Analysis of Metabolomic Reprogramming Induced by Infection with Kaposi’s Sarcoma-Associated Herpesvirus Using Untargeted Metabolomic Profiling
Abstract
1. Introduction
2. Results
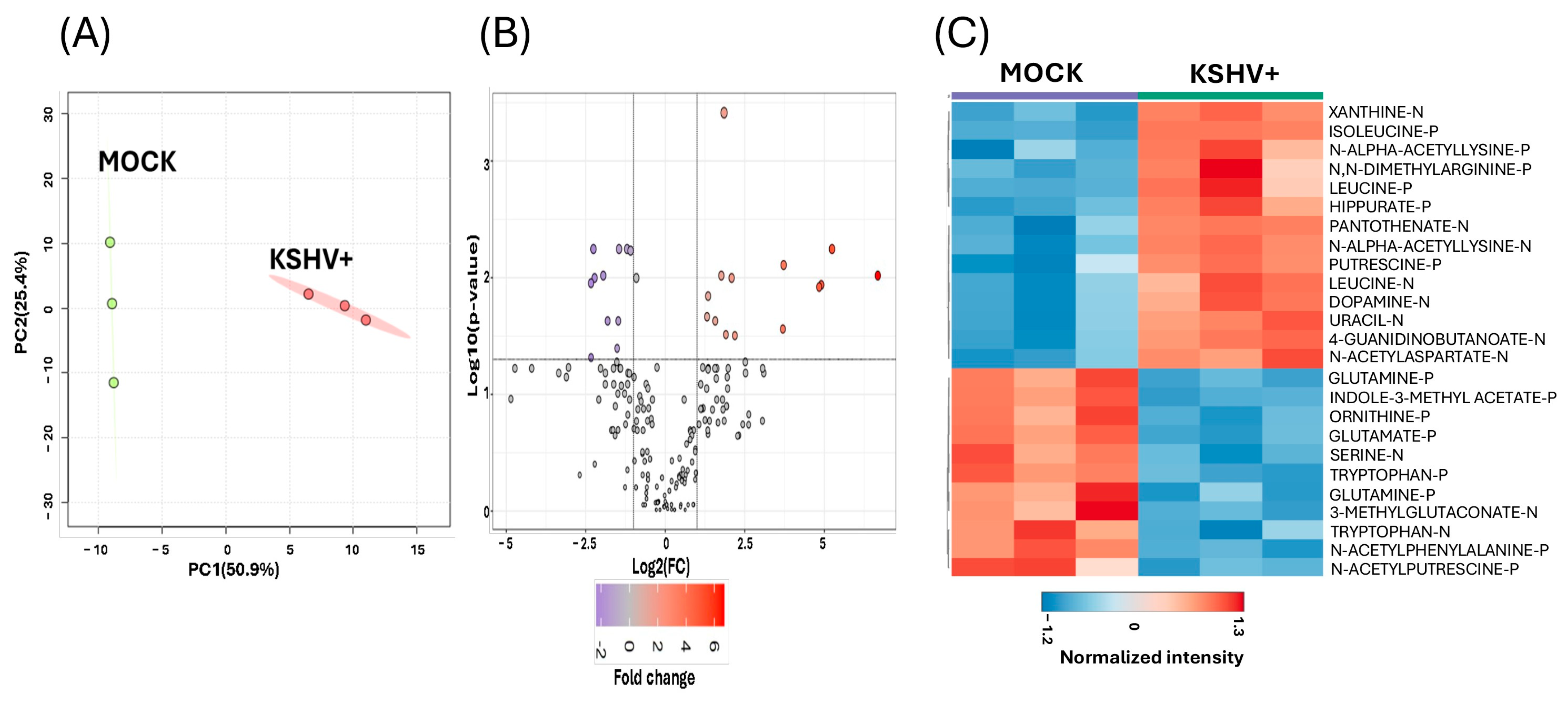
2.1. Untargeted Metabolomics Analysis of De Novo KSHV Infection in Oral Epithelial Cells
2.2. Identification of Upregulated or Downregulated Metabolites upon KSHV Infection
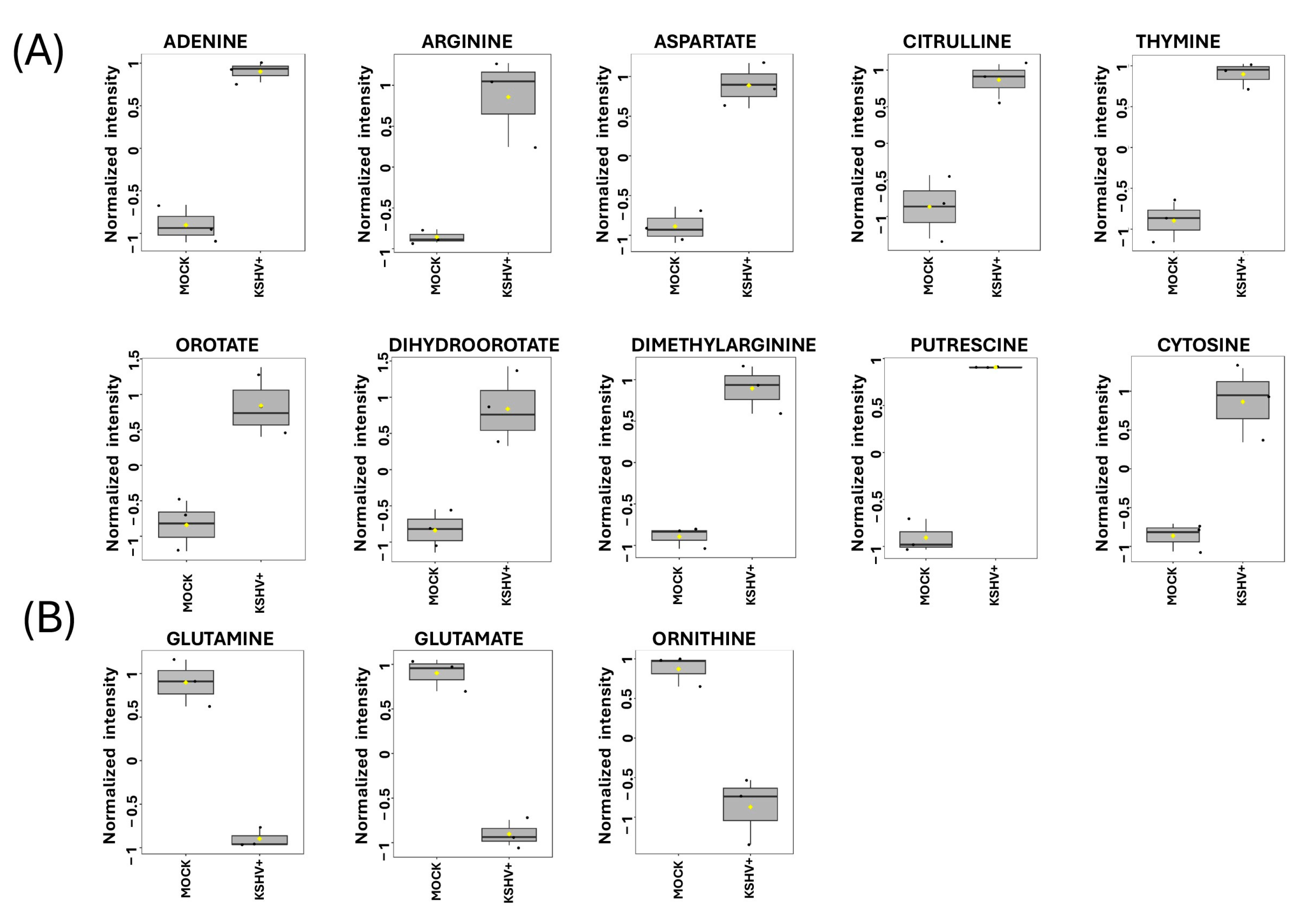
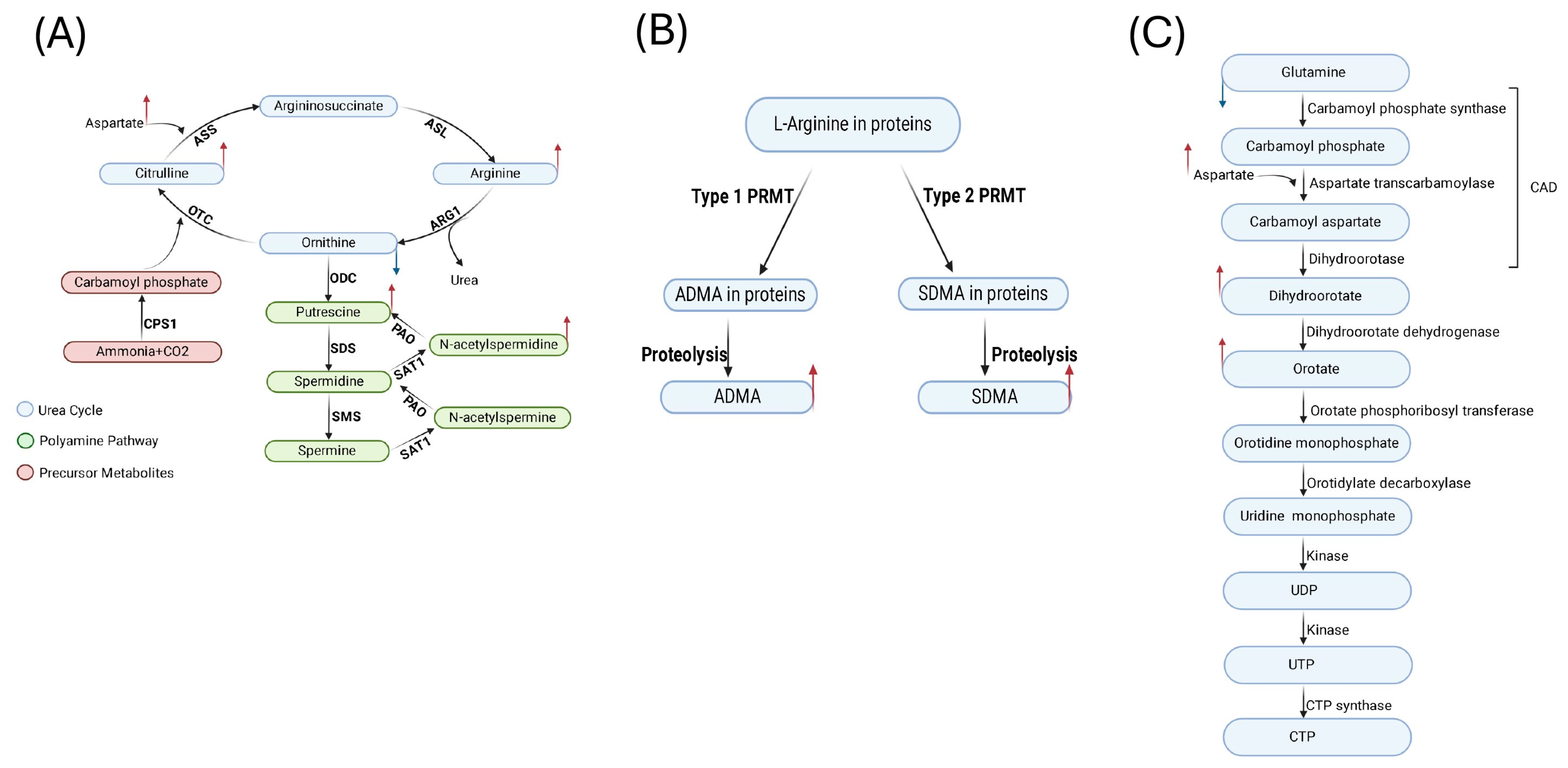
2.3. Discovering Known Metabolites Altered upon KSHV Infection
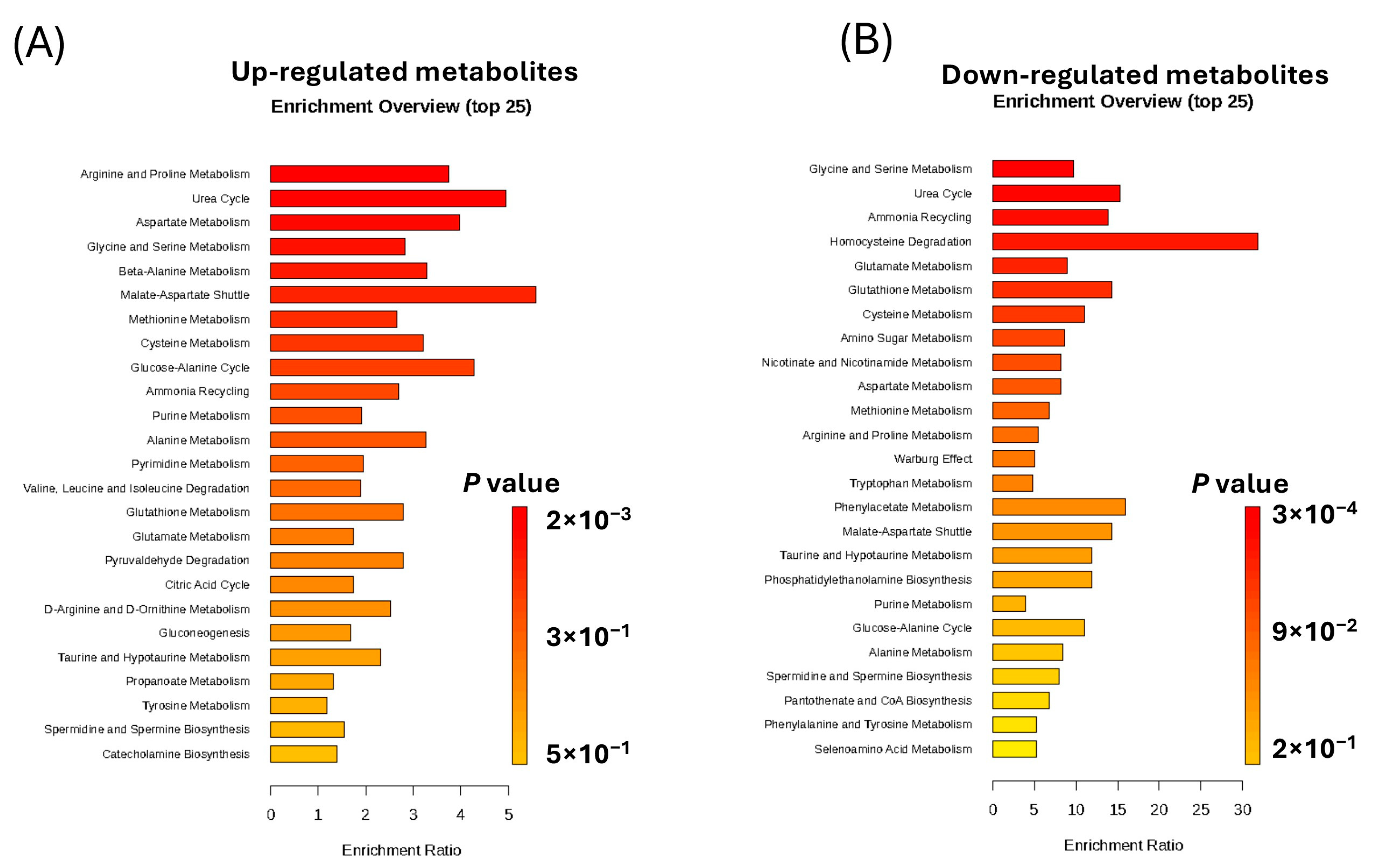
2.4. Enrichment Analysis of the Significantly Altered Metabolites in KSHV-Infected Cells
3. Discussion
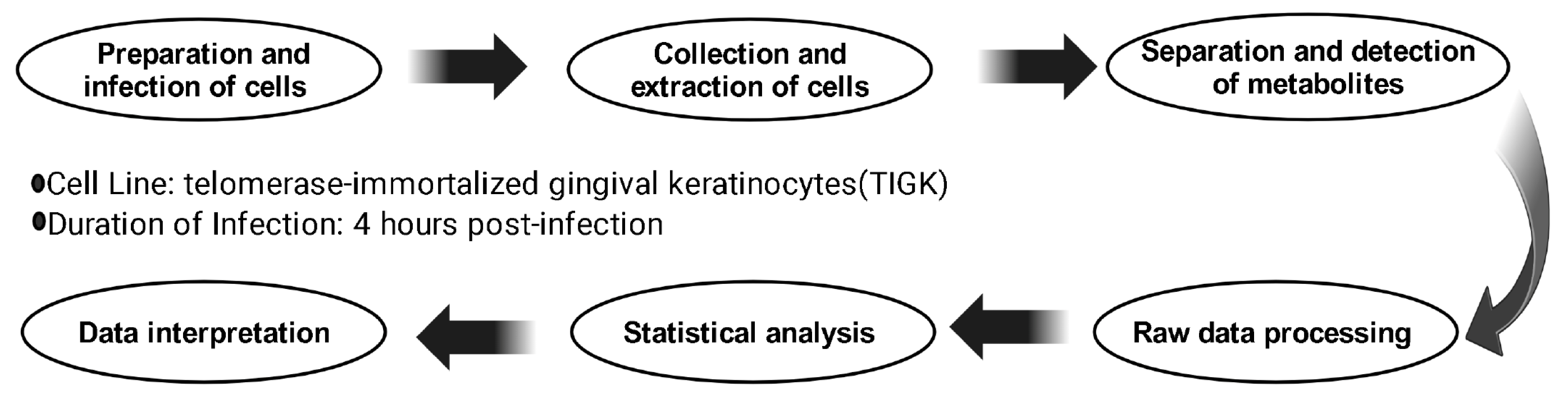
4. Materials and Methods
4.1. Cell Culture and Viral Infection
4.2. Metabolite Extraction
4.3. Analytical Instrumentation
4.4. Data Processing
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bardanzellu, F.; Fanos, V. How could metabolomics change pediatric health? Ital. J. Pediatr. 2020, 46, 37. [Google Scholar] [CrossRef] [PubMed]
- Aretz, I.; Meierhofer, D. Advantages and Pitfalls of Mass Spectrometry Based Metabolome Profiling in Systems Biology. Int. J. Mol. Sci. 2016, 17, 632. [Google Scholar] [CrossRef] [PubMed]
- Rinschen, M.M.; Ivanisevic, J.; Giera, M.; Siuzdak, G. Identification of bioactive metabolites using activity metabolomics. Nat. Rev. Mol. Cell Biol. 2019, 20, 353–367. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Iftode, N.; Rădulescu, M.A.; Aramă, Ș.S.; Aramă, V. Update on Kaposi sarcoma-associated herpesvirus (KSHV or HHV8)—Review. Rom. J. Intern. Med. 2020, 58, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, A.P.; Damania, B. AKTivation of PI3K/AKT/mTOR signaling pathway by KSHV. Front. Immunol. 2012, 3, 401. [Google Scholar] [CrossRef] [PubMed]
- Purushothaman, P.; Uppal, T.; Verma, S.C. Molecular biology of KSHV lytic reactivation. Viruses 2015, 7, 116–153. [Google Scholar] [CrossRef] [PubMed]
