Odor-Binding Protein 2 in Apis mellifera ligustica Plays Important Roles in the Response to Floral Volatiles Stimuli from Melon and Tomato Flowers
Abstract
1. Introduction
2. Results
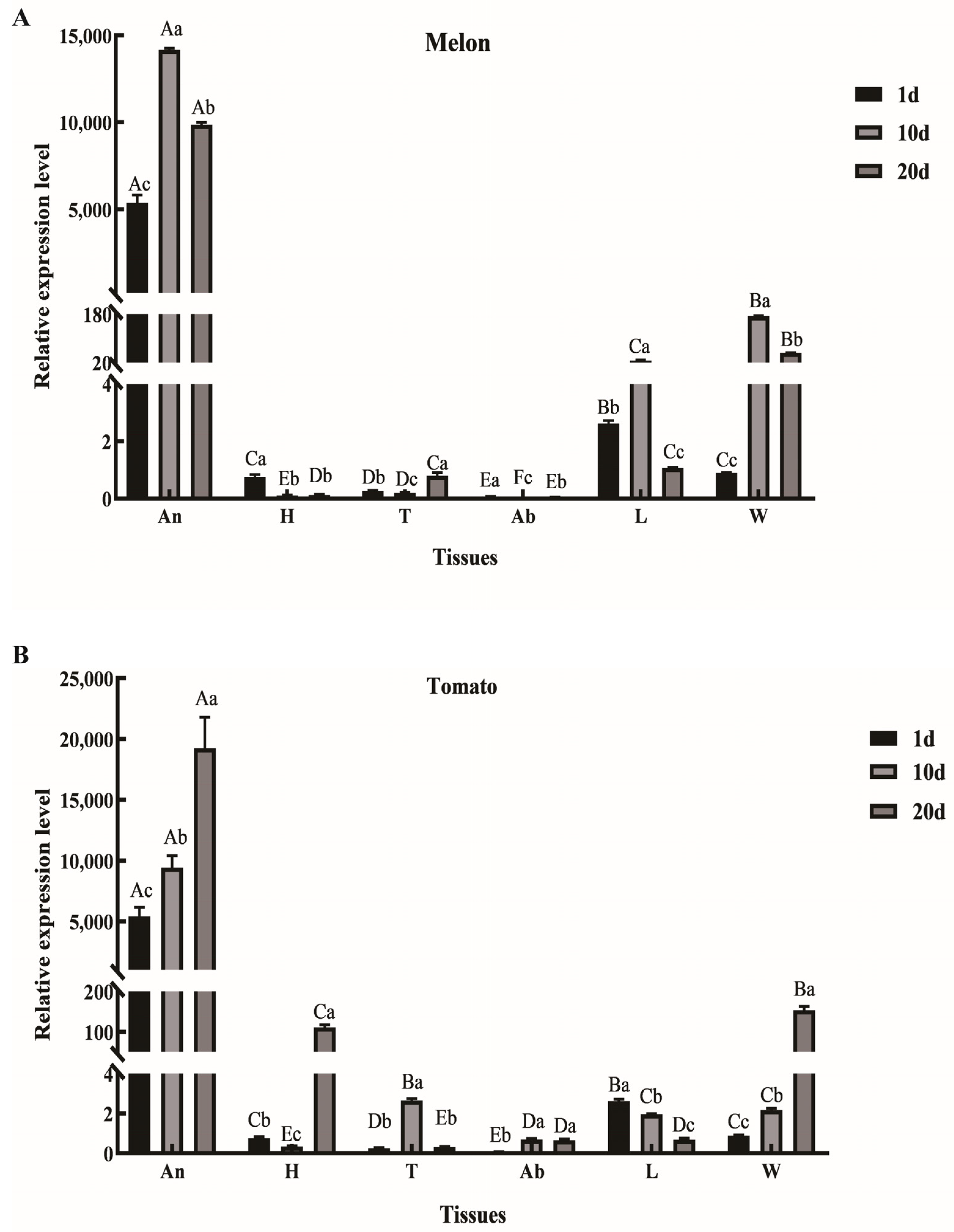
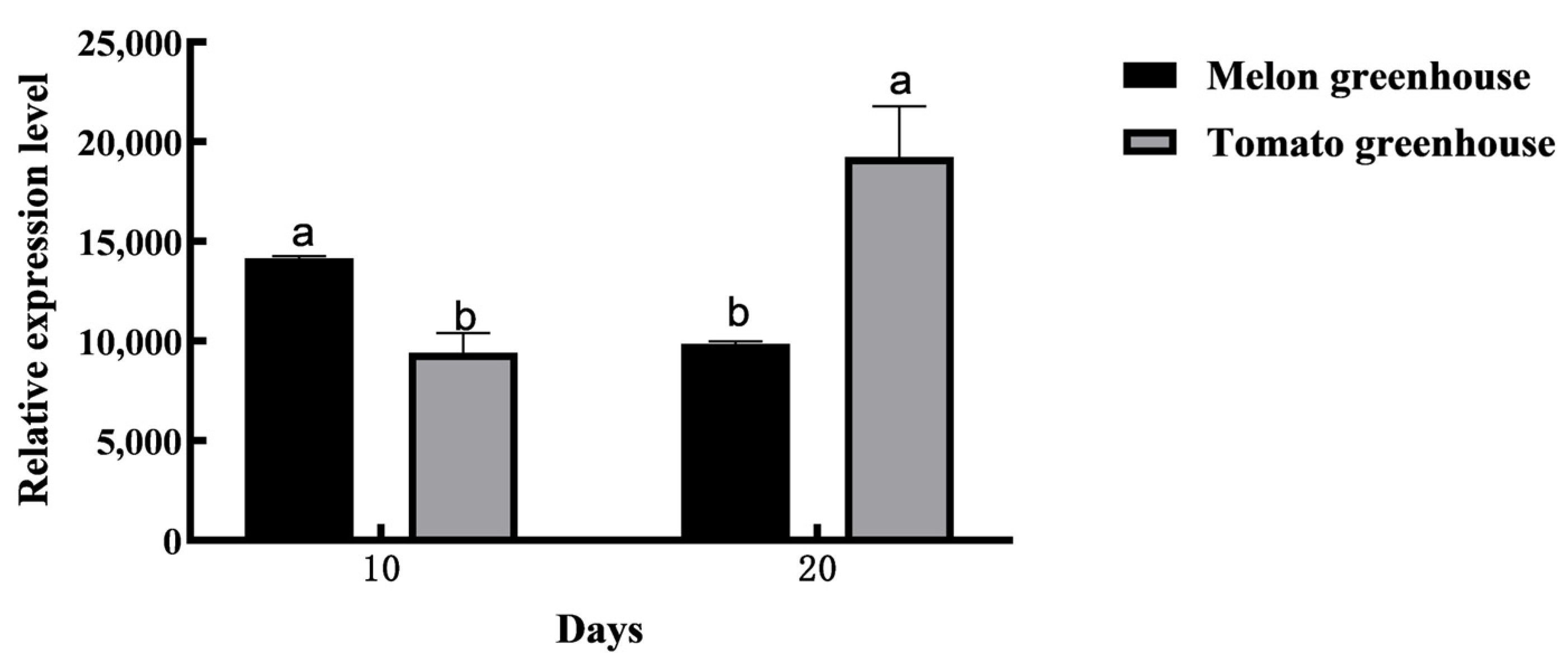
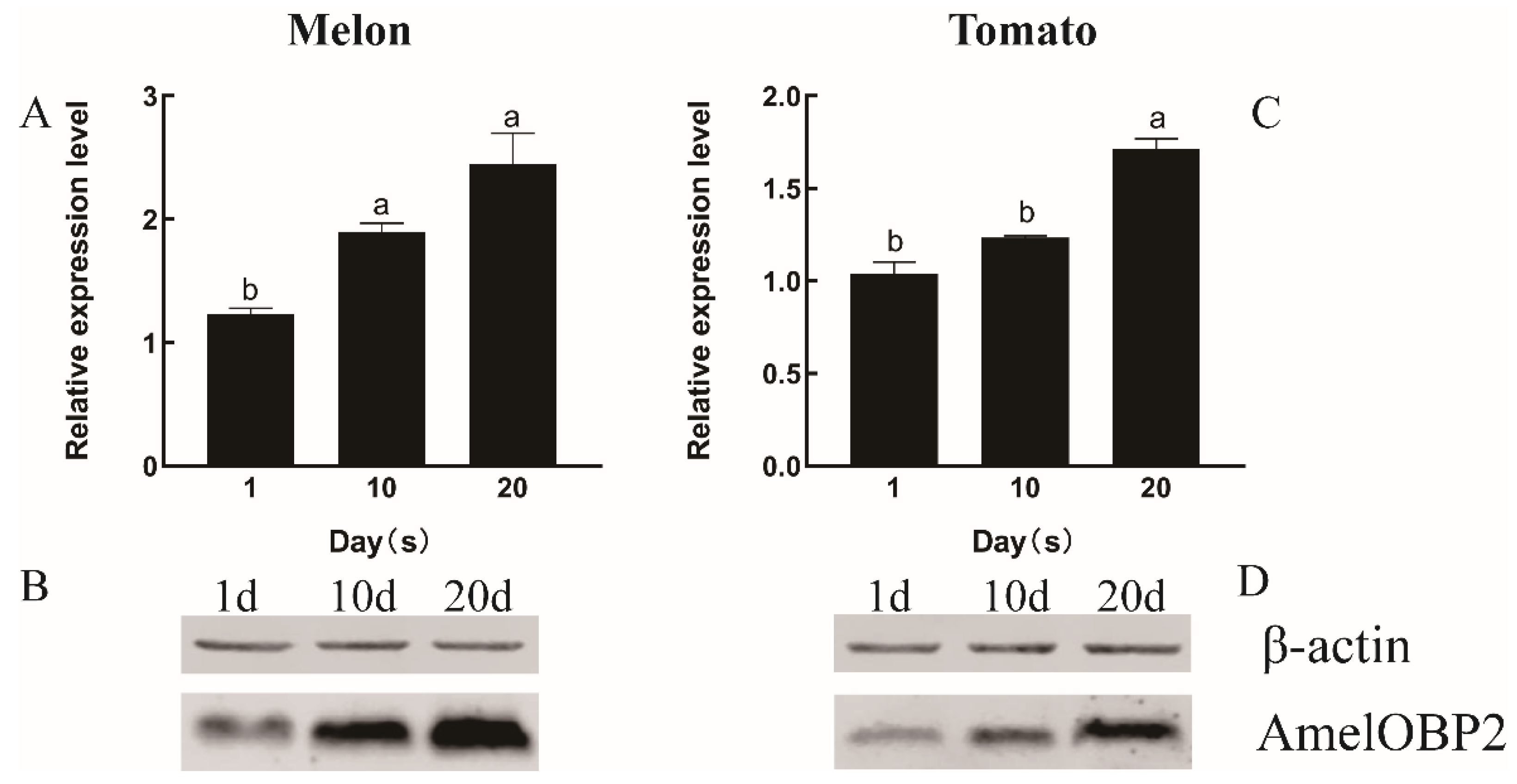
2.1. Expression Pattern Analysis of AmelOBP2
2.2. AmelOBP2 Binding Properties Analysis
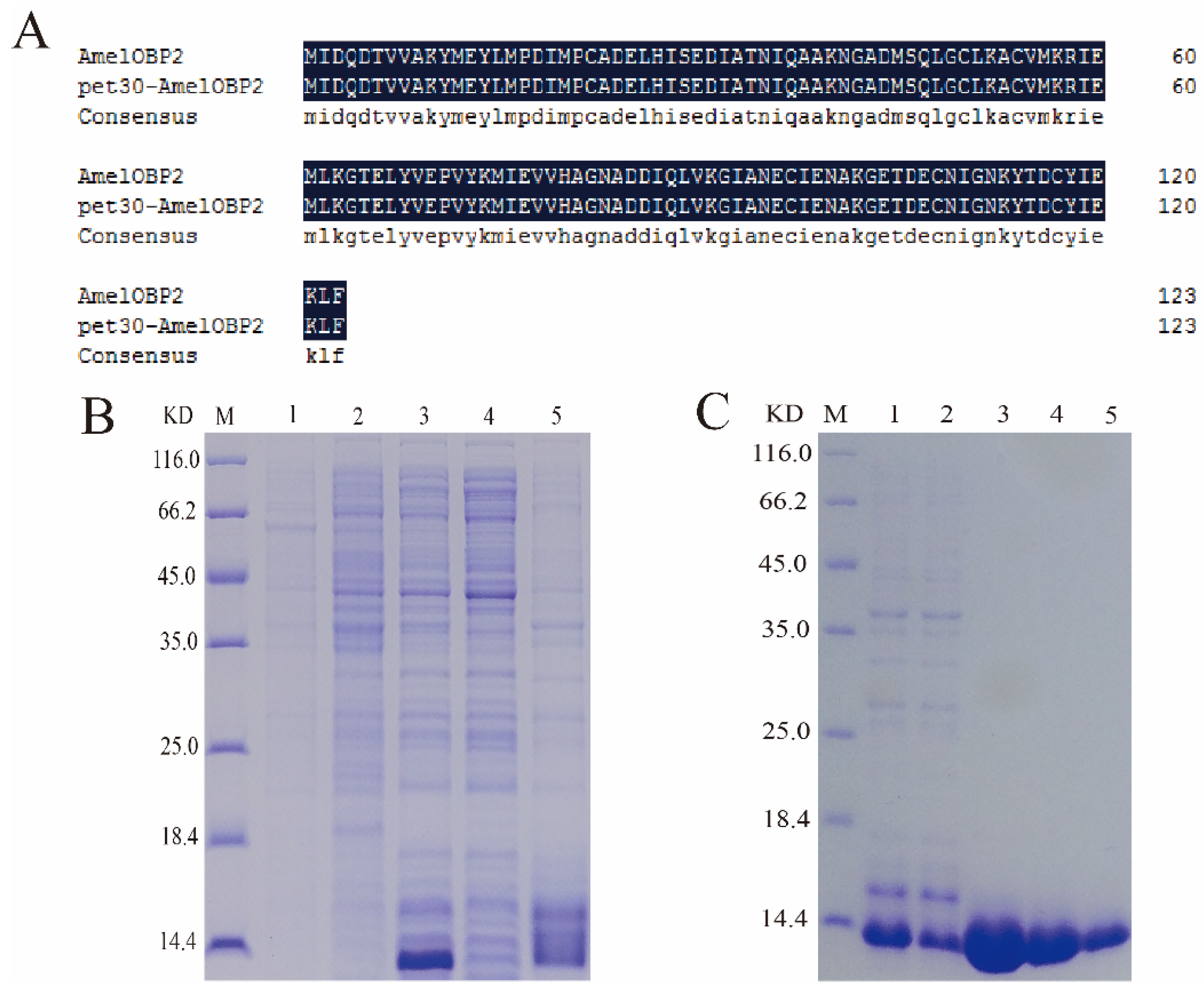
2.2.1. Recombinant AmelOBP2
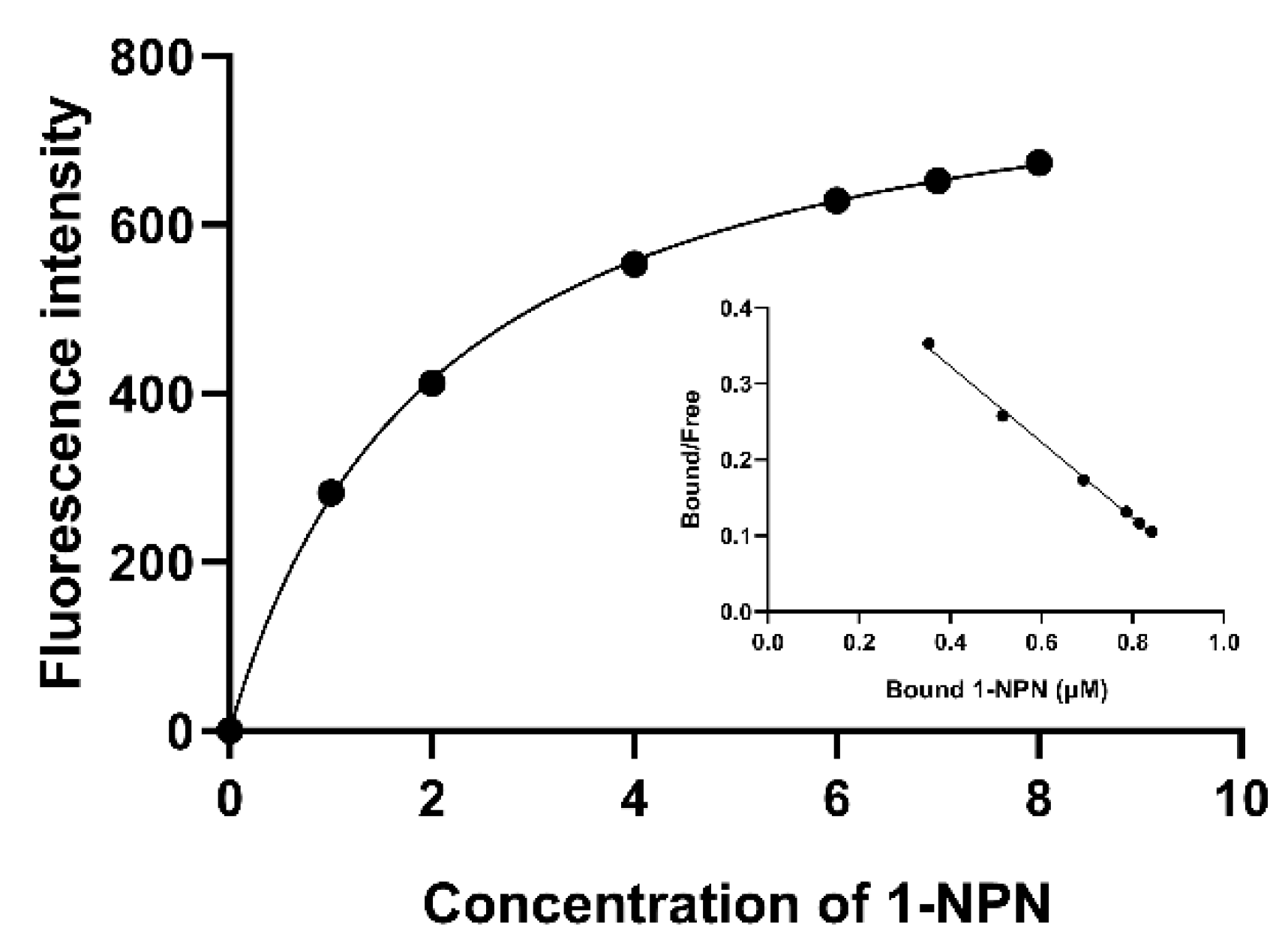
2.2.2. AmelOBP2 Binding Curve with 1-NPN
2.2.3. Ability of AmelOBP2 to Bind to Melon and Tomato Floral Volatiles
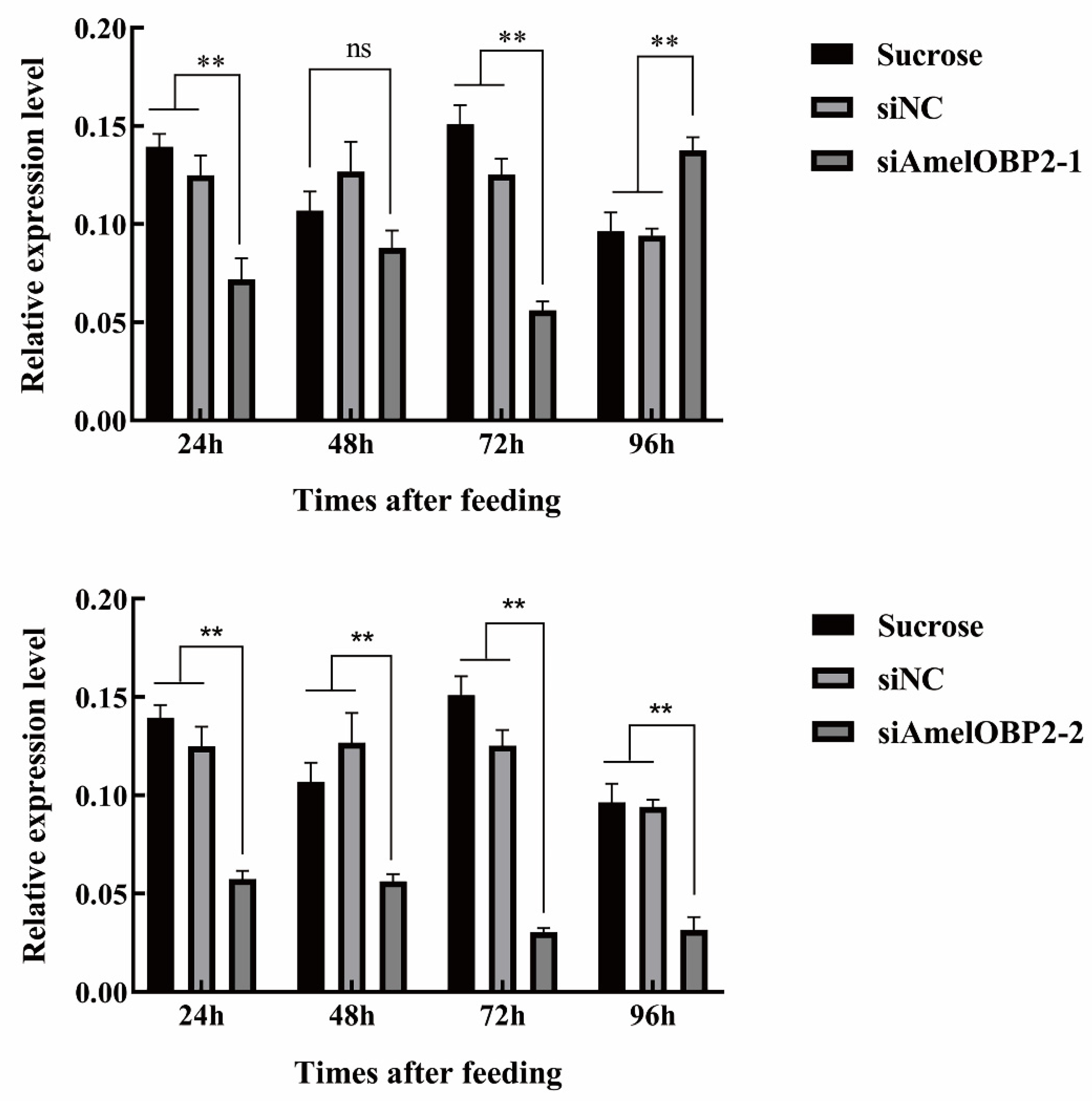
2.3. Effect of siRNA on AmelOBP2 mRNA Expression
2.4. EAG Analysis Before and After RNAi
3. Discussion
4. Materials and Methods
4.1. Insects
4.2. AmelOBP2 Expression Pattern Analysis
4.2.1. qRT-PCR
4.2.2. Purification of AmelOBP2
4.2.3. Western Blot
4.3. Fluorescence Competitive Binding Test
4.4. Detection of RNAi Silencing Effect
4.5. EAG Recording
4.6. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Reagent | System | Reaction Program |
|---|---|---|
| SYBR®Premix Ex TaqTMⅡ | 7.5 μL | Pre-denaturation: 95 °C 10 min; 40 cycles: 95 °C 60 s, 60 °C 20 s, 72 °C 30 s. Determination of solution curve: 95 °C 10 s, 40 °C 1 min, 95 °C 1 min. |
| Forward Primer, 10 μM | 0.6 μL | |
| Reverse Primer, 10 μM | 0.6 μL | |
| cDNA | 0.1 μg | |
| RNase Free dH2o | Up to15.0 μL |
| Gene Name | Primer Sequences (5’-3’) |
|---|---|
| AmelOBP2 (qRT-PCR) | F: AACACCCTCGTCACCGTTAC |
| R: CGTTTCATCACGCAGGCTTT | |
| β-actin | F: ATGCCAACACTGTCCTTTCTGG |
| R: GACCCACCAATCCATACGGA | |
| AmelOBP2 (recombinant expression) | F: CGCCATATGATGAACACCCTCGTCACCGT |
| R: CCCCTCGAGTTACGAGAACAGTTTCTCGATGT | |
| siAmelOBP2-1 | F: GCAAAGUACAUGGAGUAUUTT |
| R: AAUACUCCAUGUACUUUGCTT | |
| siAmelOBP2-2 | F: GGAUCGCGAAUGAGUGCAUTT |
| R: AUGCACUCAUUCGCGAUCCTT | |
| siNC | F: UUCUCCGAACGUGUCACGUTT |
| R: ACGUGACACGUUCGGAGAATT |
| Reagent | Dosage | Reaction Program |
|---|---|---|
| Easy Taq® DNA Polymerase | 1.0 μL | ① 95 °C, 4 min, ② 35 cycles: 94 °C, 30 s; 58 °C, 30 s; 72 °C, 30 s; |
| Forward Primer, 10 μM | 1.0 μL | |
| Reverse Primer, 10 μM | 1.0 μL | |
| cDNA | 1.0 μg | ③ 72 °C, 8 min; ④ 4 °C, ∞. |
