3. Methods for Studying Environmental Adaptation
We conducted bibliometric analysis using CiteSpace 6.3.R1, with data spanning from 2004 to 2024 [38]. A 1-year time slice was applied, focusing on “Keyword” nodes with g-index (k = 25) for collaboration networks [39]. The “Pathfinder” cutting method was used to visualize keyword co-occurrence, and the Log-Likelihood Ratio (LLR) algorithm was employed to cluster keywords [40]. This analysis identified key research themes and trends in environmental adaptation, highlighting the focus on genetic mechanisms, ecological adaptability, and climate resilience (Figure 2).
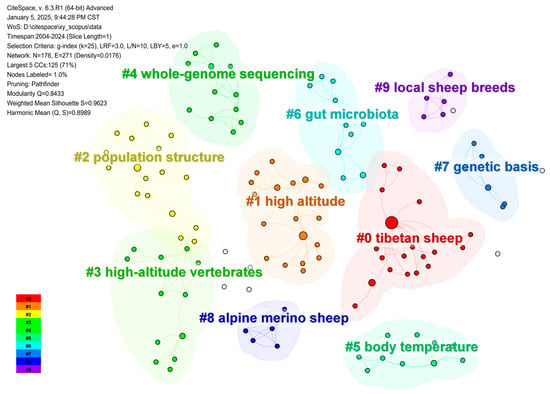
Figure 2.
Keyword co-occurrence network analysis of environmental adaptation research (2004–2024).
Whole genome sequencing (WGS) and pangenomics provide essential data for understanding environmental adaptation in sheep. Niu et al. (2024) identified 35 adaptation-related genes, including HOXA10 and JAZF1 (fat tail formation), FER and FGF5 (wool traits), and RXFP2 (horn morphology), by performing whole-genome sequencing on 266 sheep across 18 regions [41]. Missense mutations in RXFP2 and PAPSS2 were strongly linked to high-altitude adaptation, influencing skeletal morphology and metabolic processes [41]. Pangenomics provides critical insights into the interplay of core and variable genomes, where core genes ensure stability and variable genes contribute to ecological stress resistance, supporting sheep adaptation to diverse environments [12,42]. Population genetic studies using tools like ADMIXTURE and STRUCTURE have uncovered stratification among sheep populations [33,43,44]. Strong selection signals were found in EPAS1 and EGLN1 for highland sheep, particularly in Tibetan and Mongolian populations [17,45,46]. Gene flow between Xinjiang fine-wool sheep and other populations facilitated the dissemination of adaptive traits [43,45]. Contributions from wild species also enriched traits like metabolism, cold tolerance, and disease resistance. Genome-Wide Association Study (GWAS) has been pivotal in identifying the genetic bases of adaptive traits [34,43]. Studies have identified genes like ADIPOQ and TSHR linked to lipid metabolism and thermogenesis, contributing to sheep adaptation in extreme climates [47,48]. Additionally, domestication-related genes such as GDF9 and BMP15 were associated with enhanced reproductive capacity [49]. Landscape genomics integrates environmental variables with genomic data to link genes with ecological conditions [45,50,51]. Genes like EGLN1 (high-altitude) [46] and UCP1 (cold regions) show strong associations with environmental factors. Recent advancements in machine learning have facilitated the integration of environmental variables, such as temperature and precipitation, with genomic data to study adaptive traits in sheep.
Multi-omics approaches provide critical insights into sheep adaptation to environmental stresses, including temperature [32], aridity [35], UV radiation [52], and hypoxia [10]. Genomics and selection signal analyses have identified key genes (MC1R, HMOX2, BMP2 and PDGFD) linked to pigmentation [53], oxygen transport [7] and energy storage and insulation [54]. Epigenetic studies (e.g., ATAC-Seq and ChIP-Seq) have revealed the role of enhancers and promoters in regulating water retention and oxygen sensing, with the VEGFA gene implicated in adipose tissue homeostasis, which is crucial for maintaining energy balance and insulation [11]. Transcriptomic and metabolomic studies reveal that Tibetan sheep adapt to high-altitude environments by reducing lipid metabolism, enhancing cardiac function, regulating fluid balance, and boosting immunity and antioxidant capacity [55]. Microbiome studies found that Bacteroides and Prevotella enhance fiber digestion and energy production, aiding survival in arid conditions [56]. These findings highlight multi-omics as a vital tool for understanding and improving sheep’s environmental resilience.
Advanced computational tools, such as AlphaFold [57] for protein structure prediction and CRISPR-Cas9 [58] for gene validation, have revolutionized adaptation research in sheep. Integration of genomic, transcriptomic, and environmental data enables precise phenotypic predictions [54], while WGS and pangenomics uncover key adaptive mechanisms, including EGLN1 for high-altitude adaptation [17] and UCP1 for cold tolerance [18]. These advancements, combined with machine learning and multi-omics approaches, provide powerful strategies to enhance livestock resilience and sustainability under climate challenges. To further illustrate the key methodologies applied in sheep adaptation research, we summarize major research approaches and their applications in Table 1.

Table 1.
Research levels and methods in sheep studies.
4. Mechanisms of Adaptation in Sheep
4.1. Mechanisms of Hypoxia Adaptation
Sheep have developed genetic and physiological adaptations to high-altitude hypoxia, providing a valuable model for investigating mechanisms of oxygen homeostasis and hypoxia tolerance [10,83] (Table 2). Advances in genomics, transcriptomics, proteomics, and epigenetics have provided multi-level insights into these adaptive mechanisms (Figure 3). Hypoxia-inducible factors (HIFs), such as EPAS1 (HIF-2α) and HIF-1α, regulate key genes like EPO, VEGF, and PDK1, which enhance oxygen transport, angiogenesis, and energy metabolism [84]. Tibetan sheep, for instance, show strong selection signals in the EGLN1 gene, which stabilizes HIF-1α and HIF-2α, while genes like ANGPTL4 and ADAM17 contribute to vascular growth and oxygen supply [17]. Beyond genetic factors, epigenetic modifications significantly influence hypoxia adaptation. EPAS1 methylation modulates oxygen metabolism, optimizing gene expression for hypoxic conditions [85]. Additionally, histone modifications and non-coding RNAs (miRNAs, lncRNAs) regulate angiogenesis, energy metabolism, and erythropoiesis, enhancing high-altitude resilience [11,71].
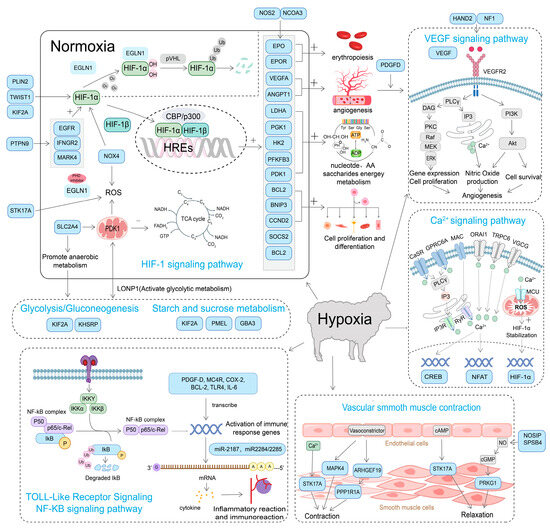
Figure 3.
Mechanisms of hypoxia adaptation in sheep.
Comparative studies with yaks (Bos grunniens) reveal convergent evolution in hypoxia-related pathways [86]. Both Tibetan sheep and yaks show selection in EPAS1, EGLN1, and PRKAA1, key regulators of the AMPK pathway for hypoxic energy metabolism [87]. However, yaks show additional enhancements in mitochondrial oxidative phosphorylation efficiency, suggesting species-specific adaptations in hypoxia tolerance [88]. Such comparisons provide an evolutionary perspective on high-altitude resilience and highlight adaptive introgression as a potential mechanism driving hypoxia adaptation in domestic sheep [89].
In response to hypoxia, genes such as SOD2 and GPX1 help reduce oxidative stress, while hemoglobin-related genes (HBA, HBB) increase oxygen-carrying capacity [17,83]. Post-translational modifications regulate hypoxia adaptation by modulating oxidative stress and oxygen transport, as seen in Tibetan sheep proteomic analysis (HBB, PRDX2, GPX1) [84]. Proteomic studies have identified critical proteins in oxygen transport, vascular development, and energy metabolism, such as HBB, PRDX2, GPX1, VEGFA, and LTBP4 [84]. Gut microbiota enhances hypoxia adaptation in Tibetan sheep by increasing Prevotellaceae-mediated volatile fatty acid production for energy metabolism [90,91,92].
Integrative multi-omics analyses have deepened our understanding of hypoxia adaptation [7]. Genetic selection in EPAS1, EGLN1, and PRKAA1 underpins genetic adaptation, while post-translational modifications and microbiota shifts enhance physiological resilience [93,94]. Future CRISPR/Cas9 and high-resolution omics studies will further refine these mechanisms for high-altitude livestock improvement.

Table 2.
Overview of known genes under local adaptation for hypoxia in sheep populations.
Table 2.
Overview of known genes under local adaptation for hypoxia in sheep populations.
| Pouplation | Genes | Function | References |
|---|---|---|---|
| Tibetan sheep | EPAS1, EGLN1, HIF1A, VEGFA, EPO, HBB | EPAS1, EGLN1, and HIF1A regulate the HIF pathway; VEGFA, HBB, HBA, and EPO enhance oxygen transport | [89,95,96] |
| Andean sheep | HMOX1, NOS3, VEGFA | HMOX1 and NOS3 modulate CO and NO signaling to regulate pulmonary vascular tone; VEGFA promotes vascular remodeling | [97,98] |
| Ethiopian sheep | PPP1R12A, RELN, PARP2, DNAH9, SDK1, ARMC3, PRDM16, COL6A3, COL25A1 | PPP1R12A, RELN, PARP2, and DNAH9 regulate respiratory system development, oxygen transport, and cellular responses to hypoxia; SDK1, ARMC3, PRDM16, COL6A3, and COL25A1 regulate oxygen transport, thermogenesis, and vascular remodeling to enhance hypoxia adaptation | [15,99] |
| Mongolian sheep | DYSF, EPAS1, JAZF1, PDGFD, NF1 | Enhance hypoxia response, vascular function, and energy metabolism for high-altitude adaptation | [33] |
4.2. Molecular Adaptations to Ultraviolet Radiation
The adaptation of sheep to UV radiation is achieved through multi-level genetic regulatory and epigenetic regulatory mechanisms. In Changthangi sheep, key genes such as TYR, TYRP1, and DCT enhance melanogenesis, effectively increasing the skin’s protection against UV radiation [100]. Epigenetic modifications such as DNA methylation in MC1R and TYR influence melanin production, regulating pigmentation patterns in high-altitude sheep [101]. Additionally, histone modifications in SLC45A2 have been linked to melanocyte differentiation, further enhancing UV protection [102]. Tibetan sheep regulate pigment deposition through MC1R and MITF, while LEF1 and GPX1 genes cooperate to enhance antioxidant capacity and repair UV-induced damage [103,104]. Studies suggest that long non-coding RNAs (lncRNAs) modulate UV response genes, influencing skin pigmentation and oxidative stress resilience [105].