- Veettil, M.V.; Bandyopadhyay, C.; Dutta, D.; Chandran, B. Interaction of KSHV with host cell surface receptors and cell entry. Viruses 2014, 6, 4024–4046. [Google Scholar] [CrossRef] [PubMed]
- Purushothaman, P.; Dabral, P.; Gupta, N.; Sarkar, R.; Verma, S.C. KSHV Genome Replication and Maintenance. Front. Microbiol. 2016, 7, 54. [Google Scholar] [CrossRef] [PubMed]
- Toth, Z.; Brulois, K.; Lee, H.-R.; Izumiya, Y.; Tepper, C.; Kung, H.-J.; Jung, J.U. Biphasic Euchromatin-to-Heterochromatin Transition on the KSHV Genome Following De Novo Infection. PLoS Pathog. 2013, 9, e1003813. [Google Scholar] [CrossRef]
- Choi, U.Y.; Lee, S.H. Understanding Metabolic Pathway Rewiring by Oncogenic Gamma Herpesvirus. J. Microbiol. Biotechnol. 2024, 34, 2143–2152. [Google Scholar] [CrossRef] [PubMed]
- Delgado, T.; Sanchez, E.L.; Camarda, R.; Lagunoff, M. Global Metabolic Profiling of Infection by an Oncogenic Virus: KSHV Induces and Requires Lipogenesis for Survival of Latent Infection. PLoS Pathog. 2012, 8, e1002866. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhu, C.; Wang, Y.; Wei, F.; Cai, Q. KSHV Reprogramming of Host Energy Metabolism for Pathogenesis. Front. Cell. Infect. Microbiol. 2021, 11, 621156. [Google Scholar] [CrossRef]
- Sanchez, E.L.; Pulliam, T.H.; Dimaio, T.A.; Thalhofer, A.B.; Delgado, T.; Lagunoff, M. Glycolysis, Glutaminolysis, and Fatty Acid Synthesis Are Required for Distinct Stages of Kaposi’s Sarcoma-Associated Herpesvirus Lytic Replication. J. Virol. 2017, 91, e02237-16. [Google Scholar] [CrossRef] [PubMed]
- Magon, K.L.; Parish, J.L. From infection to cancer: How DNA tumour viruses alter host cell central carbon and lipid metabolism. Open Biol. 2021, 11, JVI.210004. [Google Scholar] [CrossRef]
- Delgado, T.; Carroll, P.A.; Punjabi, A.S.; Margineantu, D.; Hockenbery, D.M.; Lagunoff, M. Induction of the Warburg effect by Kaposi’s sarcoma herpesvirus is required for the maintenance of latently infected endothelial cells. Proc. Natl. Acad. Sci. USA 2010, 107, 10696–10701. [Google Scholar] [CrossRef] [PubMed]