| 10×Easy Taq® Buffer | 5.0 μL | |
| 2.5 mM dNTPs | 4.0 μL | |
| ddH2o | Up to 20.0 μL |
| Reagent | Dosage | |
| Double digestion reaction 37 °C, 1 h | PCR-recovered products and pET-30a vector | 1.0 μg |
| 10 × K Buffer | 2.0 μL | |
| 0.1% BSA | 2.0 μL | |
| Nde I | 1.0 μL | |
| Xho I | 1.0 μL | |
| ddH2O | Up to 20.0 μL | |
| Ligation reaction4 °C, 24 h | pET30a | 0.1 μg |
| Target fragment digestion product | 0.1 μg | |
| T4 DNA ligase | 2.0 μL | |
| 2 × Rapid Ligation Buffer | 10.0 μL | |
| ddH2O | Up to 20.0 μL |
| Compounds | CAS Number | Purity | Manufacturer |
|---|---|---|---|
| 2-methyl-5-(1-methyl ethenyl)-cyclohexanone | 7764-50-3 | 98% | Aladdin |
| p-cymene | 99-87-6 | ≥99.5% | Aladdin |
| 1-methyl-4-(1-methyl ethenyl)-benzene | 1195-32-0 | >95% | Aladdin |
| 2,6,6-trimethyl-2,4-cycloheptadien-1-one | 503-93-5 | ≥96% | Macklin |
| β-caryophyllene | 87-44-5 | 99% | Macklin |
| gamma-terpinene | 99-85-4 | >95% | Aladdin |
| 3,7-dimethyl-1,6-octadien-3-ol | 78-70-6 | 98% | Aladdin |
| 3-methyl-6-(1-methyl ethyl)-2-cyclohexen-1-one | 89-81-6 | >94% | TCI |
| methyl benzene | 108-88-3 | 99.8% | Sigma |
| 1,3-dimethyl-benzene | 108-38-3 | >99% | Aladdin |
| β-ocimene | 13877-91-3 | >90% | Sigma |
| tetradecane | 629-59-4 | >99% | Aladdin |
| alpha-myrcene | 123-35-3 | ≥90% | Aladdin |
| 1-nonanal | 124-19-6 | 96% | Aladdin |
| 2,5-dimethyl-benzaldehyde | 5779-94-2 | 98% | Macklin |
| trans-β-ionone | 79-77-6 | >95% | TCI |
| heneicosane | 629-94-7 | >99% | Aladdin |
| 3-phenyl-2-propenal | 104-55-2 | 98% | Macklin |
| hexadecane | 544-76-3 | 99% | Macklin |
| heptadecane | 629-78-7 | ≥99% | Macklin |
| methyl palmitate | 112-39-0 | 99% | Aladdin |
| 2,2,4-trimethyl-1,3-pentanediol diisobutyrate | 6846-50-0 | 98.5% | Aladdin |
| 3-nonen-2-one | 14309-57-0 | ≥96% | Macklin |
| tetracosane | 646-31-1 | AR | Aladdin |
| e-2-decenal. | 3913-81-3 | 95% | Aladdin |
| e-2-octenal | 2548-87-0 | 95% | Aladdin |
| decanal | 112-31-2 | 97% | Aladdin |
| dodecanal | 112-54-9 | 95% | Aladdin |
| nonadecane | 629-92-5 | 98% | Macklin |
| 1,2-dimethoxy-4-(1-propenyl)-benzene | 93-16-3 | >98% | Aladdin |
| phytol | 150-86-7 | >90% | Aladdin |
| ethyl palmitate | 628-97-7 | ≥99% | Aladdin |