At the proteomic level, the SLC45A2 gene plays a crucial role in melanogenesis, further enhancing the sheep’s tolerance to UV radiation [106,107]. Ouled Jellal sheep exhibit epigenetic regulation of SDF4, which promotes cell proliferation and survival, effectively mitigating UV-induced cellular damage [108]. In Egyptian fat-tail sheep, TGM3, RAD54L, CHEK2, and MUTYH support epidermal integrity, DNA repair, and oxidative stress defense [36]. These findings highlight the coordinated regulation of genes and proteins in UV adaptation across sheep breeds. Across different sheep breeds, multiple genes contribute to UV adaptation through pigmentation, antioxidant mechanisms, and DNA repair, as summarized in Table 3.

Table 3.
Overview of known genes under local adaptation for UV in sheep populations.
4.3. Adaptation Mechanisms to Temperature Variations
4.3.1. Cold Adaptation Mechanisms
Sheep adapt to cold environments through molecular and metabolic regulation, including both UCP1-dependent and independent thermogenesis (Figure 4).
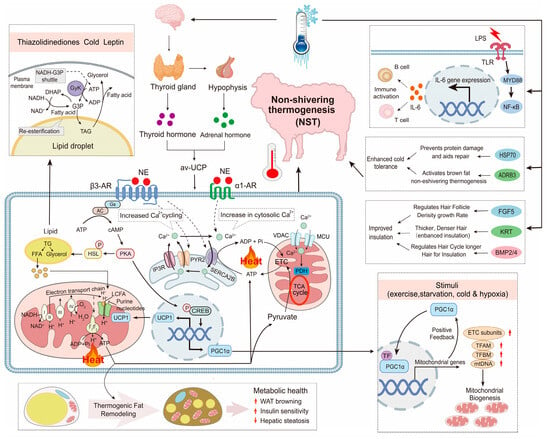
Figure 4.
Molecular mechanisms of cold adaptation in sheep. Red arrows indicate increased expression or activity levels of the corresponding components.
UCP1 in brown adipose tissue converts energy into heat to maintain body temperature, with Altay sheep primarily relying on UCP1-dependent pathways, while Hu sheep use non-UCP1 mechanisms regulated by SERCA and CKM [18]. Genes like BMPR1B and PRDM16 promote adipose browning, enhancing cold resistance [94]. Transcriptome analysis of Altay and Hu sheep under cold exposure identified PPAR (APOC3, LPL, FABP4) and cAMP (ADCY10, ADORA2a) pathways as key regulators of fatty acid metabolism and thermogenesis [110]. Additionally, BMP2 and BMP4 contribute to adipogenesis and thermogenic activation, facilitating cold adaptation in fat-storing tissues [111]. ATP2A1 and SLN were involved in calcium signaling-based non-shivering thermogenesis, further supporting energy balance and heat production under cold stress [112,113]. FGF5 regulates hair follicle cycles by promoting the transition from growth to regression, with loss-of-function mutations leading to longer wool fibers, enhancing insulation in cold-adapted sheep [114]. Cold adaptation in sheep is driven by a diverse set of genes regulating thermogenesis, lipid metabolism, and energy homeostasis across different populations (Table 4).
Cold exposure also triggers oxidative stress and immune modulation. Cold stress reduces serum immunoglobulin levels and increases pro-inflammatory cytokines (e.g., IL-6, TNF-α), reflecting suppressed immune function [115]. Cold stress upregulates HSP70 family genes (HSPA6, HSPA8), providing tissue protection and modulating inflammation [116]. Rumen microbiota, particularly Lactobacillus and Prevotella, enhance fiber digestion and SCFA production, supporting energy needs during cold seasons [117]. Host genes (FASN, CPT1A) regulate lipid metabolism and fatty acid oxidation, ensuring energy efficiency and antioxidant defense under cold stress [118]. ADRB3 also plays a role in lipid mobilization, promoting fat breakdown and thermogenic activation, further enhancing cold resistance in sheep [18].

Table 4.
Overview of known genes under local adaptation for cold in sheep populations.
Table 4.
Overview of known genes under local adaptation for cold in sheep populations.
| Pouplation | Genes | Function | References |
|---|---|---|---|
| Tibetan sheep | FKBP5, PLSCR4, CDH8, HSPA1A, HSPB1, HSPD1, HSF4 | FKBP5, PLSCR4, and CDH8 contribute to thermogenesis; HSPA1A, HSPB1, and HSPD1 enhance cold and hypoxia tolerance | [7,103] |
| Mongolian sheep | LEP, UCP1, PGC-1α, CIDEA, COX4, PM20D1 | LEP regulates metabolism; UCP1 drives WAT browning; PGC-1α enhances mitochondrial biogenesis; CIDEA and COX4 mark WAT browning; PM20D1 contributes to alternative thermogenic pathways | [20] |
| Yakut sheep | UCP1, HSP90AA1, FOXO1 | UCP1 and HSP90AA1 support thermogenesis and cold protection; FOXO1 regulates energy metabolism and antioxidant responses | [119] |
| Baikal sheep | DDB2, SOCS6 | DDB2 supports DNA repair, while SOCS6 regulates metabolism for cold adaptation | [22,23] |
| Tuva sheep | GLIS1, AADACL3, GPR179 | GLIS1 regulates cell differentiation, AADACL3 promotes fat deposition for energy storage, and GPR179 contributes to visual adaptation in cold environments | [22,23] |
| Changthangi sheep | UCP2, UCP3 | UCP2 and UCP3 enhance thermogenesis, lipid metabolism, and oxidative stress resistance | [21] |
| Altay sheep | UCP1, ADRB3, ADORA2A, ATP2A1, RYR1, IP6K1 | UCP1, ADRB3, and ADORA2A drive thermogenesis and lipid metabolism, while ATP2A1, RYR1, and IP6K1 regulate calcium signaling and energy balance, ensuring cold adaptation. | [18] |
4.3.2. Heat Adaptation Mechanisms
Heat stress triggers cellular stress responses and thermoregulation mechanisms, enabling sheep to maintain survival and productivity in high-temperature environments. Heat shock proteins (HSPs) are central to heat stress responses. HSP70 and HSP90 enhance cellular tolerance to heat stress by stabilizing proteins and inhibiting apoptosis [120,121]. Moreover, the HIF1α gene plays a key role in regulating oxygen metabolism and energy balance during heat stress in sheep, while genes such as PRLR and TNFAIP3 are involved in cellular adaptation via signaling pathways [122,123]. To mitigate heat stress, sheep upregulate glycolysis and lipid metabolism-related genes (e.g., PPARG, ACADM), providing energy to support cell survival [120]. Enhanced short-chain fatty acid (SCFA) metabolism in the rumen improves metabolic flexibility and supports adaptation to high temperatures [124]. Heat stress disrupts immune function, increasing pro-inflammatory cytokines (e.g., IL-6, IL-10) and reducing anti-inflammatory factors (e.g., TGF-β), exacerbating inflammatory responses [125]. Reactive oxygen species (ROS) accumulation induces oxidative stress, which sheep counteract by upregulating antioxidant enzymes like SOD and CAT [126]. Behavioral adaptations, such as reduced activity, seeking shade, and increased evaporative cooling, help sheep regulate body temperature under heat stress [127,128]. Heat stress impacts rumen fermentation and nutrient utilization in sheep, leading to changes in volatile fatty acid (VFA) production, which support energy metabolism and adaptation to high-temperature conditions [129,130]. The genetic basis of heat adaptation in sheep involves pathways related to thermotolerance, metabolism, and oxidative stress regulation, with population-specific variations in key adaptive genes (Table 5).

Table 5.
Overview of known genes under local adaptation for heat in sheep populations.
4.3.3. Drought Adaptation Mechanisms
Studies have shown that indigenous sheep populations in Xinjiang regulate the expression of GPX3 and GPX7 to enhance antioxidant capacity, thereby reducing water loss. Additionally, SLC4A4 and ECE1 mediate water–salt balance, optimizing renal water reabsorption to facilitate adaptation to extreme drought conditions [34]. Furthermore, Egyptian fat-tailed sheep have evolved unique metabolic adaptations, where PCK1 and ACAA2 enhance gluconeogenesis and fatty acid metabolism, improving energy efficiency while minimizing water consumption. Meanwhile, HSP70 and HSP90 function as heat shock proteins that stabilize proteins under thermal stress, ensuring cellular survival [36]. Whole-genome resequencing analyses further highlight the roles of BANK1 and TSHR in regulating energy metabolism and heat tolerance, contributing to the survival of indigenous sheep in arid environments [35]. Collectively, these genetic adaptations have played a crucial role in shaping the resilience of sheep to extreme arid conditions, providing valuable insights for breeding programs aimed at improving drought-resistant sheep populations [33]. Sheep exhibit remarkable adaptability to arid environments through a combination of genetic, physiological, and behavioral mechanisms (Table 6).

Table 6.
Overview of known genes under local adaptation for drought in sheep populations.
4.4. Integrated Environmental Adaptation Mechanisms
Sheep have developed diverse adaptations to hypoxia, cold, UV radiation, heat, and drought, enabling survival in high-altitude and arid environments. In high-altitude regions, genes such as EPAS1, EGLN1, and HIF1A enhance oxygen transport, while HSPs aid cold tolerance and MC1R, TYR regulate pigmentation for UV protection [34,131]. In deserts, HSP70 and HSP90 stabilize proteins under heat stress, AQP genes manage water retention, and FOXO1 supports fat metabolism for drought adaptation [3].
To identify genetic signatures of these adaptations, environmental–genomic approaches such as PCA and ENMs integrate climate variables with genomic data, linking key environmental factors to adaptive traits [51]. For instance, Gheyas et al. identified genomic regions associated with temperature and water scarcity adaptation. Recognizing conserved adaptive pathways, such as thyroid hormone regulation, provides insights into multi-trait adaptation [132]. The integration of multi-omics and ecological modeling facilitates the discovery of adaptive variants, supporting climate-resilient breeding strategies for improved sheep productivity under environmental stress.