- Karki, R.; Lang, S.M.; Means, R.E. The MARCH Family E3 Ubiquitin Ligase K5 Alters Monocyte Metabolism and Proliferation through Receptor Tyrosine Kinase Modulation. PLoS Pathog. 2011, 7, e1001331. [Google Scholar] [CrossRef] [PubMed]
- Bahat, A.; Macvicar, T.; Langer, T. Metabolism and Innate Immunity Meet at the Mitochondria. Front. Cell Dev. Biol. 2021, 9, 720490. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Wang, G.; Xu, Z.-G.; Tu, H.; Hu, F.; Dai, J.; Chang, Y.; Chen, Y.; Lu, Y.; Zeng, H.; et al. Lactate Is a Natural Suppressor of RLR Signaling by Targeting MAVS. Cell 2019, 178, 176–189.e15. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Li, T.; Ramos Da Silva, S.; Lee, J.-J.; Lu, C.; Eoh, H.; Jung, J.U.; Gao, S.-J. A Critical Role of Glutamine and Asparagine γ-Nitrogen in Nucleotide Biosynthesis in Cancer Cells Hijacked by an Oncogenic Virus. MBio 2017, 8, e01179-17. [Google Scholar] [CrossRef]
- Myoung, J.; Ganem, D. Infection of lymphoblastoid cell lines by Kaposi’s sarcoma-associated herpesvirus: Critical role of cell-associated virus. J. Virol. 2011, 85, 9767–9777. [Google Scholar] [CrossRef] [PubMed]
- Hassman, L.M.; Ellison, T.J.; Kedes, D.H. KSHV infects a subset of human tonsillar B cells, driving proliferation and plasmablast differentiation. J. Clin. Investig. 2011, 121, 752–768. [Google Scholar] [CrossRef] [PubMed]
- Atyeo, N.; Rodriguez, M.D.; Papp, B.; Toth, Z. Clinical Manifestations and Epigenetic Regulation of Oral Herpesvirus Infections. Viruses 2021, 13, 681. [Google Scholar] [CrossRef] [PubMed]
- Seifi, A.; Weaver, E.M.; Whipple, M.E.; Ikoma, M.; Farrenberg, J.; Huang, M.-L.; Vieira, J. The lytic activation of KSHV during keratinocyte differentiation is dependent upon a suprabasal position, the loss of integrin engagement, and calcium, but not the interaction of cadherins. Virology 2011, 410, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Duus, K.M.; Lentchitsky, V.; Wagenaar, T.; Grose, C.; Webster-Cyriaque, J. Wild-type Kaposi’s sarcoma-associated herpesvirus isolated from the oropharynx of immune-competent individuals has tropism for cultured oral epithelial cells. J. Virol. 2004, 78, 4074–4084. [Google Scholar] [CrossRef] [PubMed]