| 1,3-bis (1,1-dimethyl ethyl)-benzene | 1014-60-4 | >98% | Aladdin |
| benzenepropanal | 104-53-0 | 95% | Aladdin |
| benzaldehyde | 100-52-7 | ≥99.5% | Aladdin |
| benzeneacetaldehyde | 122-78-1 | 95% | Aladdin |
| e-2-hexenal | 6728-26-3 | 98% | Aladdin |
References
- Burkle, L.A.; Glenny, W.R.; Runyon, J.B. Intraspecific and interspecific variation in floral volatiles over time. Plant Ecol. 2020, 221, 529–544. [Google Scholar] [CrossRef]
- Vega-Polanco, M.; Solís-Montero, L.; Rojas, J.C.; Cruz-López, L.; Alavez-Rosas, D.; Vallejo-Marín, M. Intraspecific variation of scent and its impact on pollinators’ preferences. AoB Plants 2023, 15, plad049. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.W.; Milet-Pinheiro, P.; Gerlach, G.; Ayasse, M.; Nunes, C.E.P.; Alves-dos-Santos, I.; Ramírez, S.R. Macroevolution of floral scent chemistry across radiations of male euglossine bee-pollinated plants Macroevolución de olores florales a través de radiaciones de plantas polinizadas por abejas euglosinas machos Macroevolução dos voláteis florais em radiações de plantas polinizadas por machos de abelhas Euglossini. Evolution 2024, 78, 98–110. [Google Scholar] [PubMed]
- Howell, A.D.; Alarcón, R. Osmia bees (Hymenoptera: Megachilidae) can detect nectar-rewarding flowers using olfactory cues. Anim. Behav. 2007, 74, 199–205. [Google Scholar] [CrossRef]
- Wright, G.A.; Schiestl, F.P. The evolution of floral scent: The influence of olfactory learning by insect pollinators on the honest signalling of floral rewards. Funct. Ecol. 2009, 23, 841–851. [Google Scholar] [CrossRef]
- Chapurlat, E.; Ågren, J.; Anderson, J.; Friberg, M.; Sletvold, N. Conflicting selection on floral scent emission in the orchid Gymnadenia conopsea. New Phytol. 2019, 222, 2009–2022. [Google Scholar] [CrossRef]
- Martínez-Martínez, C.A.; Cordeiro, G.D.; Martins, H.O.J.; Kobal, R.O.A.C.; Milet-Pinheiro, P.; Stanton, M.A.; Franco, E.L.; Krug, C.; Mateus, S.; Schlindwein, C.; et al. Floral Volatiles: A Promising Method to Access the Rare Nocturnal and Crepuscular Bees. Front. Ecol. Evol. 2021, 9, 676743. [Google Scholar] [CrossRef]