5. Main Findings and Discussion
5.1. Identification and Functional Analysis of Adaptive Genes
Sheep exhibit diverse genetic adaptations to extreme environments, driven by natural selection and the genetic variability within key adaptive genes. Our analysis highlights how genetic variability in critical genes across various environmental stressors, including hypoxia, cold, drought, heat, and UV radiation, enables sheep to adapt to different environments. These genetic differences are essential for enabling populations to thrive in specific ecological niches, as they influence the functional outcomes of adaptation. In high-altitude hypoxic conditions, EPAS1, HIF1A, and EGLN1 regulate the HIF pathway, which is responsible for enhancing oxygen transport and metabolic adaptation [89,95,96]. The genetic variability within these genes allows sheep populations to have different responses to varying levels of oxygen, which is particularly critical in high-altitude environments. For instance, EPAS1 has undergone convergent evolution across multiple high-altitude domestic species, including Tibetan sheep, yaks, Tibetan cattle, and Tibetan pigs, showing that genetic diversity within this gene contributes to the adaptability of different species to low-oxygen environments. This genetic variability provides the flexibility to modulate hypoxia-related pathways in different ways, enhancing survival and reproductive success at different altitudes [86]. In cold environments, UCP1, UCP2, and UCP3 facilitate thermogenesis [20,119], and the genetic variability in these genes influences the effectiveness of non-shivering thermogenesis across sheep breeds. Similarly, LEP and PGC-1α [20] support lipid metabolism, with their genetic variations determining the extent of cold tolerance in various populations. Similar mechanisms are observed in reindeer (Rangifer tarandus) and muskox (Ovibos moschatus), which also rely on UCP1-mediated non-shivering thermogenesis to withstand extreme cold [133]. However, Tibetan sheep exhibit unique lipid metabolism adaptations, resembling those of yaks rather than other sheep breeds [134]. In arid environments, GPX3, GPX7, ANXA6, PTGS2, CPB1, and CPVL regulate water–salt metabolism and oxidative stress resistance, ensuring efficient water usage [34]. Comparable adaptations occur in dromedary camels, where AQP genes enhance water retention, while Kazakh sheep and Bactrian camels share selection signals in SLC4A4, aiding in sodium balance under drought conditions. Additionally, in hot environments, HSP70 and HSP90 mitigate oxidative damage and support heat stress resistance [28]. These genes are also critical for cattle, where strong selection in HSP90AA1 enables thermotolerance [3]. Notably, Egyptian fat-tailed sheep exhibit higher HSP expression, similar to that of desert-adapted goats, enhancing cellular stress tolerance [135]. For UV adaptation, genes such as MC1R, MITF, and GPX1 help protect against UV damage and maintain skin integrity [104,109]. Comparable pigmentation adaptations are seen in horses and cattle, where ASIP and MC1R variants contribute to coat color variation under high UV exposure [136]. Tibetan sheep exhibit strong ASIP selection, resembling the dark pigmentation patterns of Tibetan cattle and goats [137]. Reproductive adaptation is vital for survival at high altitudes, where PAPPA and BMPR1B regulate follicle growth and litter size. The genetic variability in these genes allows Tibetan sheep to optimize reproductive success under hypoxic conditions. Specifically, PAPPA influences dominant follicle development, and BMPR1B undergoes splicing and genetic variations that help improve reproductive success in response to environmental challenges [94]. Multi-omics analyses further reveal cell-type-specific expression and epigenetic modifications, highlighting key mechanisms of reproductive adaptability under hypoxia conditions [3]. These adaptive mechanisms, driven by genetic variability within key genes and their regulatory pathways, are critical for sheep survival across diverse environments. The presence of genetic diversity within these genes offers valuable targets for molecular breeding programs aimed at improving climate resilience and enhancing productivity across different ecological conditions.
5.2. Integration of Signaling Pathways
Cooperated signaling pathways play a crucial role in sheep adaptation to diverse environmental stressors. The HIF signaling pathway is central to high-altitude adaptation, with EPAS1, EGLN1, and HIF1A enhancing oxygen transport and mitochondrial efficiency [87,138], similar to yaks [88]. Additionally, the VEGF and PPAR signaling pathway further promotes survival at high altitudes [6]. To combat cold and UV-induced oxidative stress, the NRF2 antioxidant pathway mitigates ROS damage, while Ca2⁺ signaling regulates heat shock protein (HSP) responses, mitochondrial function, and energy production, supporting temperature stress resilience [139]. In cold climates, UCP1-dependent thermogenesis, modulated by calcium and cAMP pathways, facilitates heat production and metabolic adaptation, a mechanism also observed in reindeer and yak [18]. In hot and arid environments, PPAR signaling enhances lipid oxidation and water conservation, optimizing metabolic efficiency for drought resilience [140,141,142]. AMPK signaling maintains energy homeostasis, a crucial mechanism for both high-altitude and arid-adapted breeds [143]. Immune resilience is essential for sheep to survive in extreme climates. JAK/STAT and NF-κB pathways regulate adaptive immune responses, with selection signals in Tibetan and Andean sheep suggesting immunogenetic modifications to counteract hypoxia-induced immunosuppression [144,145]. Meanwhile, mTOR signaling governs autophagy and cell proliferation under nutrient limitations, a key factor in resource-scarce environments such as high-altitude grasslands and semi-arid regions [68,146]. These integrated signaling networks collectively optimize stress response, metabolic efficiency, and physiological adaptation, ensuring sheep survival and productivity across extreme environments. Future research should explore single-cell transcriptomics and CRISPR-based functional validation to elucidate species-specific signaling adaptations and enhance livestock resilience through genetic selection.
5.3. Epigenetic and Microbiome Regulation
Epigenetic modifications and gut microbiota composition are key to sheep adaptation. High-altitude hypoxia or drought induces DNA methylation and histone modifications, regulating stress–response genes. Demethylation of HIF1A and EPAS1 enhances oxygen transport and metabolism [13]. Methylation of PPARGC1A and GDF9 influences follicular development and ovulation, adjusting reproductive strategies to environmental stress [147]. Similarly, histone acetylation in BMP15 and FSHR modulates hormone signaling, promoting prolificacy in high-fecundity breeds [148]. The gut microbiome adapts to optimize nutrient absorption and metabolism. At high altitudes, Lactobacillus and Bacteroides enhance digestion and SCFA production [64], improving energy efficiency under hypoxia. SCFAs also act as epigenetic modulators, regulating metabolism and immune responses [67]. In arid conditions, microbial shifts maintain intestinal integrity and aid water conservation. The combined regulation of gene expression through epigenetics and microbiome interactions, such as through PPARG and FASN, optimizes energy efficiency and oxidative stress resistance, highlighting the integrated role of these systems in supporting sheep’s survival under harsh environmental conditions [62,63].
6. Conclusions and Future Perspectives
Sheep are highly adaptable to extreme environments, including high-altitude hypoxia, cold, and drought. Key genes such as EPAS1 and HIF1A enhance hypoxia tolerance, while UCP1 and HSP70 facilitate cold adaptation through thermogenesis and stress responses. In drought conditions, genes like GPX3 and SLC4A4 improve water reabsorption. However, environmental adaptation in sheep involves complex, multi-level regulatory networks, not just single-gene effects. While genomics and epigenetics have provided insights, functional validation of candidate genes and environmental factors remains a challenge.
Future research should explore unique adaptive mechanisms in sheep that contribute to their survival in extreme environments. For instance, how high-altitude sheep develop metabolic strategies to combat hypoxia beyond the known HIF pathway, or how drought-resistant breeds optimize renal function for water conservation. Understanding these species-specific adaptations can provide deeper insight into evolutionary biology and improve breeding programs. Additionally, integrating genomics, transcriptomics, metabolomics, and epigenomics can help construct dynamic regulatory networks to uncover novel adaptation strategies. Gene-editing technologies like CRISPR/Cas9, AI, and protein structure prediction tools (e.g., AlphaFold) can further refine our understanding of these mechanisms. Time series studies and comparative genomics across species will help reveal shared adaptive strategies and their evolutionary significance.
In livestock production, genomic selection (GS) and marker-assisted selection (MAS) techniques can accelerate the breeding of sheep adapted to extreme environments, supporting sustainable agriculture and food security. Studies on indigenous sheep genetic variation also offer insights into conservation and productivity optimization. As climate change poses new challenges, these research efforts will not only improve livestock management but also contribute to the broader understanding of biological adaptation.
Author Contributions
Conceptualization, X.G. and X.K.; literature review and investigation, L.Z. and L.T.; writing—original draft preparation, L.Z.; writing—review and editing, X.G., W.D. and X.K.; supervision, X.K.; project administration, X.G. and W.D.; additional contributions, K.Z. and H.N. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (32160771 and 32302707), the Yunnan Provincial Agricultural Union Foundation (202101BD070001-006), Major Science and Technology Projects in Yunnan Province (202202AE090005), the National Key Research and Development Program of China (grant number 2022YFD1100408), and the “Xingdian Talent” Industry Innovation Talent Program in Yunnan Province (XDYC-CYCX2022-0029).
Acknowledgments
In this section, we can acknowledge any support given that is not covered by the author’s contribution or funding sections. This may include administrative and technical support, or donations in kind (e.g., materials used for experiments).
