MiR-21-5p and miR-223-3p as Treatment Response Biomarkers in Pediatric Eosinophilic Esophagitis
Abstract
1. Introduction
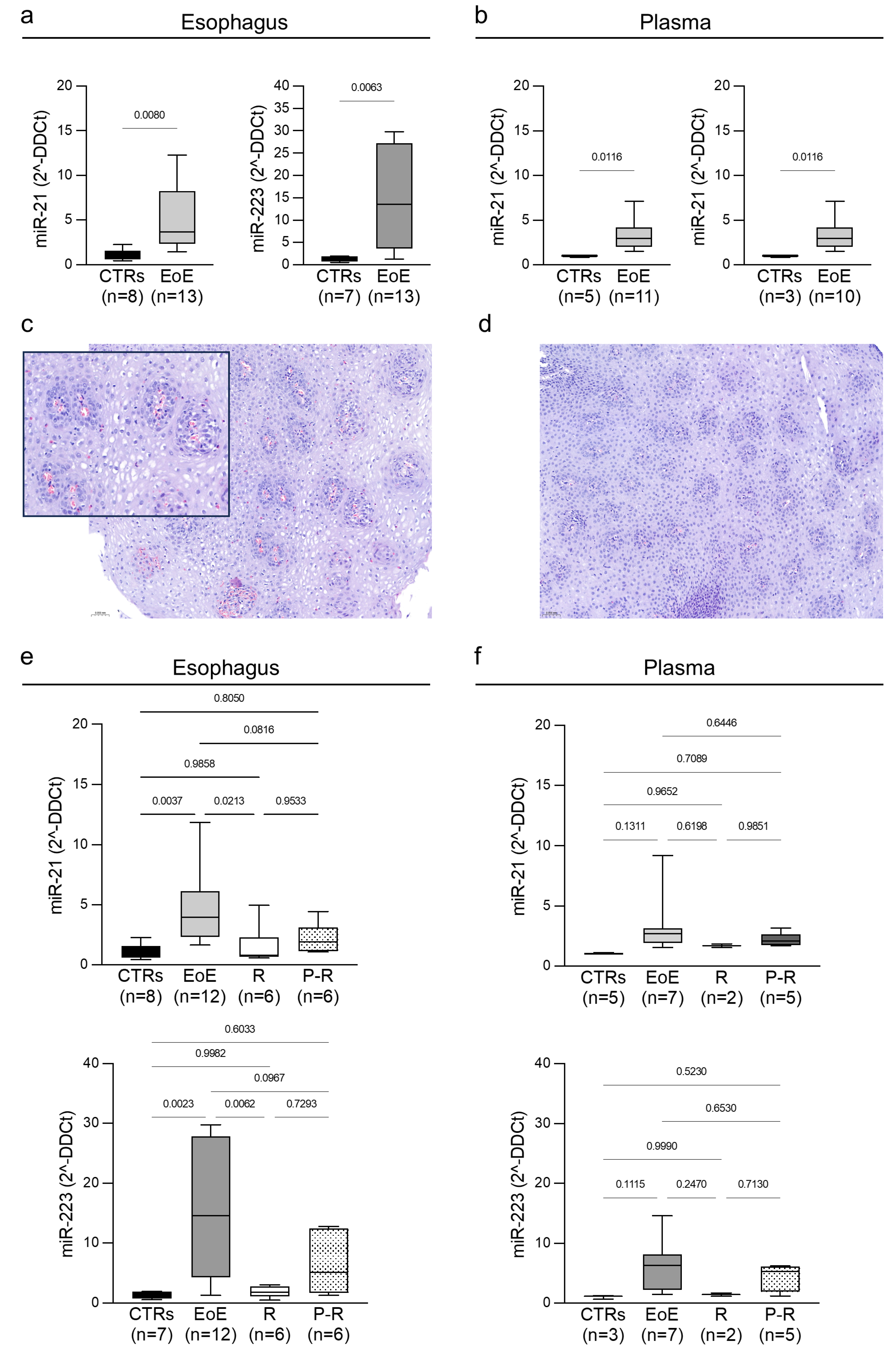
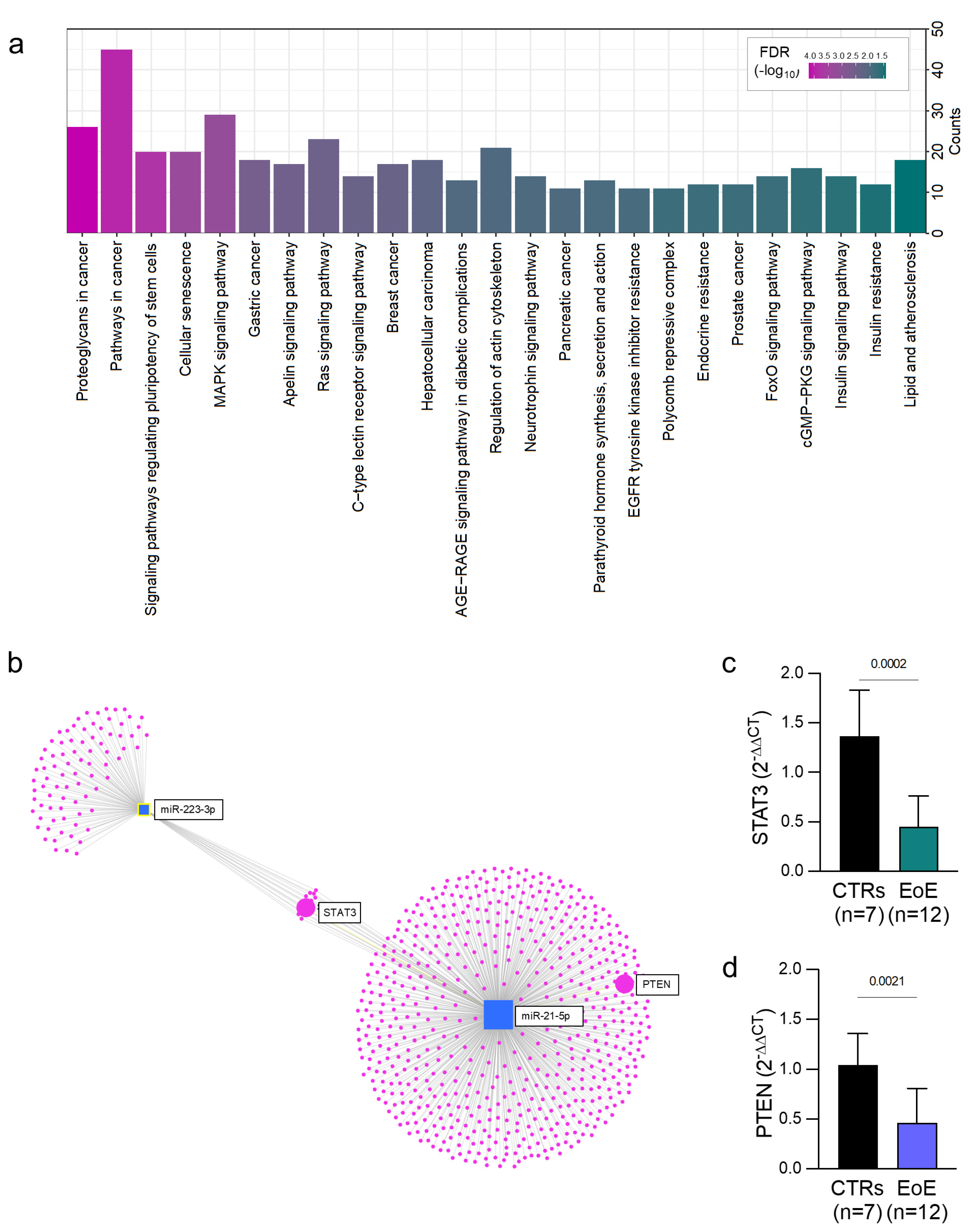
2. Results
3. Discussion
4. Materials and Methods
4.1. Study Design, Patients, and Data Collection
4.2. RNA Isolation and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
4.3. Bioinformatic Analysis
4.4. Histological Analysis
4.5. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CCS | corticosteroids |
| EoE | eosinophilic esophagitis |
| FDR | false discovery rate |
| H/E | hematoxylin/eosin |
| HPF | high-power field |
| miRNA | MicroRNA |
| PPI | proton pump inhibitors |
| qRT-PCR | quantitative real-time polymerase chain reaction |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| David | Database for Annotation, Visualization, and Integrated Discovery |
References
- Furuta, G.T.; Katzka, D.A. Eosinophilic Esophagitis. N. Engl. J. Med. 2015, 373, 1640–1648. [Google Scholar] [PubMed]
- Spergel, J.M.; Brown-Whitehorn, T.A.; Muir, A.; Liacouras, C.A. Medical algorithm: Diagnosis and treatment of eosinophilic esophagitis in children. Allergy 2020, 75, 1522–1524. [Google Scholar] [PubMed]
- Dellon, E.S.; Hirano, I. Epidemiology and Natural History of Eosinophilic Esophagitis. Gastroenterology 2018, 154, 319–332.e3. [Google Scholar]
- Gupta, S.K. Noninvasive markers of eosinophilic esophagitis. Gastrointest. Endosc. Clin. N. Am. 2008, 18, 157–167. [Google Scholar] [PubMed]
- Ho, P.T.B.; Clark, I.M.; Le, L.T.T. MicroRNA-Based Diagnosis and Therapy. Int. J. Mol. Sci. 2022, 23, 7167. [Google Scholar] [CrossRef]
- Tarallo, A.; Carissimo, A.; Gatto, F.; Nusco, E.; Toscano, A.; Musumeci, O.; Coletta, M.; Karali, M.; Acampora, E.; Damiano, C.; et al. microRNAs as biomarkers in Pompe disease. Genet. Med. 2019, 21, 591–600. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar]
- Allegra, A.; Alonci, A.; Campo, S.; Penna, G.; Petrungaro, A.; Gerace, D.; Musolino, C. Circulating microRNAs: New biomarkers in diagnosis, prognosis and treatment of cancer (review). Int. J. Oncol. 2012, 41, 1897–1912. [Google Scholar]