- Twidle, A.M.; Mas, F.; Harper, A.R.; Horner, R.M.; Welsh, T.J.; Suckling, D.M. Kiwifruit Flower Odor Perception and Recognition by Honey Bees, Apis mellifera. J. Agric. Food Chem. 2015, 63, 5597–5602. [Google Scholar] [CrossRef]
- Neiswander, R.B. Pollination of Greenhouse Tomatoes by Honey Bees. J. Econ. Entomol. 1956, 49, 436–437. [Google Scholar] [CrossRef]
- Dos Santos, S.A.; Roselino, A.C.; Hrncir, M.; Bego, L.R. Pollination of tomatoes by the stingless bee Melipona quadrifasciata and the honey bee Apis mellifera (Hymenoptera, Apidae). Genet. Mol. Res. 2009, 8, 751–757. [Google Scholar] [CrossRef]
- Potts, S.G.; Imperatriz-Fonseca, V.; Ngo, H.T.; Aizen, M.A.; Biesmeijer, J.C.; Breeze, T.D.; Dicks, L.V.; Garibaldi, L.A.; Hill, R.; Settele, J.; et al. Safeguarding pollinators and their values to human well-being. Nature 2016, 540, 220–229. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Shan, S.; Gu, S.; Huang, X.; Li, Z.; Khashaveh, A.; Zhang, Y. Prior experience with food reward influences the behavioral responses of the honeybee Apis mellifera and the bumblebee Bombus lantschouensis to tomato floral scent. Insects 2020, 11, 884. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, J.; Shen, J.; Zhao, H.; Ma, W.; Jiang, Y. Differences in EAG Response and Behavioral Choices between Honey Bee and Bumble Bee to Tomato Flower Volatiles. Insects 2022, 13, 987. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, J.; Gao, F.; Chen, M.; Jiang, Y.; Zhao, H.; Ma, W. Electrophysiological and Behavioral Responses of Apis mellifera and Bombus terrestris to Melon Flower Volatiles. Insects 2022, 13, 973. [Google Scholar]
- Zhang, L.; Jiang, H.-Q.; Wu, F.; Wen, P.; Qing, J.; Song, X.-M.; Li, H.-L. Eastern honeybee Apis cerana sense cold flowering plants by increasing the static binding affinity of odorant-binding protein to cold floral volatiles from loquats. Int. J. Biol. Macromol. 2023, 232, 123227. [Google Scholar]
- Leal, W.S. Odorant reception in insects: Roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 2013, 58, 373–391. [Google Scholar] [CrossRef]
- Zhao, H.; Peng, Z.; Du, Y.; Xu, K.; Guo, L.; Yang, S.; Ma, W.; Jiang, Y. Comparative antennal transcriptome of Apis cerana cerana from four developmental stages. Gene 2018, 660, 102–108. [Google Scholar]