- Casper, C.; Krantz, E.; Selke, S.; Kuntz, S.R.; Wang, J.; Huang, M.L.; Pauk, J.S.; Corey, L.; Wald, A. Frequent and asymptomatic oropharyngeal shedding of human herpesvirus 8 among immunocompetent men. J. Infect. Dis. 2007, 195, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Vieira, J.; Huang, M.L.; Koelle, D.M.; Corey, L. Transmissible Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) in saliva of men with a history of Kaposi’s sarcoma. J. Virol. 1997, 71, 7083–7087. [Google Scholar] [CrossRef] [PubMed]
- Atyeo, N.; Chae, M.Y.; Toth, Z.; Sharma, A.; Papp, B. Kaposi’s Sarcoma-Associated Herpesvirus Immediate Early Proteins Trigger FOXQ1 Expression in Oral Epithelial Cells, Engaging in a Novel Lytic Cycle-Sustaining Positive Feedback Loop. J. Virol. 2023, 97, e01696-22. [Google Scholar] [CrossRef] [PubMed]
- Wan, Q.; Tavakoli, L.; Wang, T.-Y.; Tucker, A.J.; Zhou, R.; Liu, Q.; Feng, S.; Choi, D.; He, Z.; Gack, M.U.; et al. Hijacking of nucleotide biosynthesis and deamidation-mediated glycolysis by an oncogenic herpesvirus. Nat. Commun. 2024, 15, 1442. [Google Scholar] [CrossRef] [PubMed]
- Choi, U.Y.; Lee, J.J.; Park, A.; Zhu, W.; Lee, H.R.; Choi, Y.J.; Yoo, J.S.; Yu, C.; Feng, P.; Gao, S.J.; et al. Oncogenic human herpesvirus hijacks proline metabolism for tumorigenesis. Proc. Natl. Acad. Sci. USA 2020, 117, 8083–8093. [Google Scholar] [CrossRef] [PubMed]
- Chandel, N.S. Amino Acid Metabolism. Cold Spring Harb. Perspect. Biol. 2021, 13, a040584. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.K.; Bose, D.; Robertson, E.S. HIF1alpha-Regulated Expression of the Fatty Acid Binding Protein Family Is Important for Hypoxic Reactivation of Kaposi’s Sarcoma-Associated Herpesvirus. J. Virol. 2021, 95, e02063-20. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-Z.; Yan, X.-H. Sensors for the mTORC1 pathway regulated by amino acids. J. Zhejiang Univ.-Sci. B 2019, 20, 699–712. [Google Scholar] [CrossRef] [PubMed]