- Markey, G.E.; Donohoe, C.L.; McNamee, E.N.; Masterson, J.C. MicroRNA dysregulation and therapeutic opportunities in esophageal diseases. Am. J. Physiol. Gastrointest. Liver Physiol. 2023, 325, G1–G13. [Google Scholar]
- Jhaveri, P.B.; Lambert, K.A.; Bogale, K.; Lehman, E.; Alexander, C.; Ishmael, F.; Jhaveri, P.N.; Hicks, S.D. Salivary microRNAs in pediatric eosinophilic esophagitis. Allergy Asthma Proc. 2023, 44, 145–152. [Google Scholar]
- Votto, M.; Strisciuglio, C.; Indolfi, C.; Marseglia, G.L.; Miraglia Del Giudice, M.; Licari, A. Correspondence to “Salivary Immunoinflammatory Proteins Identify Children with Eosinophilic Esophagitis”. Allergy 2024, 79, 3176–3177. [Google Scholar] [PubMed]
- Zahm, A.M.; Menard-Katcher, C.; Benitez, A.J.; Tsoucas, D.M.; Le Guen, C.L.; Hand, N.J.; Friedman, J.R. Pediatric eosinophilic esophagitis is associated with changes in esophageal microRNAs. Am. J. Physiol. Gastrointest. Liver Physiol. 2014, 307, G803–G812. [Google Scholar]
- Cañas, J.A.; Tabares, A.; Barbero, C.; García-Sánchez, D.; Sastre, B.; Rodrigo-Muñoz, J.M.; Mahíllo-Fernández, I.; Rayo, A.; Borrell, B.; Cilleruelo, M.L.; et al. Proton-pump Inhibitor Response Prediction Using Esophageal microRNAs in Children With Eosinophilic Esophagitis. J. Pediatr. Gastroenterol. Nutr. 2020, 71, 755–763. [Google Scholar]
- Lu, T.X.; Sherrill, J.D.; Wen, T.; Plassard, A.J.; Besse, J.A.; Abonia, J.P.; Franciosi, J.P.; Putnam, P.E.; Eby, M.; Martin, L.J.; et al. MicroRNA signature in patients with eosinophilic esophagitis, reversibility with glucocorticoids, and assessment as disease biomarkers. J. Allergy Clin. Immunol. 2012, 129, 1064–1075.e9. [Google Scholar]
- Zou, K.; Dong, H.; Li, M.; Zhang, Y.; Zhang, K.; Song, D.; Chu, C. Comprehensive analysis of transcriptome-wide N6-methyladenosine methylomes in the Barrett’s esophagus in rats. Genomics 2023, 115, 110687. [Google Scholar] [PubMed]