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| HIF-1α | Hypoxia-Inducible Factor 1-Alpha |
| EPAS1 | Endothelial PAS Domain Protein 1 |
| UCP1 | Uncoupling Protein 1 |
| SLC4A4 | Solute Carrier Family 4 Member 4 |
| GPX3 | Glutathione Peroxidase 3 |
| SOCS2 | Suppressor of Cytokine Signaling 2 |
| SOD1 | Superoxide Dismutase 1 |
| GPX4 | Glutathione Peroxidase 4 |
| HSP70 | Heat Shock Protein 70 |
| BMP2 | Bone Morphogenetic Protein 2 |
| BMP4 | Bone Morphogenetic Protein 4 |
| VEGFA | Vascular Endothelial Growth Factor A |
| EGLN1 | Egl-9 Family Hypoxia-Inducible Factor 1 |
| PPARGC1A | Peroxisome Proliferator-Activated Receptor Gamma Coactivator 1-Alpha |
| GDF9 | Growth Differentiation Factor 9 |
| BMPR1B | Bone Morphogenetic Protein Receptor Type 1B |
| FSHR | Follicle-Stimulating Hormone Receptor |
| MC1R | Melanocortin 1 Receptor |
| MITF | Microphthalmia-Associated Transcription Factor |
| LEF1 | Lymphoid Enhancer-Binding Factor 1 |
| PRDM16 | PR/SET Domain 16 |
| ATP2A1 | Sarcoplasmic/Endoplasmic Reticulum Calcium ATPase 1 |
| ADRB3 | Beta-3 Adrenergic Receptor |
| PPARγ | Peroxisome Proliferator-Activated Receptor Gamma |
| AMPK | AMP-Activated Protein Kinase |
| JAK/STAT | Janus Kinase/Signal Transducers and Activators of Transcription |
| NF-κB | Nuclear Factor Kappa B |
| mTOR | Mechanistic Target of Rapamycin |
| AQP | Aquaporin |
| FOXO1 | Forkhead Box O1 |
| NOS3 | Nitric Oxide Synthase 3 |
| PARP2 | Poly(ADP-Ribose) Polymerase 2 |
| DNAH9 | Dynein Axonemal Heavy Chain 9 |
| SDK1 | Sidekick Cell Adhesion Molecule 1 |
| ARMC3 | Armadillo Repeat Containing 3 |
| PRDM16 | PR/SET Domain 16 |
| COL6A3 | Collagen Type VI Alpha 3 Chain |
| COL25A1 | Collagen Type XXV Alpha 1 Chain |
| HSPA1A | Heat Shock Protein Family A (Hsp70) Member 1A |
| HSPB1 | Heat Shock Protein Family B (Small) Member 1 |
| HSPD1 | Heat Shock Protein Family D (Hsp60) Member 1 |
| HSF4 | Heat Shock Transcription Factor 4 |
| PMEL | Premelanosome Protein |
| MLANA | Melan-A |
| DDB2 | Damage-Specific DNA Binding Protein 2 |
| SOCS6 | Suppressor of Cytokine Signaling 6 |
| GLIS1 | GLIS Family Zinc Finger 1 |
| AADACL3 | Arylacetamide Deacetylase Like 3 |
| GPR179 | G Protein-Coupled Receptor 179 |
References
- Daly, K.G.; Mullin, V.E.; Hare, A.J.; Halpin, Á.; Mattiangeli, V.; Teasdale, M.D.; Rossi, C.; Geiger, S.; Krebs, S.; Medugorac, I.; et al. Ancient Genomics and the Origin, Dispersal, and Development of Domestic Sheep. Science 2025, 387, 492–497. [Google Scholar] [CrossRef]
- Deng, J.; Xie, X.-L.; Wang, D.-F.; Zhao, C.; Lv, F.-H.; Li, X.; Yang, J.; Yu, J.-L.; Shen, M.; Gao, L.; et al. Paternal Origins and Migratory Episodes of Domestic Sheep. Curr. Biol. 2020, 30, 4085–4095.e6. [Google Scholar] [CrossRef] [PubMed]
- Alberto, F.J.; Boyer, F.; Orozco-terWengel, P.; Streeter, I.; Servin, B.; de Villemereuil, P.; Benjelloun, B.; Librado, P.; Biscarini, F.; Colli, L.; et al. Convergent Genomic Signatures of Domestication in Sheep and Goats. Nat. Commun. 2018, 9, 813. [Google Scholar] [CrossRef] [PubMed]
- Chessa, B.; Pereira, F.; Arnaud, F.; Amorim, A.; Goyache, F.; Mainland, I.; Kao, R.R.; Pemberton, J.M.; Beraldi, D.; Stear, M.J.; et al. Revealing the History of Sheep Domestication Using Retrovirus Integrations. Science 2009, 324, 532–536. [Google Scholar] [CrossRef]
- Zeder, M.A. Domestication and Early Agriculture in the Mediterranean Basin: Origins, Diffusion, and Impact. Proc. Natl. Acad. Sci. USA 2008, 105, 11597. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Yuan, C.; An, X.; Guo, T.; Zhang, W.; Lu, Z.; Liu, J. Genome-Wide Selection Signals Reveal Candidate Genes Associated with Plateau Adaptation in Tibetan Sheep. Animals 2024, 14, 3212. [Google Scholar] [CrossRef]
- Zhang, W.; Yuan, C.; An, X.; Guo, T.; Wei, C.; Lu, Z.; Liu, J. Genomic Insights into Tibetan Sheep Adaptation to Different Altitude Environments. Int. J. Mol. Sci. 2024, 25, 12394. [Google Scholar] [CrossRef]
- Li, X.; Yang, J.; Shen, M.; Xie, X.-L.; Liu, G.-J.; Xu, Y.-X.; Lv, F.-H.; Yang, H.; Yang, Y.-L.; Liu, C.-B.; et al. Whole-Genome Resequencing of Wild and Domestic Sheep Identifies Genes Associated with Morphological and Agronomic Traits. Nat. Commun. 2020, 11, 2815. [Google Scholar] [CrossRef]
- Caiye, Z.; Song, S.; Li, M.; Huang, X.; Luo, Y.; Fang, S. Genome-Wide DNA Methylation Analysis Reveals Different Methylation Patterns in Chinese Indigenous Sheep with Different Type of Tail. Front. Vet. Sci. 2023, 10, 1125262. [Google Scholar] [CrossRef]
- Yan, Z.; Yang, J.; Wei, W.-T.; Zhou, M.-L.; Mo, D.-X.; Wan, X.; Ma, R.; Wu, M.-M.; Huang, J.-H.; Liu, Y.-J.; et al. A Time-Resolved Multi-Omics Atlas of Transcriptional Regulation in Response to High-Altitude Hypoxia across Whole-Body Tissues. Nat. Commun. 2024, 15, 3970. [Google Scholar] [CrossRef]
- Zhang, D.; Cheng, J.; Li, X.; Huang, K.; Yuan, L.; Zhao, Y.; Xu, D.; Zhang, Y.; Zhao, L.; Yang, X.; et al. Comprehensive Multi-Tissue Epigenome Atlas in Sheep: A Resource for Complex Traits, Domestication, and Breeding. iMeta 2024, 3, e254. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Lu, Y.; Gao, Z.; Yue, D.; Hong, J.; Wu, J.; Xi, D.; Deng, W.; Chong, Y. Pan-Omics in Sheep: Unveiling Genetic Landscapes. Animals 2024, 14, 273. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Chen, B.; Langda, S.; Pu, P.; Zhu, X.; Zhou, S.; Kalds, P.; Zhang, K.; Bhati, M.; Leonard, A.; et al. Multi-Omic Analyses Shed Light on The Genetic Control of High-Altitude Adaptation in Sheep. Genom. Proteom. Bioinform. 2024, 22, qzae030. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhao, Y.; Li, C.; Wang, L.; Tian, F.; Jin, H. Physiological, Immune Response, Antioxidant Capacity and Lipid Metabolism Changes in Grazing Sheep during the Cold Season. Animals 2022, 12, 2332. [Google Scholar] [CrossRef]
- Edea, Z.; Dadi, H.; Dessie, T.; Kim, K.-S. Genomic Signatures of High-Altitude Adaptation in Ethiopian Sheep Populations. Genes Genom. 2019, 41, 973–981. [Google Scholar] [CrossRef]
- Zhu, M.; Yang, Y.; Yang, H.; Zhao, Z.; Zhang, H.; Blair, H.T.; Zheng, W.; Wang, M.; Fang, C.; Yu, Q.; et al. Whole-Genome Resequencing of the Native Sheep Provides Insights into the Microevolution and Identifies Genes Associated with Reproduction Traits. BMC Genom. 2023, 24, 392. [Google Scholar] [CrossRef]
- Wei, C.; Wang, H.; Liu, G.; Zhao, F.; Kijas, J.W.; Ma, Y.; Lu, J.; Zhang, L.; Cao, J.; Wu, M.; et al. Genome-Wide Analysis Reveals Adaptation to High Altitudes in Tibetan Sheep. Sci. Rep. 2016, 6, 26770. [Google Scholar] [CrossRef]
- Jiao, D.; Ji, K.; Liu, H.; Wang, W.; Wu, X.; Zhou, J.; Zhang, Y.; Zhou, H.; Hickford, J.G.H.; Degen, A.A.; et al. Transcriptome Analysis Reveals Genes Involved in Thermogenesis in Two Cold-Exposed Sheep Breeds. Genes 2021, 12, 375. [Google Scholar] [CrossRef]
- Wang, H.; Zhong, L.; Dong, Y.; Meng, L.; Ji, C.; Luo, H.; Fu, M.; Qi, Z.; Mi, L. Whole-Genome Resequencing Reveals Domestication and Signatures of Selection in Ujimqin, Sunit, and Wu Ranke Mongolian Sheep Breeds. Anim. Biosci. 2022, 35, 1303–1313. [Google Scholar] [CrossRef]
- Zhang, Y.-M. Role of White Adipose Tissue Browning in Cold Seasonal Acclimation in Grazing Mongolian Sheep (Ovis aries). J. Therm. Biol. 2022, 109, 103333. [Google Scholar] [CrossRef]
- Ahmad, S.F.; Mehrotra, A.; Charles, S.; Ganai, N.A. Analysis of Selection Signatures Reveals Important Insights into the Adaptability of High-Altitude Indian Sheep Breed Changthangi. Gene 2021, 799, 145809. [Google Scholar] [CrossRef]
- Yudin, N.S.; Larkin, D.M. Candidate Genes for Domestication and Resistance to Cold Climate According to Whole Genome Sequencing Data of Russian Cattle and Sheep Breeds. Vavilovskii Zhurnal Genet. Sel. 2023, 27, 463–470. [Google Scholar] [CrossRef]
- Sweet-Jones, J.; Yurchenko, A.A.; Igoshin, A.V.; Yudin, N.S.; Swain, M.T.; Larkin, D.M. Resequencing and Signatures of Selection Scan in Two Siberian Native Sheep Breeds Point to Candidate Genetic Variants for Adaptation and Economically Important Traits. Anim. Genet. 2021, 52, 126–131. [Google Scholar] [CrossRef] [PubMed]