- Xu, P.; Atkinson, R.; Jones, D.N.; Smith, D.P. Drosophila OBP LUSH is required for activity of pheromone-sensitive neurons. Neuron 2005, 45, 193–200. [Google Scholar] [CrossRef]
- Zeng, B.; Ye, Y.; Ma, J.; Song, J. Juvenile hormone upregulates sugarbabe for vitellogenesis and egg development in the migratory locust Locusta migratoria. Arch. Insect Biochem. Physiol. 2021, 106, e21742. [Google Scholar] [CrossRef]
- Abendroth, J.A.; Moural, T.W.; Wei, H.; Zhu, F. Roles of insect odorant binding proteins in communication and xenobiotic adaptation. Front. Insect Sci. 2023, 3, 1274197. [Google Scholar]
- Pesenti, M.E.; Spinelli, S.; Bezirard, V.; Briand, L.; Pernollet, J.-C.; Tegoni, M.; Cambillau, C. Structural basis of the honey bee PBP pheromone and pH-induced conformational change. J. Mol. Biol. 2008, 380, 158–169. [Google Scholar] [PubMed]
- Weng, C.; Fu, Y.; Jiang, H.; Zhuang, S.; Li, H. Binding interaction between a queen pheromone component HOB and pheromone binding protein ASP1 of Apis cerana. Int. J. Biol. Macromol. 2015, 72, 430–436. [Google Scholar] [PubMed]
- Briand, L.; Nespoulous, C.; Huet, J.C.; Takahashi, M.; Pernollet, J.C. Ligand binding and physico-chemical properties of ASP2, a recombinant odorant-binding protein from honeybee (Apis mellifera L.). Eur. J. Biochem. 2001, 268, 752–760. [Google Scholar]
- Li, H.L.; Song, X.M.; Wu, F.; Qiu, Y.L.; Fu, X.B.; Zhang, L.Y.; Tan, J. Chemical structure of semiochemicals and key binding sites together determine the olfactory functional modes of odorant-binding protein 2 in Eastern honey bee, Apis cerana. Int. J. Biol. Macromol. 2020, 145, 876–884. [Google Scholar] [CrossRef]
- Iovinella, I.; Dani, F.R.; Niccolini, A.; Sagona, S.; Michelucci, E.; Gazzano, A.; Turillazzi, S.; Felicioli, A.; Pelosi, P. Differential Expression of Odorant-Binding Proteins in the Mandibular Glands of the Honey Bee According to Caste and Age. J. Proteome Res. 2011, 10, 3439–3449. [Google Scholar] [CrossRef]
- Zhao, H.; Peng, Z.; Huang, L.; Zhao, S.; Liu, M. Expression Profile and Ligand Screening of a Putative Odorant-Binding Protein, AcerOBP6, from the Asian Honeybee. Insects 2021, 12, 955. [Google Scholar] [CrossRef]
- Larter, N.K.; Sun, J.S.; Carlson, J.R. Organization and function of Drosophila odorant binding proteins. eLife 2016, 5, e20242. [Google Scholar] [CrossRef]
