Genome-Wide Identification and Expression Profiling of Dehydration-Responsive Element-Binding Family Genes in Flax (Linum usitatissimum L.)
Abstract
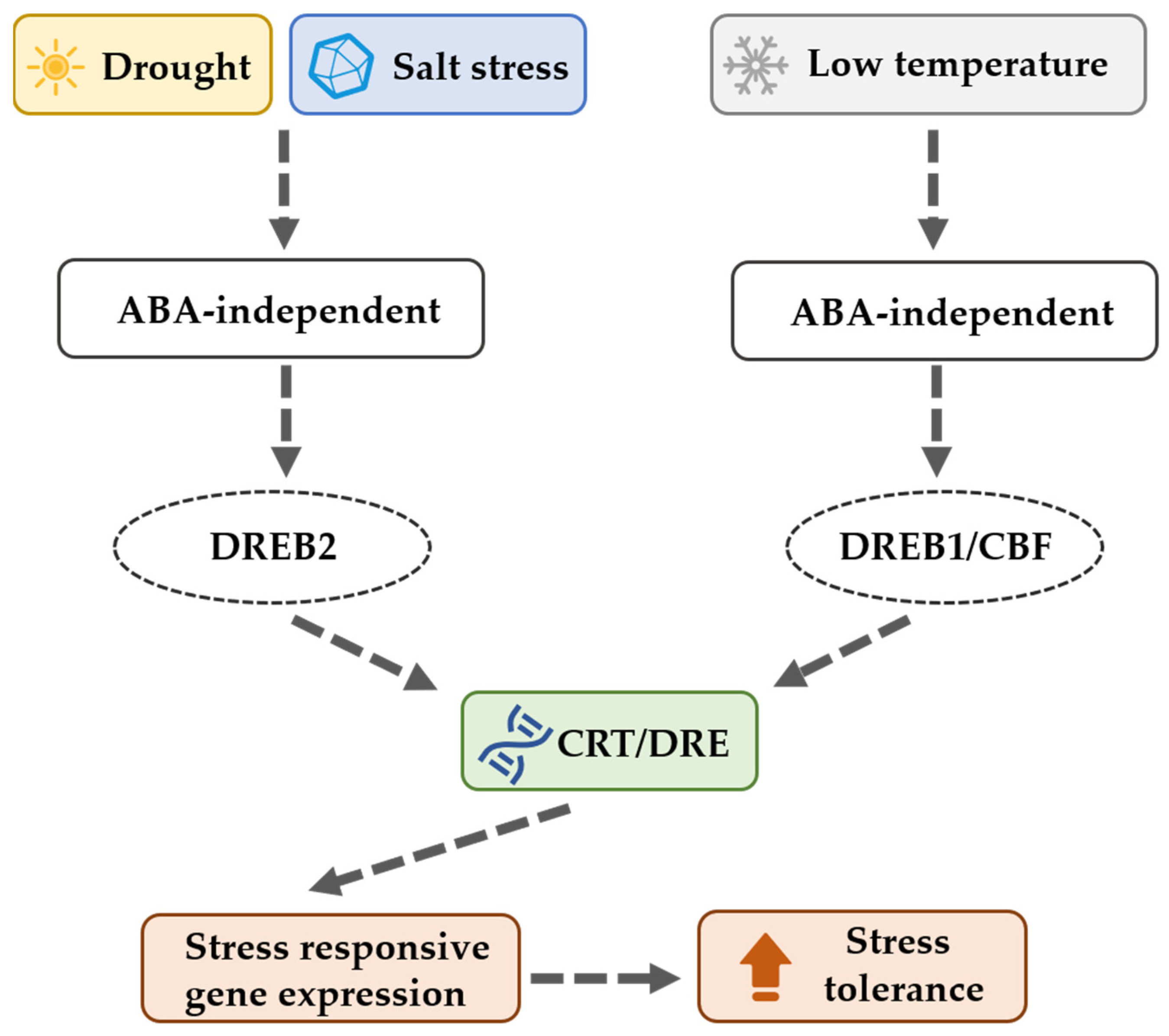
1. Introduction
2. Results
2.1. Identification of DREB Genes in Flax
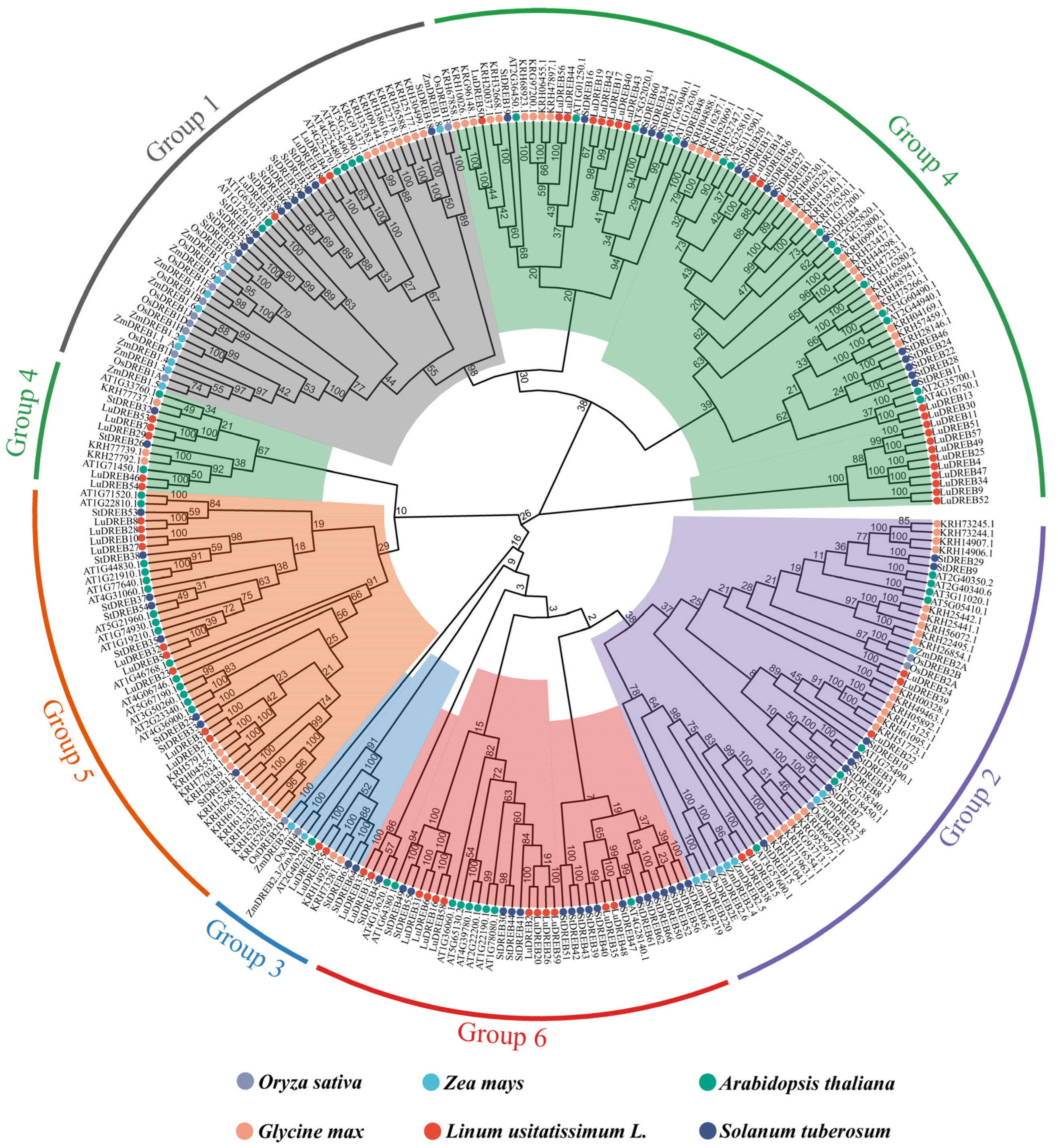