- Baazaoui, I.; Bedhiaf-Romdhani, S.; Mastrangelo, S.; Ciani, E. Genome-Wide Analyses Reveal Population Structure and Identify Candidate Genes Associated with Tail Fatness in Local Sheep from a Semi-Arid Area. Animal 2021, 15, 100193. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.X.; Feng, X.P.; Wang, H.L.; Meng, C.H.; Zhang, J.; Qian, Y.; Zhong, J.F.; Cao, S.X. Transcriptome Analysis Reveals Corresponding Genes and Key Pathways Involved in Heat Stress in Hu Sheep. Cell Stress Chaperones 2019, 24, 1045–1054. [Google Scholar] [CrossRef]
- Ahbara, A.M.; Musa, H.H.; Robert, C.; Abebe, A.; Al-Jumaili, A.S.; Kebede, A.; Latairish, S.; Agoub, M.O.; Clark, E.; Hanotte, O.; et al. Natural Adaptation and Human Selection of Northeast African Sheep Genomes. Genomics 2022, 114, 110448. [Google Scholar] [CrossRef]
- Jaiswal, L.; De, S.; Singh, R.K.; Baithalu, R.K. Molecular Characterization and Protein Structure Prediction of Heat Shock Transcriptional Factors in Goat (Capra hircus) and Sheep (Ovis aries). Anim. Biotechnol. 2020, 31, 432–439. [Google Scholar] [CrossRef]
- Singh, K.M.; Singh, S.; Ganguly, I.; Nachiappan, R.K.; Ganguly, A.; Venkataramanan, R.; Chopra, A.; Narula, H.K. Association of Heat Stress Protein 90 and 70 Gene Polymorphism with Adaptability Traits in Indian Sheep (Ovis aries). Cell Stress Chaperones 2017, 22, 675–684. [Google Scholar] [CrossRef]
- Aboul-Naga, A.M.; Alsamman, A.M.; El Allali, A.; Elshafie, M.H.; Abdelal, E.S.; Abdelkhalek, T.M.; Abdelsabour, T.H.; Mohamed, L.G.; Hamwieh, A. Genome-Wide Analysis Identified Candidate Variants and Genes Associated with Heat Stress Adaptation in Egyptian Sheep Breeds. Front. Genet. 2022, 13, 898522. [Google Scholar] [CrossRef]
- Li, Y.; Kong, L.; Deng, M.; Lian, Z.; Han, Y.; Sun, B.; Guo, Y.; Liu, G.; Liu, D. Heat Stress-Responsive Transcriptome Analysis in the Liver Tissue of Hu Sheep. Genes 2019, 10, 395. [Google Scholar] [CrossRef]
- Song, Y.; Zhao, X.; Aihemaiti, A.; Haire, A.; Gao, Y.; Niu, C.; Yang, P.; Liu, G.; Jia, G.; Wusiman, A. The Mechanism of Heat Stress Resistance During Spermatogenesis in Turpan Black Sheep. Front. Vet. Sci. 2022, 9, 846981. [Google Scholar] [CrossRef] [PubMed]
- Patiabadi, Z.; Razmkabir, M.; EsmailizadehKoshkoiyeh, A.; Moradi, M.H.; Rashidi, A.; Mahmoudi, P. Whole-Genome Scan for Selection Signature Associated with Temperature Adaptation in Iranian Sheep Breeds. PLoS ONE 2024, 19, e0309023. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.; Wang, H.; Liu, G.; Lu, J.; Yuan, Z.; Li, T.; Liu, E.; Lu, Z.; Du, L.; Wei, C. Whole-Genome Resequencing of Chinese Indigenous Sheep Provides Insight into the Genetic Basis Underlying Climate Adaptation. Genet. Sel. Evol. 2024, 56, 26. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Li, W.-R.; Lv, F.-H.; He, S.-G.; Tian, S.-L.; Peng, W.-F.; Sun, Y.-W.; Zhao, Y.-X.; Tu, X.-L.; Zhang, M.; et al. Whole-Genome Sequencing of Native Sheep Provides Insights into Rapid Adaptations to Extreme Environments. Mol. Biol. Evol. 2016, 33, 2576–2592. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, J.; Tuersuntuoheti, M.; Zhou, W.; Han, Z.; Li, X.; Yang, R.; Zhang, L.; Zheng, L.; Liu, S. Landscape Genomics Reveals Adaptive Divergence of Indigenous Sheep in Different Ecological Environments of Xinjiang, China. Sci. Total Environ. 2023, 904, 166698. [Google Scholar] [CrossRef]
- Mwacharo, J.M.; Kim, E.-S.; Elbeltagy, A.R.; Aboul-Naga, A.M.; Rischkowsky, B.A.; Rothschild, M.F. Genomic Footprints of Dryland Stress Adaptation in Egyptian Fat-Tail Sheep and Their Divergence from East African and Western Asia Cohorts. Sci. Rep. 2017, 7, 17647. [Google Scholar] [CrossRef] [PubMed]
- Guo, T.; Zhao, H.; Yuan, C.; Huang, S.; Zhou, S.; Lu, Z.; Niu, C.; Liu, J.; Zhu, S.; Yue, Y.; et al. Selective Sweeps Uncovering the Genetic Basis of Horn and Adaptability Traits on Fine-Wool Sheep in China. Front. Genet. 2021, 12, 604235. [Google Scholar] [CrossRef]
- Chen, C. Searching for Intellectual Turning Points: Progressive Knowledge Domain Visualization. Proc. Natl. Acad. Sci. USA 2004, 101 (Suppl. 1), 5303–5310. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Ibekwe-SanJuan, F.; Hou, J. The Structure and Dynamics of Cocitation Clusters: A Multiple-Perspective Cocitation Analysis. J. Am. Soc. Inf. Sci. Technol. 2010, 61, 1386–1409. [Google Scholar] [CrossRef]
- Chen, C.; Song, M. Visualizing a Field of Research: A Methodology of Systematic Scientometric Reviews. PLoS ONE 2019, 14, e0223994. [Google Scholar] [CrossRef]
- Niu, Y.; Li, Y.; Zhao, Y.; He, X.; Zhao, Q.; Pu, Y.; Ma, Y.; Jiang, L. Whole-Genome Sequencing Identifies Functional Genes for Environmental Adaptation in Chinese Sheep. J. Genet. Genom. 2024, 51, 1278–1285. [Google Scholar] [CrossRef]
- Woolley, S.A.; Salavati, M.; Clark, E.L. Recent Advances in the Genomic Resources for Sheep. Mamm. Genome 2023, 34, 545–558. [Google Scholar] [CrossRef] [PubMed]
- Kijas, J.W.; Lenstra, J.A.; Hayes, B.; Boitard, S.; Porto Neto, L.R.; San Cristobal, M.; Servin, B.; McCulloch, R.; Whan, V.; Gietzen, K.; et al. Genome-Wide Analysis of the World’s Sheep Breeds Reveals High Levels of Historic Mixture and Strong Recent Selection. PLoS Biol. 2012, 10, e1001258. [Google Scholar] [CrossRef]
- Zhao, C.; Wang, D.; Yang, C.; Chen, Y.; Teng, J.; Zhang, X.; Cao, Z.; Wei, X.; Ning, C.; Yang, Q.; et al. Population Structure and Breed Identification of Chinese Indigenous Sheep Breeds Using Whole Genome SNPs and InDels. Genet. Sel. Evol. 2024, 56, 60. [Google Scholar] [CrossRef]
- Lv, F.-H.; Agha, S.; Kantanen, J.; Colli, L.; Stucki, S.; Kijas, J.W.; Joost, S.; Li, M.-H.; Ajmone Marsan, P. Adaptations to Climate-Mediated Selective Pressures in Sheep. Mol. Biol. Evol. 2014, 31, 3324–3343. [Google Scholar] [CrossRef] [PubMed]
- Buroker, N.E.; Ning, X.-H.; Zhou, Z.-N.; Li, K.; Cen, W.-J.; Wu, X.-F.; Zhu, W.-Z.; Scott, C.R.; Chen, S.-H. EPAS1 and EGLN1 Associations with High Altitude Sickness in Han and Tibetan Chinese at the Qinghai–Tibetan Plateau. Blood Cells Mol. Dis. 2012, 49, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Qiao, L.; An, L.; Wang, W.; Liu, J.; Ren, Y.; Pan, Y.; Jing, J.; Liu, W. Transcriptome Analysis of Adipose Tissues from Two Fat-Tailed Sheep Breeds Reveals Key Genes Involved in Fat Deposition. BMC Genom. 2018, 19, 338. [Google Scholar] [CrossRef]
- Fonseca, P.A.S.; Suárez-Vega, A.; Arranz, J.J.; Gutiérrez-Gil, B. Integration of Selective Sweeps across the Sheep Genome: Understanding the Relationship Between Production and Adaptation Traits. Genet. Sel. Evol. 2024, 56, 40. [Google Scholar] [CrossRef]
- Hanrahan, J.P.; Gregan, S.M.; Mulsant, P.; Mullen, M.; Davis, G.H.; Powell, R.; Galloway, S.M. Mutations in the Genes for Oocyte-Derived Growth Factors GDF9 and BMP15 Are Associated with Both Increased Ovulation Rate and Sterility in Cambridge and Belclare Sheep (Ovis aries). Biol. Reprod. 2004, 70, 900–909. [Google Scholar] [CrossRef]
- Joost, S.; Bonin, A.; Bruford, M.W.; Després, L.; Conord, C.; Erhardt, G.; Taberlet, P. A Spatial Analysis Method (SAM) to Detect Candidate Loci for Selection: Towards a Landscape Genomics Approach to Adaptation. Mol. Ecol. 2007, 16, 3955–3969. [Google Scholar] [CrossRef]
- Rellstab, C.; Gugerli, F.; Eckert, A.J.; Hancock, A.M.; Holderegger, R. A Practical Guide to Environmental Association Analysis in Landscape Genomics. Mol. Ecol. 2015, 24, 4348–4370. [Google Scholar] [CrossRef] [PubMed]
- Chedid, M.; Jaber, L.S.; Giger-Reverdin, S.; Duvaux-Ponter, C.; Hamadeh, S.K. Review: Water Stress in Sheep Raised Under Arid Conditions. Can. J. Anim. Sci. 2014, 94, 243–257. [Google Scholar] [CrossRef]
- Gebreselassie, G.; Liang, B.; Berihulay, H.; Islam, R.; Abied, A.; Jiang, L.; Zhao, Z.; Ma, Y. Genomic Mapping Identifies Two Genetic Variants in the MC1R Gene for Coat Colour Variation in Chinese Tan Sheep. PLoS ONE 2020, 15, e0235426. [Google Scholar] [CrossRef]