- Hull, J.J.; Perera, O.P.; Snodgrass, G.L. Cloning and expression profiling of odorant-binding proteins in the tarnished plant bug, Lygus lineolaris. Insect Mol. Biol. 2014, 23, 78–97. [Google Scholar] [CrossRef]
- Vogt, R.G.; Riddiford, L.M. Pheromone binding and inactivation by moth antennae. Nature 1981, 293, 161–163. [Google Scholar] [CrossRef]
- Zhang, X.Q.; Yan, Q.; Li, L.L.; Xu, J.W.; Mang, D.; Wang, X.L.; Hoh, H.H.; Ye, J.; Ju, Q.; Ma, Y.; et al. Different binding properties of two general-odorant binding proteins in Athetis lepigone with sex pheromones, host plant volatiles and insecticides. Pestic. Biochem. Physiol. 2020, 164, 173–182. [Google Scholar] [CrossRef]
- Hua, J.F.; Zhang, S.; Cui, J.J.; Wang, D.J.; Wang, C.Y.; Luo, J.Y.; Lv, L.M. Identification and binding characterization of three odorant binding proteins and one chemosensory protein from Apolygus lucorum (Meyer-Dur). J. Chem. Ecol. 2012, 38, 1163–1170. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.Z.; Liu, J.T.; Zhou, J.J.; Wang, Q.; Dong, J.Z.; Zhang, Y.J.; Li, X.C.; Li, J.; Gu, S.H. Expressional and functional comparisons of two general odorant binding proteins in Agrotis ipsilon. Insect Biochem. Mol. Biol. 2018, 98, 34–47. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Xu, K.; Zhao, H.; Jiang, Y.; Li, H. Identification and functional characterization of AcerOBP15 from Apis cerana cerana (Hymenoptera: Apidae). Apidologie 2021, 52, 668–683. [Google Scholar] [CrossRef]
- Borchers, A.; Pieler, T. Programming pluripotent precursor cells derived from Xenopus embryos to generate specific tissues and organs. Genes 2022, 1, 413–426. [Google Scholar] [CrossRef]
- Ben-Shahar, Y. The foraging gene, behavioral plasticity, and honeybee division of labor. J. Comp. Physiol. A 2005, 191, 987–994. [Google Scholar] [CrossRef]
- Li, H.L.; Zhang, Y.L.; Gao, Q.K.; Cheng, J.A.; Lou, B.G. Molecular Identification of cDNA, Immunolocalization, and Expression of a Putative Odorant-Binding Protein from an Asian Honey Bee, Apis cerana cerana. J. Chem. Ecol. 2008, 34, 1593–1601. [Google Scholar] [CrossRef]