- Barik, S. The Uniqueness of Tryptophan in Biology: Properties, Metabolism, Interactions and Localization in Proteins. Int. J. Mol. Sci. 2020, 21, 8776. [Google Scholar] [CrossRef] [PubMed]
- Kincaid, J.W.; Berger, N.A. NAD metabolism in aging and cancer. Exp. Biol. Med. 2020, 245, 1594–1614. [Google Scholar] [CrossRef]
- Keshet, R.; Szlosarek, P.; Carracedo, A.; Erez, A. Rewiring urea cycle metabolism in cancer to support anabolism. Nat. Rev. Cancer 2018, 18, 634–645. [Google Scholar] [CrossRef] [PubMed]
- Spinelli, J.B.; Yoon, H.; Ringel, A.E.; Jeanfavre, S.; Clish, C.B.; Haigis, M.C. Metabolic recycling of ammonia via glutamate dehydrogenase supports breast cancer biomass. Science 2017, 358, 941–946. [Google Scholar] [CrossRef] [PubMed]
- Privatt, S.R.; Braga, C.P.; Johnson, A.; Lidenge, S.J.; Berry, L.; Ngowi, J.R.; Ngalamika, O.; Chapple, A.G.; Mwaiselage, J.; Wood, C.; et al. Comparative polar and lipid plasma metabolomics differentiate KSHV infection and disease states. Cancer Metab. 2023, 11, 13. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Pulido, Y.E.; Mounce, B.C. Good cop, bad cop: Polyamines play both sides in host immunity and viral replication. Semin. Cell Dev. Biol. 2023, 146, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Gibson, W.; Roizman, B. Compartmentalization of Spermine and Spermidine in the Herpes Simplex Virion. Proc. Natl. Acad. Sci. USA 1971, 68, 2818–2821. [Google Scholar] [CrossRef] [PubMed]
- Choi, U.Y.; Lee, J.J.; Park, A.; Jung, K.L.; Lee, S.-A.; Choi, Y.J.; Lee, H.-R.; Lai, C.-J.; Eoh, H.; Jung, J.U. Herpesvirus-induced spermidine synthesis and eIF5A hypusination for viral episomal maintenance. Cell Rep. 2022, 40, 111234. [Google Scholar] [CrossRef] [PubMed]