- Dellon, E.S.; Liacouras, C.A.; Molina-Infante, J.; Furuta, G.T.; Spergel, J.M.; Zevit, N.; Spechler, S.J.; Attwood, S.E.; Straumann, A.; Aceves, S.S.; et al. Updated International Consensus Diagnostic Criteria for Eosinophilic Esophagitis: Proceedings of the AGREE Conference. Gastroenterology 2018, 155, 1022–1033.e10. [Google Scholar] [PubMed]
- Wu, Y.; Song, Y.; Xiong, Y.; Wang, X.; Xu, K.; Han, B.; Bai, Y.; Li, L.; Zhang, Y.; Zhou, L. MicroRNA-21 (Mir-21) Promotes Cell Growth and Invasion by Repressing Tumor Suppressor PTEN in Colorectal Cancer. Cell. Physiol. Biochem. 2017, 43, 945–958. [Google Scholar]
- Li, Z.; Dong, X.; Wang, Z.; Liu, W.; Deng, N.; Ding, Y.; Tang, L.; Hla, T.; Zeng, R.; Li, L.; et al. Regulation of PTEN by Rho small GTPases. Nat. Cell Biol. 2005, 7, 399–404. [Google Scholar]
- Yang, X.O.; Panopoulos, A.D.; Nurieva, R.; Chang, S.H.; Wang, D.; Watowich, S.S.; Dong, C. STAT3 regulates cytokine-mediated generation of inflammatory helper T cells. J. Biol. Chem. 2007, 282, 9358–9363. [Google Scholar] [CrossRef]
- Wang, J.; Shen, Y.; Li, C.; Liu, C.; Wang, Z.H.; Li, Y.S.; Ke, X.; Hu, G.H. IL-37 attenuates allergic process via STAT6/STAT3 pathways in murine allergic rhinitis. Int. Immunopharmacol. 2019, 69, 27–33. [Google Scholar] [CrossRef]
- Meng, F.; Henson, R.; Wehbe-Janek, H.; Ghoshal, K.; Jacob, S.T.; Patel, T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 2007, 133, 647–658. [Google Scholar] [PubMed]
- Chang, L.; Zhou, G.; Soufan, O.; Xia, J. miRNet 2.0: Network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res. 2020, 48, W244–W251. [Google Scholar] [PubMed]
- Hasan, S.H.; Taylor, S.; Garg, S.; Buras, M.R.; Doyle, A.D.; Bauer, C.S.; Wright, B.L.; Schroeder, S. Diagnosis of Pediatric Non-Esophageal Eosinophilic Gastrointestinal Disorders by Eosinophil Peroxidase Immunohistochemistry. Pediatr. Dev. Pathol. 2021, 24, 513–522. [Google Scholar] [PubMed]
| EOE Patients n = 13 | |
|---|---|
| Sex males, n (%) | 8 (61) |
| Age years, median ± SD | 13 ± 4.9 |
| History of atopy, n (%) | 9 (69) |
| 3 (27.1) |
| 5 (38.4) |