- Yang, C. The development and lives of male and worker bees of Apis cerana cerana. J. Honey Bee 1998, 8, 16. [Google Scholar]
- Mohammedi, A.; Crauser, D.; Paris, A.; Le Conte, Y. Effect of a brood pheromone on honeybee hypopharyngeal glands. Comptes Rendus De L’academie Des Sciences. Ser. III Sci. De La Vie 1996, 319, 769–772. [Google Scholar]
- Mohammedi, A.; Paris, A.; Crauser, D.; Le Conte, Y. Effect of aliphatic esters on ovary development of queenless bees (Apis mellifera L.). Naturwissenschaften 1998, 85, 455–458. [Google Scholar] [CrossRef]
- Pankiw, T.; Page Jr, R.E.; Kim Fondrk, M. Brood pheromone stimulates pollen foraging in honey bees (Apis mellifera). Behav. Ecol. Sociobiol. 1998, 44, 193–198. [Google Scholar] [CrossRef]
- Zhang, X.M. Floral volatile sesquiterpenes of Elsholtzia rugulosa (Lamiaceae) selectively attract Asian honey bees. J. Appl. Entomol. 2018, 142, 359–362. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
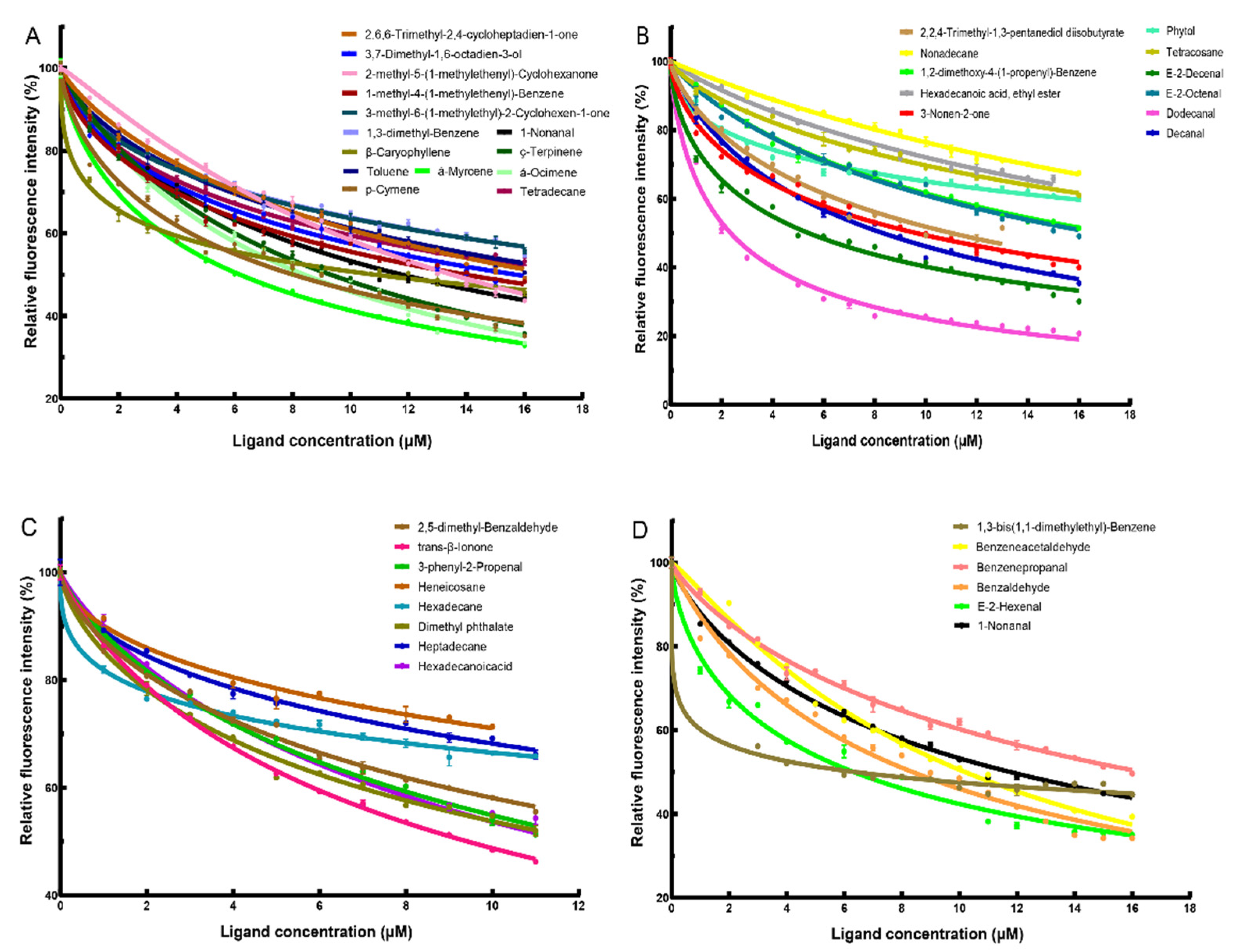
| Category | Ligand Names | IC50 (μM) | Ki (μM) |
|---|---|---|---|
| Melon floral volatiles | dodecanal | 2.35 | 1.05 |
| e-2-decenal | 5.41 | 2.42 | |
| e-2-hexenal | 6.33 | 2.83 | |
| 1,3-bis (1,1-dimethyl ethyl)-benzene | 6.37 | 2.85 | |
| decanal | 8.32 | 3.72 | |
| benzaldehyde | 8.37 | 3.74 | |
| trans-β-ionone | 9.43 | 4.21 | |
| 3-nonen-2-one | 9.63 | 4.30 | |
| benzeneacetaldehyde | 10.19 | 4.55 | |
| 2,2,4-trimethyl-1,3-pentanediol diisobutyrate | 11.01 | 4.92 | |
| 1-nonanal | 11.77 | 5.26 | |
| hexadecenoic acid | 11.85 | 5.29 | |
| dimethyl phthalate | 12.40 | 5.54 | |
| 3-phenyl-2-propenal | 12.74 | 5.69 | |
| 2,5-dimethyl-benzaldehyde | 15.94 | 7.12 | |
| benzenepropanal | 16.34 | 7.30 | |
| e-2-octenal | 16.68 | 7.45 | |
| 1,2-dimethoxy-4-(1-propenyl)-benzene | 17.20 | 7.68 | |
| hexadecanoic acid, ethyl ester | 28.77 | 12.85 | |
| tetracosane | 29.60 | 13.22 | |
| nonadecane | 32.96 | 14.72 | |
| phytol | 36.30 | 16.22 | |
| heptadecane | 37.33 | 16.68 | |
| heneicosane | 48.57 | 21.70 | |
| hexadecane | 68.59 | 30.64 | |
| Tomato floral volatiles | alpha-myrcene | 6.18 | 2.76 |
| p-cymene | 8.02 | 3.58 | |
| β-ocimene | 8.40 | 3.75 | |
| gamma-terpinene | 9.31 | 4.16 | |
| β-caryophyllene | 10.83 | 4.84 | |
| 1-nonanal | 11.77 | 5.26 | |