- Fiches, G.N.; Wu, Z.; Zhou, D.; Biswas, A.; Li, T.-W.; Kong, W.; Jean, M.; Santoso, N.G.; Zhu, J. Polyamine biosynthesis and eIF5A hypusination are modulated by the DNA tumor virus KSHV and promote KSHV viral infection. PLoS Pathog. 2022, 18, e1010503. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Mao, Y.; Zhao, L.; Li, L.; Wu, J.; Zhao, M.; Du, W.; Yu, L.; Jiang, P. p53 regulation of ammonia metabolism through urea cycle controls polyamine biosynthesis. Nature 2019, 567, 253–256. [Google Scholar] [CrossRef] [PubMed]
- Morales, Y.; Cáceres, T.; May, K.; Hevel, J.M. Biochemistry and regulation of the protein arginine methyltransferases (PRMTs). Arch. Biochem. Biophys. 2016, 590, 138–152. [Google Scholar] [CrossRef] [PubMed]
- Tain, Y.L.; Hsu, C.N. Toxic Dimethylarginines: Asymmetric Dimethylarginine (ADMA) and Symmetric Dimethylarginine (SDMA). Toxins 2017, 9, 92. [Google Scholar] [CrossRef] [PubMed]
- Bedford, M.T. Arginine methylation at a glance. J. Cell Sci. 2007, 120, 4243–4246. [Google Scholar] [CrossRef] [PubMed]
- Bedford, M.T.; Clarke, S.G. Protein Arginine Methylation in Mammals: Who, What, and Why. Mol. Cell 2009, 33, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Hartley, A.-V.; Lu, T. Modulating the modulators: Regulation of protein arginine methyltransferases by post-translational modifications. Drug Discov. Today 2020, 25, 1735–1743. [Google Scholar] [CrossRef] [PubMed]
- Kurz, K.; Teerlink, T.; Sarcletti, M.; Weiss, G.; Zangerle, R.; Fuchs, D. Plasma concentrations of the cardiovascular risk factor asymmetric dimethylarginine (ADMA) are increased in patients with HIV-1 infection and correlate with immune activation markers. Pharmacol. Res. 2009, 60, 508–514. [Google Scholar] [CrossRef] [PubMed]
- Karacaer, C.; Yaylacı, S.; Demirci, T.; Cekic, D.; Suner, K.O.; Cokluk, E.; Varım, C. Association of mortality and endothelial dysfunction with serum ADMA level in COVID-19 patients. Pak. J. Med. Sci. 2022, 38, 1808. [Google Scholar] [CrossRef] [PubMed]