- Xu, Y.-X.; Wang, B.; Jing, J.-N.; Ma, R.; Luo, Y.-H.; Li, X.; Yan, Z.; Liu, Y.-J.; Gao, L.; Ren, Y.-L.; et al. Whole-Body Adipose Tissue Multi-Omic Analyses in Sheep Reveal Molecular Mechanisms Underlying Local Adaptation to Extreme Environments. Commun. Biol. 2023, 6, 159. [Google Scholar] [CrossRef]
- Li, X.; Han, B.; Liu, D.; Wang, S.; Wang, L.; Pei, Q.; Zhang, Z.; Zhao, J.; Huang, B.; Zhang, F.; et al. Whole-Genome Resequencing to Investigate the Genetic Diversity and Mechanisms of Plateau Adaptation in Tibetan Sheep. J. Anim. Sci. Biotechnol. 2024, 15, 164. [Google Scholar] [CrossRef]
- Betancur-Murillo, C.L.; Aguilar-Marín, S.B.; Jovel, J. Prevotella: A Key Player in Ruminal Metabolism. Microorganisms 2022, 11, 1. [Google Scholar] [CrossRef]
- Yang, Z. AlphaFold2 and Its Applications in the Fields of Biology and Medicine. Signal Transduct. Target. Ther. 2023, 8, 115. [Google Scholar]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Zhang, D.; Yuan, C.; An, X.; Guo, T.; Lu, Z.; Liu, J. Transcriptome and Metabolome Revealed the Effects of Hypoxic Environment on Ovarian Development of Tibetan Sheep. Genomics 2025, 117, 110973. [Google Scholar] [CrossRef]
- Tian, D.; Zhang, W.; Wang, L.; Qi, J.; Xu, T.; Zuo, M.; Han, B.; Li, X.; Zhao, K. Proteo-Transcriptomic Profiles Reveal Genetic Mechanisms Underlying Primary Hair Follicle Development in Coarse Sheep Fetal Skin. J. Proteom. 2025, 310, 105327. [Google Scholar] [CrossRef]
- Zhao, B.; Luo, H.; Huang, X.; Wei, C.; Di, J.; Tian, Y.; Fu, X.; Li, B.; Liu, G.E.; Fang, L.; et al. Integration of a Single-Step Genome-Wide Association Study with a Multi-Tissue Transcriptome Analysis Provides Novel Insights into the Genetic Basis of Wool and Weight Traits in Sheep. Genet. Sel. Evol. 2021, 53, 56. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Li, F.; Wei, Z.; Ei-Samahy, M.A.; Feng, X.; Yang, F.; Wang, F. Integrative Genome-Wide DNA Methylome and Transcriptome Analysis of Ovaries from Hu Sheep with High and Low Prolific. Front. Cell Dev. Biol. 2022, 10, 820558. [Google Scholar] [CrossRef]
- Zhao, B.; Luo, H.; He, J.; Huang, X.; Chen, S.; Fu, X.; Zeng, W.; Tian, Y.; Liu, S.; Li, C.; et al. Comprehensive Transcriptome and Methylome Analysis Delineates the Biological Basis of Hair Follicle Development and Wool-Related Traits in Merino Sheep. BMC Biol. 2021, 19, 197. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Liu, Y.-J.; Wei, W.-T.; Huang, Q.-X.; Zhao, L.-P.; Luo, L.-Y.; Zhu, Q.; Zhang, L.; Chen, Y.; Ren, Y.-L.; et al. Single-Cell Transcriptome and Metagenome Profiling Reveals the Genetic Basis of Rumen Functions and Convergent Developmental Patterns in Ruminants. Genome Res. 2023, 33, 1690–1707. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.; Fei, X.; Li, T.; Lu, Z.; Chu, M.; Di, R.; He, X.; Wang, X.; Wei, C. Transcriptome Study Digs out BMP2 Involved in Adipogenesis in Sheep Tails. BMC Genom. 2022, 23, 457. [Google Scholar] [CrossRef]
- Su, Y.; He, S.; Chen, Q.; Zhang, H.; Huang, C.; Zhao, Q.; Pu, Y.; He, X.; Jiang, L.; Ma, Y.; et al. Integrative ATAC-Seq and RNA-Seq Analysis of Myogenic Differentiation of Ovine Skeletal Muscle Satellite Cell. Genomics 2024, 116, 110851. [Google Scholar] [CrossRef]
- He, H.; Fang, C.; Liu, L.; Li, M.; Liu, W. Environmental Driving of Adaptation Mechanism on Rumen Microorganisms of Sheep Based on Metagenomics and Metabolomics Data Analysis. Int. J. Mol. Sci. 2024, 25, 10957. [Google Scholar] [CrossRef]
- Tian, D.; Han, B.; Li, X.; Liu, D.; Zhou, B.; Zhao, C.; Zhang, N.; Wang, L.; Pei, Q.; Zhao, K. Genetic Diversity and Selection of Tibetan Sheep Breeds Revealed by Whole-Genome Resequencing. Anim. Biosci. 2023, 36, 991–1002. [Google Scholar] [CrossRef]
- Jiang, J.; Cao, Y.; Shan, H.; Wu, J.; Song, X.; Jiang, Y. The GWAS Analysis of Body Size and Population Verification of Related SNPs in Hu Sheep. Front. Genet. 2021, 12, 642552. [Google Scholar] [CrossRef]
- Bao, G.; Li, S.; Zhao, F.; Wang, J.; Liu, X.; Hu, J.; Shi, B.; Wen, Y.; Zhao, L.; Luo, Y. Comprehensive Transcriptome Analysis Reveals the Role of lncRNA in Fatty Acid Metabolism in the Longissimus Thoracis Muscle of Tibetan Sheep at Different Ages. Front. Nutr. 2022, 9, 847077. [Google Scholar] [CrossRef]
- Wang, F.; Liu, J.; Zeng, Q.; Zhuoga, D. Comparative Analysis of Long Noncoding RNA and mRNA Expression Provides Insights into Adaptation to Hypoxia in Tibetan Sheep. Sci. Rep. 2022, 12, 6597. [Google Scholar] [CrossRef]
- Fei, X.; Jin, M.; Wang, Y.; Li, T.; Lu, Z.; Yuan, Z.; Wang, H.; Lu, J.; Quan, K.; Di, R.; et al. Transcriptome Reveals Key microRNAs Involved in Fat Deposition Between Different Tail Sheep Breeds. PLoS ONE 2022, 17, e0264804. [Google Scholar] [CrossRef]
- He, Z.; Li, S.; Zhao, F.; Sun, H.; Hu, J.; Wang, J.; Liu, X.; Li, M.; Zhao, Z.; Luo, Y. LncRNA and Protein Expression Profiles Reveal Heart Adaptation to High-Altitude Hypoxia in Tibetan Sheep. Int. J. Mol. Sci. 2023, 25, 385. [Google Scholar] [CrossRef]
- Miao, X.; Luo, Q. Genome-Wide Transcriptome Analysis between Small-Tail Han Sheep and the Surabaya Fur Sheep Using High-Throughput RNA Sequencing. Reproduction 2013, 145, 587–596. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Jin, X.; Hao, Z.; Wang, J.; Hu, J.; Liu, X.; Li, S.; Zhao, F.; Li, M.; Zhao, Z.; et al. Identification and Screening of Circular RNAs During Adipogenic Differentiation of Ovine Preadipocyte by RNA-Seq. J. Anim. Sci. 2024, 102, skae042. [Google Scholar] [CrossRef] [PubMed]
- Tian, D.; Pei, Q.; Jiang, H.; Guo, J.; Ma, X.; Han, B.; Li, X.; Zhao, K. Comprehensive Analysis of the Expression Profiles of mRNA, lncRNA, circRNA, and miRNA in Primary Hair Follicles of Coarse Sheep Fetal Skin. BMC Genom. 2024, 25, 574. [Google Scholar] [CrossRef]
- Chen, S.; Liu, S.; Shi, S.; Jiang, Y.; Cao, M.; Tang, Y.; Li, W.; Liu, J.; Fang, L.; Yu, Y.; et al. Comparative Epigenomics Reveals the Impact of Ruminant-Specific Regulatory Elements on Complex Traits. BMC Biol. 2022, 20, 273. [Google Scholar] [CrossRef]
- Chemonges, S.; Gupta, R.; Mills, P.C.; Kopp, S.R.; Sadowski, P. Characterisation of the Circulating Acellular Proteome of Healthy Sheep Using LC-MS/MS-Based Proteomics Analysis of Serum. Proteome Sci. 2016, 15, 11. [Google Scholar] [CrossRef]
- Coutu, A.N.; Taurozzi, A.J.; Mackie, M.; Jensen, T.Z.T.; Collins, M.J.; Sealy, J. Palaeoproteomics Confirm Earliest Domesticated Sheep in Southern Africa ca. 2000 BP. Sci. Rep. 2021, 11, 6631. [Google Scholar] [CrossRef]
- Han, J.; Guo, T.; Yue, Y.; Lu, Z.; Liu, J.; Yuan, C.; Niu, C.; Yang, M.; Yang, B. Quantitative Proteomic Analysis Identified Differentially Expressed Proteins with Tail/Rump Fat Deposition in Chinese Thin- and Fat-Tailed Lambs. PLoS ONE 2021, 16, e0246279. [Google Scholar] [CrossRef]
- Sousa, S.D.; Lucini, L.; Ajmone-Marsan, P.; van Tilburg, M.F.; Moura, A.A. Untargeted Metabolomic Profiling of Accessory Sex Gland Fluid from Morada Nova Rams. Mol. Reprod. Dev. 2020, 87, 409–418. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Meng, Z.; Deng, J.; Sun, X.; Liu, T.; Tang, Y.; Zhang, Z.; Liu, Y.; Zhu, W. Metabonomic Identification of Serum Biomarkers Related to Heat Stress Tolerance of Sheep. Anim. Sci. J. 2022, 93, e13792. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Zhao, F.; Hu, J.; Wang, J.; Liu, X.; Zhao, Z.; Xi, Q.; Sun, H.; Li, S.; Luo, Y. Physiology and Transcriptomics Analysis Reveal the Contribution of Lungs on High-Altitude Hypoxia Adaptation in Tibetan Sheep. Front. Physiol. 2022, 13, 885444. [Google Scholar] [CrossRef]
- Zhao, P.; Li, S.; He, Z.; Zhao, F.; Wang, J.; Liu, X.; Li, M.; Hu, J.; Zhao, Z.; Luo, Y. Physiology and Proteomic Basis of Lung Adaptation to High-Altitude Hypoxia in Tibetan Sheep. Animals. 2022, 12, 2134. [Google Scholar] [CrossRef] [PubMed]
- Xi, Q.; Zhao, F.; Hu, J.; Wang, J.; Liu, X.; Dang, P.; Luo, Y.; Li, S. Expression and Variations in EPAS1 Associated with Oxygen Metabolism in Sheep. Genes 2022, 13, 1871. [Google Scholar] [CrossRef]