| 3 (27.1) |
| Comorbidities (%) | 5 (38.5) |
| 3 (23) |
| 4 (30.8) |
| EoE treatment, n (%) | |
| 4 (30.7) |
| 7 (53.8) |
| 2 (15.3) |
| Eos/HPF (mean) | |
| T0 biopsy | 33.75 |
| T1 biopsy | 13.23 |
| EREFS (mean) | |
| T0 | 2.07 |
| T1 | 1.53 |
| Esophageal miR-223-3p | Estimate | p Value |
|---|---|---|
| (Intercept) | −24.595264 | 0.2176 |
| EC-T0_Blood | 0.011237 | 0.2752 |
| EC-T0_Esophagus | 1.015288 | 0.0318 * |
| Allergy | 6.610019 | 0.4009 |
| EREFS-T0 | −8.176028 | 0.07 |
| CD | 11.834543 | 0.272 |
| DM1 | 7.98814 | 0.3675 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tarallo, A.; Casertano, M.; Valanzano, A.; Cenni, S.; Creoli, M.; Russo, G.; Damiano, C.; Carissimo, A.; Cioce, A.; Martinelli, M.; et al. MiR-21-5p and miR-223-3p as Treatment Response Biomarkers in Pediatric Eosinophilic Esophagitis. Int. J. Mol. Sci. 2025, 26, 3111. https://doi.org/10.3390/ijms26073111
Tarallo A, Casertano M, Valanzano A, Cenni S, Creoli M, Russo G, Damiano C, Carissimo A, Cioce A, Martinelli M, et al. MiR-21-5p and miR-223-3p as Treatment Response Biomarkers in Pediatric Eosinophilic Esophagitis. International Journal of Molecular Sciences. 2025; 26(7):3111. https://doi.org/10.3390/ijms26073111
Chicago/Turabian StyleTarallo, Antonietta, Marianna Casertano, Anna Valanzano, Sabrina Cenni, Mara Creoli, Giuseppina Russo, Carla Damiano, Annamaria Carissimo, Alessandro Cioce, Massimo Martinelli, and et al. 2025. "MiR-21-5p and miR-223-3p as Treatment Response Biomarkers in Pediatric Eosinophilic Esophagitis" International Journal of Molecular Sciences 26, no. 7: 3111. https://doi.org/10.3390/ijms26073111
APA StyleTarallo, A., Casertano, M., Valanzano, A., Cenni, S., Creoli, M., Russo, G., Damiano, C., Carissimo, A., Cioce, A., Martinelli, M., Miele, E., Staiano, A., Iafusco, D., Parenti, G., & Strisciuglio, C. (2025). MiR-21-5p and miR-223-3p as Treatment Response Biomarkers in Pediatric Eosinophilic Esophagitis. International Journal of Molecular Sciences, 26(7), 3111. https://doi.org/10.3390/ijms26073111