- Hannemann, J.; Balfanz, P.; Schwedhelm, E.; Hartmann, B.; Ule, J.; Müller-Wieland, D.; Dahl, E.; Dreher, M.; Marx, N.; Böger, R. Elevated serum SDMA and ADMA at hospital admission predict in-hospital mortality of COVID-19 patients. Sci. Rep. 2021, 11, 9895. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.W.; Cho, Y.; Bae, G.-U.; Kim, S.-N.; Kim, Y.K. Protein arginine methyltransferases: Promising targets for cancer therapy. Exp. Mol. Med. 2021, 53, 788–808. [Google Scholar] [CrossRef] [PubMed]
- Niu, D.; Ma, Y.; Ren, P.; Chang, S.; Li, C.; Jiang, Y.; Han, C.; Lan, K. Methylation of KSHV vCyclin by PRMT5 contributes to cell cycle progression and cell proliferation. PLoS Pathog. 2024, 20, e1012535. [Google Scholar] [CrossRef] [PubMed]
- Strahan, R.C.; McDowell-Sargent, M.; Uppal, T.; Purushothaman, P.; Verma, S.C. KSHV encoded ORF59 modulates histone arginine methylation of the viral genome to promote viral reactivation. PLoS Pathog. 2017, 13, e1006482. [Google Scholar] [CrossRef]
- Robinson, A.D.; Eich, M.-L.; Varambally, S. Dysregulation of de novo nucleotide biosynthetic pathway enzymes in cancer and targeting opportunities. Cancer Lett. 2020, 470, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Villa, E.; Ali, E.; Sahu, U.; Ben-Sahra, I. Cancer Cells Tune the Signaling Pathways to Empower de Novo Synthesis of Nucleotides. Cancers 2019, 11, 688. [Google Scholar] [CrossRef] [PubMed]
- Qin, C.; Rao, Y.; Yuan, H.; Wang, T.-Y.; Zhao, J.; Espinosa, B.; Liu, Y.; Zhang, S.; Savas, A.C.; Liu, Q.; et al. SARS-CoV-2 couples evasion of inflammatory response to activated nucleotide synthesis. Proc. Natl. Acad. Sci. USA 2022, 119, e2122897119. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Tian, M.; Zhang, S.; Delfarah, A.; Gao, R.; Rao, Y.; Savas, A.C.; Lu, A.; Bubb, L.; Lei, X.; et al. Deamidation Shunts RelA from Mediating Inflammation to Aerobic Glycolysis. Cell Metab. 2020, 31, 937–955.e7. [Google Scholar] [CrossRef] [PubMed]