- Wu, D.-D.; Yang, C.-P.; Wang, M.-S.; Dong, K.-Z.; Yan, D.-W.; Hao, Z.-Q.; Fan, S.-Q.; Chu, S.-Z.; Shen, Q.-S.; Jiang, L.-P.; et al. Convergent Genomic Signatures of High-Altitude Adaptation Among Domestic Mammals. Natl. Sci. Rev. 2020, 7, 952–963. [Google Scholar] [CrossRef]
- Hu, X.-J.; Yang, J.; Xie, X.-L.; Lv, F.-H.; Cao, Y.-H.; Li, W.-R.; Liu, M.-J.; Wang, Y.-T.; Li, J.-Q.; Liu, Y.-G.; et al. The Genome Landscape of Tibetan Sheep Reveals Adaptive Introgression from Argali and the History of Early Human Settlements on the Qinghai–Tibetan Plateau. Mol. Biol. Evol. 2019, 36, 283–303. [Google Scholar] [CrossRef]
- Qi, X.; Zhang, Q.; He, Y.; Yang, L.; Zhang, X.; Shi, P.; Yang, L.; Liu, Z.; Zhang, F.; Liu, F.; et al. The Transcriptomic Landscape of Yaks Reveals Molecular Pathways for High Altitude Adaptation. Genome Biol. Evol. 2019, 11, 72–85. [Google Scholar] [CrossRef]
- Zhao, P.; Ma, X.; Ren, J.; Zhang, L.; Min, Y.; Li, C.; Lu, Y.; Ma, Y.; Hou, M.; Jia, H. Variations in HBA Gene Contribute to High-Altitude Hypoxia Adaptation via Affected O2 Transfer in Tibetan Sheep. Front. Zool. 2024, 21, 30. [Google Scholar] [CrossRef]
- Li, B.; Jia, G.; Wen, D.; Zhao, X.; Zhang, J.; Xu, Q.; Zhao, X.; Jiang, N.; Liu, Z.; Wang, Y. Rumen Microbiota of Indigenous and Introduced Ruminants and Their Adaptation to the Qinghai–Tibetan Plateau. Front. Microbiol. 2022, 13, 1027138. [Google Scholar] [CrossRef]
- Fan, Q.; Cui, X.; Wang, Z.; Chang, S.; Wanapat, M.; Yan, T.; Hou, F. Rumen Microbiota of Tibetan Sheep (Ovis aries) Adaptation to Extremely Cold Season on the Qinghai-Tibetan Plateau. Front. Vet. Sci. 2021, 8, 673822. [Google Scholar] [CrossRef]
- Chen, Q.; Sha, Y.; Liu, X.; He, Y.; Chen, X.; Yang, W.; Gao, M.; Huang, W.; Wang, J.; He, J.; et al. Unique Rumen Micromorphology and Microbiota–Metabolite Interactions: Features and Strategies for Tibetan Sheep Adaptation to the Plateau. Front. Microbiol. 2024, 15, 1471732. [Google Scholar] [CrossRef]
- Tan, W.; Proudfoot, C.; Lillico, S.G.; Whitelaw, C.B.A. Gene Targeting, Genome Editing: From Dolly to Editors. Transgenic Res. 2016, 25, 273–287. [Google Scholar] [CrossRef]
- Han, B.; Tian, D.; Li, X.; Liu, S.; Tian, F.; Liu, D.; Wang, S.; Zhao, K. Multiomics Analyses Provide New Insight into Genetic Variation of Reproductive Adaptability in Tibetan Sheep. Mol. Biol. Evol. 2024, 41, msae058. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Xi, Q.; He, Z.; Sun, H.; Li, S. Expression and Variations in EPO Associated with Oxygen Metabolism in Tibetan Sheep. Animals 2024, 14, 535. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Liu, M.; Mfoundou, J.D.L.; Wang, X. Expression and Distribution Patterns of VEGF, TGF-Β1 and HIF-1α in the Ovarian Follicles of Tibetan Sheep. Vet. Med. Sci. 2022, 8, 2223–2229. [Google Scholar] [CrossRef]
- Llanos, A.J.; Ebensperger, G.; Herrera, E.A.; Reyes, R.V.; Cabello, G.; Díaz, M.; Giussani, D.A.; Parer, J.T. The Heme Oxygenase-Carbon Monoxide System in the Regulation of Cardiorespiratory Function at High Altitude. Respir. Physiol. Neurobiol. 2012, 184, 186–191. [Google Scholar] [CrossRef]
- Parraguez, V.H.; Atlagich, M.A.; Urquieta, B.; Galleguillos, M.; De Los Reyes, M.; Kooyman, D.L.; Araneda, S.; Raggi, L.A. Expression of Vascular Endothelial Growth Factor and Endothelial Nitric Oxide Synthase Is Increased in the Placenta of Sheep at High Altitude in the Andes. Can. J. Vet. Res. 2010, 74, 193–199. [Google Scholar]
- Wiener, P.; Robert, C.; Ahbara, A.; Salavati, M.; Abebe, A.; Kebede, A.; Wragg, D.; Friedrich, J.; Vasoya, D.; Hume, D.A.; et al. Whole-Genome Sequence Data Suggest Environmental Adaptation of Ethiopian Sheep Populations. Genome Biol. Evol. 2021, 13, evab014. [Google Scholar] [CrossRef]
- Vasu, M.; Ahlawat, S.; Chhabra, P.; Sharma, U.; Arora, R.; Sharma, R.; Mir, M.A.; Singh, M.K. Genetic Insights into Fiber Quality, Coat Color and Adaptation in Changthangi and Muzzafarnagri Sheep: A Comparative Skin Transcriptome Analysis. Gene 2024, 891, 147826. [Google Scholar] [CrossRef]
- Zhang, W.; Jin, M.; Lu, Z.; Li, T.; Wang, H.; Yuan, Z.; Wei, C. Whole Genome Resequencing Reveals Selection Signals Related to Wool Color in Sheep. Animals 2023, 13, 3265. [Google Scholar] [CrossRef] [PubMed]
- Le, L.; Escobar, I.E.; Ho, T.; Lefkovith, A.J.; Latteri, E.; Haltaufderhyde, K.D.; Dennis, M.K.; Plowright, L.; Sviderskaya, E.V.; Bennett, D.C.; et al. SLC45A2 Protein Stability and Regulation of Melanosome pH Determine Melanocyte Pigmentation. Mol. Biol. Cell 2020, 31, 2687–2702. [Google Scholar] [CrossRef] [PubMed]
- An, L.; Li, Y.; Yaq, L.; Wang, Y.; Dai, Q.; Du, S.; Ru, Y.; Zhoucuo, Q.; Wang, J. Transcriptome Analysis Reveals Molecular Regulation Mechanism of Tibet Sheep Tolerance to High Altitude Oxygen Environment. Anim. Biotechnol. 2023, 34, 5097–5112. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Luosang, C.; Yuan, C.; Guo, T.; Wei, C.; Liu, J.; Lu, Z. Selection Signatures of Wool Color in Gangba Sheep Revealed by Genome-Wide SNP Discovery. BMC Genom. 2024, 25, 606. [Google Scholar] [CrossRef]
- Liang, X.; Zhang, C.; Shen, L.; Ding, L.; Guo, H. Role of Non-Coding RNAs in UV-induced Radiation Effects (Review). Exp. Ther. Med. 2024, 27, 262. [Google Scholar] [CrossRef]
- Wang, H.; Xue, L.; Li, Y.; Zhao, B.; Chen, T.; Liu, Y.; Chang, L.; Wang, J. Distribution and Expression of SLC45A2 in the Skin of Sheep with Different Coat Colors. Folia Histochem. Cytobiol. 2016, 54, 143–150. [Google Scholar] [CrossRef]
- Tearle, R.G.; Chen, T.; Brien, F.D. A 3-Bp Deletion in the SLC45A2 Gene Is Associated with Loss of Fleece Pigmentation in Black-Fleeced Suffolk Sheep. Anim. Genet. 2025, 56, e13495. [Google Scholar] [CrossRef]
- Ouhrouch, A.; Boitard, S.; Boyer, F.; Servin, B.; Da Silva, A.; Pompanon, F.; Haddioui, A.; Benjelloun, B. Genomic Uniqueness of Local Sheep Breeds From Morocco. Front. Genet. 2021, 12, 723599. [Google Scholar] [CrossRef]
- Yang, G.L.; Fu, D.L.; Lang, X.; Ylan, Y.F.; Luo, Y.Z. Genetic Variation of 5 SNPs of MC1R Gene in Chinese Indigenous Sheep Breeds. Genetika 2014, 50, 1188–1199. [Google Scholar] [CrossRef]
- Ji, K.; Jiao, D.; Yang, G.; Degen, A.A.; Zhou, J.; Liu, H.; Wang, W.; Cong, H. Transcriptome Analysis Revealed Potential Genes Involved in Thermogenesis in Muscle Tissue in Cold-Exposed Lambs. Front. Genet. 2022, 13, 1017458. [Google Scholar] [CrossRef]
- Young, J.M.; Juengel, J.L.; Dodds, K.G.; Laird, M.; Dearden, P.K.; McNeilly, A.S.; McNatty, K.P.; Wilson, T. The Activin Receptor-like Kinase 6 Booroola Mutation Enhances Suppressive Effects of Bone Morphogenetic Protein 2 (BMP2), BMP4, BMP6 and Growth and Differentiation Factor-9 on FSH Release from Ovine Primary Pituitary Cell Cultures. J. Endocrinol. 2008, 196, 251–261. [Google Scholar] [CrossRef]
- Bal, N.C.; Periasamy, M. Uncoupling of Sarcoendoplasmic Reticulum Calcium ATPase Pump Activity by Sarcolipin as the Basis for Muscle Non-Shivering Thermogenesis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2020, 375, 20190135. [Google Scholar] [CrossRef]
- Wang, S.; Gopinath, T.; Larsen, E.K.; Weber, D.K.; Walker, C.; Uddigiri, V.R.; Mote, K.R.; Sahoo, S.K.; Periasamy, M.; Veglia, G. Structural Basis for Sarcolipin’s Regulation of Muscle Thermogenesis by the Sarcoplasmic Reticulum Ca2+-ATPase. Sci. Adv. 2021, 7, eabi7154. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; He, S.; Liu, M.; Liu, G.; Yuan, Z.; Liu, C.; Zhang, X.; Zhang, N.; Li, W. Molecular Cloning, Characterization, and Expression of Sheep FGF5 Gene. Gene 2015, 555, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Xu, Y.; Jin, X.; Wang, Z.; Mao, C.; Guo, S.; Yan, S.; Shi, B. Influence of Cold Environments on Growth, Antioxidant Status, Immunity and Expression of Related Genes in Lambs. Animals 2022, 12, 2535. [Google Scholar] [CrossRef] [PubMed]