| 2-methyl-5-(1-methyl ethenyl)-cyclohexanone | 13.50 | 6.03 | |
| 1-methyl-4-(1-methyl ethenyl)-benzene | 13.95 | 6.23 | |
| 3,7-dimethyl-1,6-octadien-3-ol | 15.70 | 7.01 | |
| 2,6,6-trimethyl-2,4-cyclopentadiene-1-one | 17.02 | 7.60 | |
| tetradecane | 18.05 | 8.06 | |
| methyl benzene | 18.57 | 8.30 | |
| 1,3-dimethyl-benzene | 24.64 | 11.01 | |
| 3-methyl-6-(1-methyl ethyl)-2-cyclohexen-1-one | 25.03 | 11.18 |
| Compounds | EAG Relative Value (mV) | Silencing Efficiency (%) | ||
|---|---|---|---|---|
| CK | NC | RNAi | ||
| e-2-hexenal | 4.91 ± 0.09 a | 4.81 ± 0.11 a | 3.21 ± 0.01 b | 34.64% |
| 1-nonanal | 2.96 ± 0.05 a | 2.86 ± 0.08 a | 1.85 ± 0.17 b | 37.28% |
| e-2-octenal | 3.40 ± 0.01 a | 3.22 ± 0.10 a | 1.52 ± 0.05 b | 55.20% |
| 2-methyl-5-(1-methyl ethenyl)-cyclohexanone | 1.11 ± 0.17 | 1.14 ± 0.03 | 0.75 ± 0.13 | 32.48% |
| methyl benzene | 0.74 ± 0.06 | 0.79 ± 0.08 | 0.56 ± 0.04 | 28.76% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Ma, W.; Zhang, Y.; Lu, S.; Zhang, C.; Zhao, H.; Jiang, Y. Odor-Binding Protein 2 in Apis mellifera ligustica Plays Important Roles in the Response to Floral Volatiles Stimuli from Melon and Tomato Flowers. Int. J. Mol. Sci. 2025, 26, 3176. https://doi.org/10.3390/ijms26073176
Zhang J, Ma W, Zhang Y, Lu S, Zhang C, Zhao H, Jiang Y. Odor-Binding Protein 2 in Apis mellifera ligustica Plays Important Roles in the Response to Floral Volatiles Stimuli from Melon and Tomato Flowers. International Journal of Molecular Sciences. 2025; 26(7):3176. https://doi.org/10.3390/ijms26073176
Chicago/Turabian StyleZhang, Jiangchao, Weihua Ma, Yue Zhang, Surong Lu, Chaoying Zhang, Huiting Zhao, and Yusuo Jiang. 2025. "Odor-Binding Protein 2 in Apis mellifera ligustica Plays Important Roles in the Response to Floral Volatiles Stimuli from Melon and Tomato Flowers" International Journal of Molecular Sciences 26, no. 7: 3176. https://doi.org/10.3390/ijms26073176
APA StyleZhang, J., Ma, W., Zhang, Y., Lu, S., Zhang, C., Zhao, H., & Jiang, Y. (2025). Odor-Binding Protein 2 in Apis mellifera ligustica Plays Important Roles in the Response to Floral Volatiles Stimuli from Melon and Tomato Flowers. International Journal of Molecular Sciences, 26(7), 3176. https://doi.org/10.3390/ijms26073176