- Schmid, R.; Heuckeroth, S.; Korf, A.; Smirnov, A.; Myers, O.; Dyrlund, T.S.; Bushuiev, R.; Murray, K.J.; Hoffmann, N.; Lu, M.; et al. Integrative analysis of multimodal mass spectrometry data in MZmine 3. Nat. Biotechnol. 2023, 41, 447–449. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Diedrich, J.; Chu, Y.-Y.; Yates, J.R. Extracting Accurate Precursor Information for Tandem Mass Spectra by RawConverter. Anal. Chem. 2015, 87, 11361–11367. [Google Scholar] [CrossRef] [PubMed]
- Patterson, R.E.; Kirpich, A.S.; Koelmel, J.P.; Kalavalapalli, S.; Morse, A.M.; Cusi, K.; Sunny, N.E.; McIntyre, L.M.; Garrett, T.J.; Yost, R.A. Improved experimental data processing for UHPLC–HRMS/MS lipidomics applied to nonalcoholic fatty liver disease. Metabolomics 2017, 13, 142. [Google Scholar] [CrossRef]
- Pang, Z.; Chong, J.; Zhou, G.; David, A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.-É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alfaez, A.; Christopher, M.W.; Garrett, T.J.; Papp, B. Analysis of Metabolomic Reprogramming Induced by Infection with Kaposi’s Sarcoma-Associated Herpesvirus Using Untargeted Metabolomic Profiling. Int. J. Mol. Sci. 2025, 26, 3109. https://doi.org/10.3390/ijms26073109
Alfaez A, Christopher MW, Garrett TJ, Papp B. Analysis of Metabolomic Reprogramming Induced by Infection with Kaposi’s Sarcoma-Associated Herpesvirus Using Untargeted Metabolomic Profiling. International Journal of Molecular Sciences. 2025; 26(7):3109. https://doi.org/10.3390/ijms26073109
Chicago/Turabian StyleAlfaez, Abdulkarim, Michael W. Christopher, Timothy J. Garrett, and Bernadett Papp. 2025. "Analysis of Metabolomic Reprogramming Induced by Infection with Kaposi’s Sarcoma-Associated Herpesvirus Using Untargeted Metabolomic Profiling" International Journal of Molecular Sciences 26, no. 7: 3109. https://doi.org/10.3390/ijms26073109
APA StyleAlfaez, A., Christopher, M. W., Garrett, T. J., & Papp, B. (2025). Analysis of Metabolomic Reprogramming Induced by Infection with Kaposi’s Sarcoma-Associated Herpesvirus Using Untargeted Metabolomic Profiling. International Journal of Molecular Sciences, 26(7), 3109. https://doi.org/10.3390/ijms26073109