- Rawash, R.A.A.; Sharaby, M.A.; Hassan, G.E.-D.A.; Elkomy, A.E.; Hafez, E.E.; Hafsa, S.H.A.; Salem, M.M.I. Expression Profiling of HSP 70 and Interleukins 2, 6 and 12 Genes of Barki Sheep during Summer and Winter Seasons in Two Different Locations. Int. J. Biometeorol. 2022, 66, 2047–2053. [Google Scholar] [CrossRef]
- Zhang, L.; Ren, W.; Bi, Y.; Zhang, J.; Cheng, Y.; Xu, X. Effects of Different Feeding Patterns on the Rumen Bacterial Community of Tan Lambs, Based on High-Throughput Sequencing of 16S rRNA Amplicons. Front. Microbiol. 2023, 14, 1228935. [Google Scholar] [CrossRef]
- Mi, H.; Hu, F.; Gebeyew, K.; Cheng, Y.; Du, R.; Gao, M.; He, Z.; Tan, Z. Genome Wide Transcriptome Analysis Provides Bases on Hepatic Lipid Metabolism Disorder Affected by Increased Dietary Grain Ratio in Fattening Lambs. BMC Genom. 2023, 24, 364. [Google Scholar] [CrossRef]
- Zhang, X.; Ge, L.; Jin, G.; Liu, Y.; Yu, Q.; Chen, W.; Chen, L.; Dong, T.; Miyagishima, K.J.; Shen, J.; et al. Cold-Induced FOXO1 Nuclear Transport Aids Cold Survival and Tissue Storage. Nat. Commun. 2024, 15, 2859. [Google Scholar] [CrossRef]
- Lu, Z.; Chu, M.; Li, Q.; Jin, M.; Fei, X.; Ma, L.; Zhang, L.; Wei, C. Transcriptomic Analysis Provides Novel Insights into Heat Stress Responses in Sheep. Animals 2019, 9, 387. [Google Scholar] [CrossRef]
- McManus, C.M.; Faria, D.A.; Lucci, C.M.; Louvandini, H.; Pereira, S.A.; Paiva, S.R. Heat Stress Effects on Sheep: Are Hair Sheep More Heat Resistant? Theriogenology 2020, 155, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.; Hu, J.; Wang, J.; Liu, X.; Li, S.; Luo, Y. Effect of Glycolysis and Heat Shock Proteins on Hypoxia Adaptation of Tibetan Sheep at Different Altitude. Gene 2021, 803, 145893. [Google Scholar] [CrossRef] [PubMed]
- Marai, I.F.M.; El-Darawany, A.A.; Fadiel, A.; Abdel-Hafez, M.A.M. Physiological Traits as Affected by Heat Stress in Sheep—A Review. Small Rumin. Res. 2007, 71, 1–12. [Google Scholar] [CrossRef]
- Amini, A.; Pirmohammadi, R.; Khalilvandi, H.; Mazaheri-Khameneh, R. Effects of Heat Stress on in Vivo and in Vitro Ruminal Metabolism in Fat-Tailed Ewes. Anim. Prod. Sci. 2022, 62, 860–869. [Google Scholar] [CrossRef]
- Most, M.S.; Yates, D.T. Inflammatory Mediation of Heat Stress-Induced Growth Deficits in Livestock and Its Potential Role as a Target for Nutritional Interventions: A Review. Animals 2021, 11, 3539. [Google Scholar] [CrossRef]
- Shi, L.; Xu, Y.; Mao, C.; Wang, Z.; Guo, S.; Jin, X.; Yan, S.; Shi, B. Effects of Heat Stress on Antioxidant Status and Immune Function and Expression of Related Genes in Lambs. Int. J. Biometeorol. 2020, 64, 2093–2104. [Google Scholar] [CrossRef]
- van Wettere, W.H.E.J.; Kind, K.L.; Gatford, K.L.; Swinbourne, A.M.; Leu, S.T.; Hayman, P.T.; Kelly, J.M.; Weaver, A.C.; Kleemann, D.O.; Walker, S.K. Review of the Impact of Heat Stress on Reproductive Performance of Sheep. J. Anim. Sci. Biotechnol. 2021, 12, 26. [Google Scholar] [CrossRef]
- Berihulay, H.; Abied, A.; He, X.; Jiang, L.; Ma, Y. Adaptation Mechanisms of Small Ruminants to Environmental Heat Stress. Animals 2019, 9, 75. [Google Scholar] [CrossRef]
- Belhadj Slimen, I.; Najar, T.; Ghram, A.; Abdrrabba, M. Heat Stress Effects on Livestock: Molecular, Cellular and Metabolic Aspects, a Review. J. Anim. Physiol. Anim. Nutr. 2016, 100, 401–412. [Google Scholar] [CrossRef]
- Kim, H.R.; Seong, P.; Seol, K.-H.; Park, J.-E.; Kim, H.; Park, W.; Cho, J.H.; Lee, S.D. Effects of Heat Stress on Growth Performance, Physiological Responses, and Carcass Traits in Broilers. J. Therm. Biol. 2024, 127, 103994. [Google Scholar] [CrossRef]
- Wei, C.; Wang, H.; Liu, G.; Wu, M.; Cao, J.; Liu, Z.; Liu, R.; Zhao, F.; Zhang, L.; Lu, J.; et al. Genome-Wide Analysis Reveals Population Structure and Selection in Chinese Indigenous Sheep Breeds. BMC Genom. 2015, 16, 194. [Google Scholar] [CrossRef]
- Tsartsianidou, V.; Sánchez-Molano, E.; Kapsona, V.V.; Basdagianni, Z.; Chatziplis, D.; Arsenos, G.; Triantafyllidis, A.; Banos, G. A Comprehensive Genome-Wide Scan Detects Genomic Regions Related to Local Adaptation and Climate Resilience in Mediterranean Domestic Sheep. Genet. Sel. Evol. 2021, 53, 90. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Li, X.; Wu, Z.; Zhang, G.; Wang, N.; Dou, M.; Liu, S.; Yang, C.; Meng, G.; Sun, H.; et al. Convergent Molecular Evolution of Thermogenesis and Circadian Rhythm in Arctic Ruminants. Proc. R. Soc. B Biol. Sci. 2023, 290, 20230538. [Google Scholar] [CrossRef]
- Ahmad, H.I.; Mahmood, S.; Hassan, M.; Sajid, M.; Ahmed, I.; Shokrollahi, B.; Shahzad, A.H.; Abbas, S.; Raza, S.; Komal, K.; et al. Genomic Insights into Yak (Bos grunniens) Adaptations for Nutrient Assimilation in High-Altitudes. Sci. Rep. 2024, 14, 5650. [Google Scholar]
- Arero, G.B.; Ozmen, O. Effects of Heat Stress on Reproduction and Gene Expression in Sheep. Anim. Reprod. 2025, 22, e20240067. [Google Scholar] [CrossRef]
- Rochus, C.M.; Westberg Sunesson, K.; Jonas, E.; Mikko, S.; Johansson, A.M. Mutations in ASIP and MC1R: Dominant Black and Recessive Black Alleles Segregate in Native Swedish Sheep Populations. Anim. Genet. 2019, 50, 712–717. [Google Scholar] [CrossRef]
- Bertolini, F.; Moscatelli, G.; Schiavo, G.; Bovo, S.; Ribani, A.; Ballan, M.; Bonacini, M.; Prandi, M.; Dall’Olio, S.; Fontanesi, L. Signatures of Selection Are Present in the Genome of Two Close Autochthonous Cattle Breeds Raised in the North of Italy and Mainly Distinguished for Their Coat Colours. J. Anim. Breed. Genet. 2022, 139, 307–319. [Google Scholar] [CrossRef]
- Lee, F.S. Hypoxia Inducible Factor Pathway Proteins in High-Altitude Mammals. Trends Biochem. Sci. 2024, 49, 79–92. [Google Scholar] [CrossRef]
- Li, Y.; Yang, Y.; Yu, B.; Gao, R.; Wang, X. Transcriptome and Metabolome Analyses Reveal High-Altitude Adaptation Mechanism of Epididymis Sperm Maturation in Tibetan Sheep. Animals 2024, 14, 3117. [Google Scholar] [CrossRef]
- Wang, Y.; Li, X.; Cao, Y.; Xiao, C.; Liu, Y.; Jin, H.; Cao, Y. Effect of the ACAA1 Gene on Preadipocyte Differentiation in Sheep. Front. Genet. 2021, 12, 649140. [Google Scholar] [CrossRef]
- Chen, B.; Yuan, C.; Guo, T.; Liu, J.; Yang, B.; Lu, Z. Molecular Mechanism of m6A Methylation Modification Genes METTL3 and FTO in Regulating Heat Stress in Sheep. Int. J. Mol. Sci. 2023, 24, 11926. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Wang, L.; Xiao, C.; Zhou, M.; Li, M.; Li, H. miR136 Regulates Proliferation and Differentiation of Small Tail Han Sheep Preadipocytes. Adipocyte 2023, 12, 2173966. [Google Scholar] [CrossRef]
- Huang, Z.; Wang, J.; Qi, D.; Li, X.; Wang, J.; Zhou, J.; Ruan, Y.; Laer, Y.; Baqian, Z.; Yang, C. Uncovering the Genetic Diversity and Adaptability of Butuo Black Sheep through Whole-Genome Re-Sequencing. PLoS ONE 2024, 19, e0303419. [Google Scholar] [CrossRef] [PubMed]
- He, T.; Guo, W.; Yang, G.; Su, H.; Dou, A.; Chen, L.; Ma, T.; Su, J.; Liu, M.; Su, B.; et al. A Single-Cell Atlas of an Early Mongolian Sheep Embryo. Vet. Sci. 2023, 10, 543. [Google Scholar] [CrossRef]
- Gera, R.; Arora, R.; Chhabra, P.; Sharma, U.; Parsad, R.; Ahlawat, S.; Mir, M.A.; Singh, M.K.; Kumar, R. Exploring Transcriptomic Mechanisms Underlying Pulmonary Adaptation to Diverse Environments in Indian Rams. Mol. Biol. Rep. 2024, 51, 1111. [Google Scholar] [CrossRef] [PubMed]
- Viola, I.; Accornero, P.; Manenti, I.; Miretti, S.; Baratta, M.; Toschi, P. mTOR Is an Essential Gate in Adapting the Functional Response of Ovine Trophoblast Cells Under Stress-Inducing Environments. Placenta 2024, 158, 14–22. [Google Scholar] [CrossRef]
- Pan, Z.; Wang, X.; Di, R.; Liu, Q.; Hu, W.; Cao, X.; Guo, X.; He, X.; Lv, S.; Li, F.; et al. A 5-Methylcytosine Site of Growth Differentiation Factor 9 (GDF9) Gene Affects Its Tissue-Specific Expression in Sheep. Animals 2018, 8, 200. [Google Scholar] [CrossRef]
- Wang, W.; Liu, S.; Li, F.; Pan, X.; Li, C.; Zhang, X.; Ma, Y.; La, Y.; Xi, R.; Li, T. Polymorphisms of the Ovine BMPR-IB, BMP-15 and FSHR and Their Associations with Litter Size in Two Chinese Indigenous Sheep Breeds. Int. J. Mol. Sci. 2015, 16, 11385. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).