2.2. Phylogenetic Analysis of LuDREB Proteins
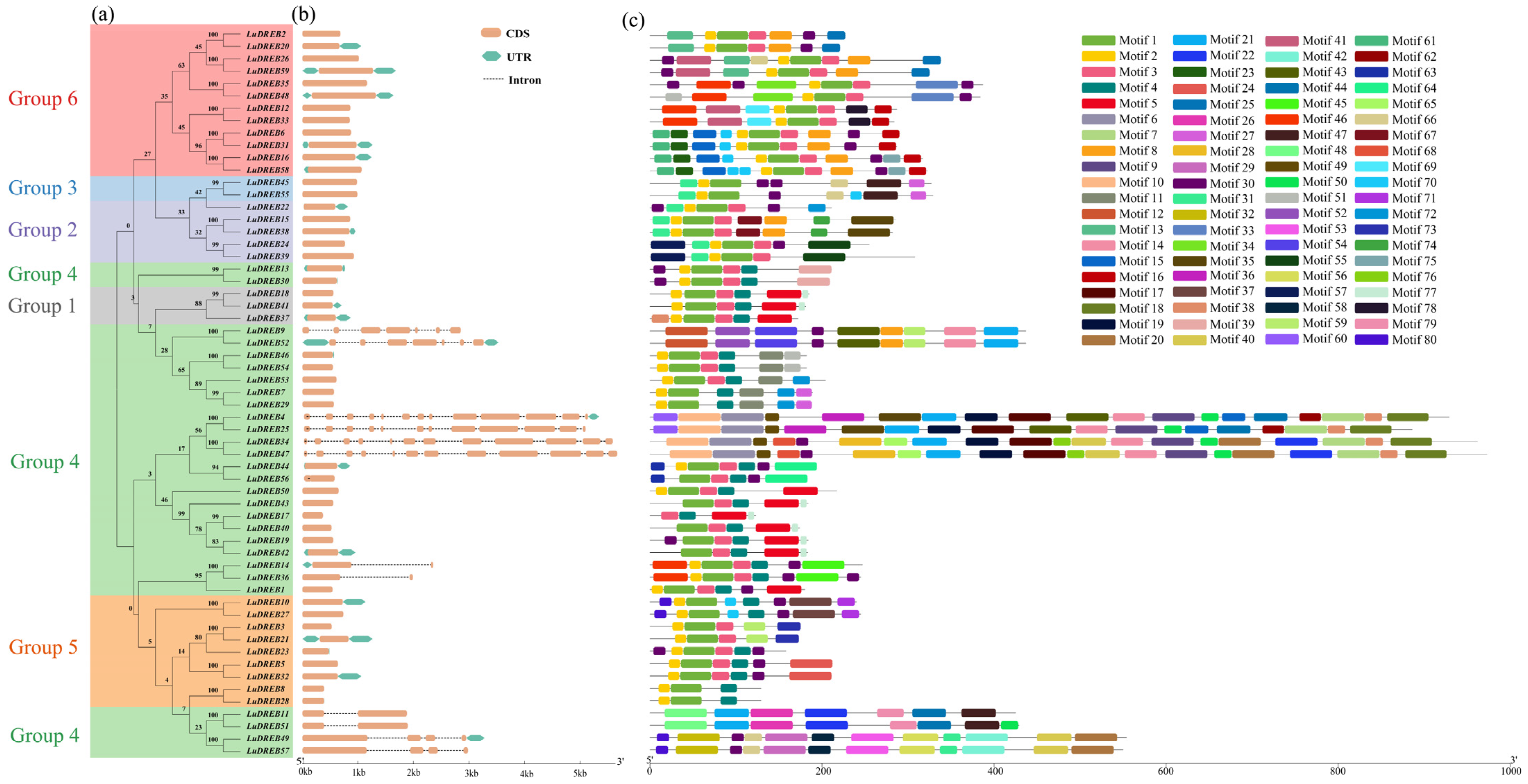
2.3. Sequence Analysis of Flax DREBs
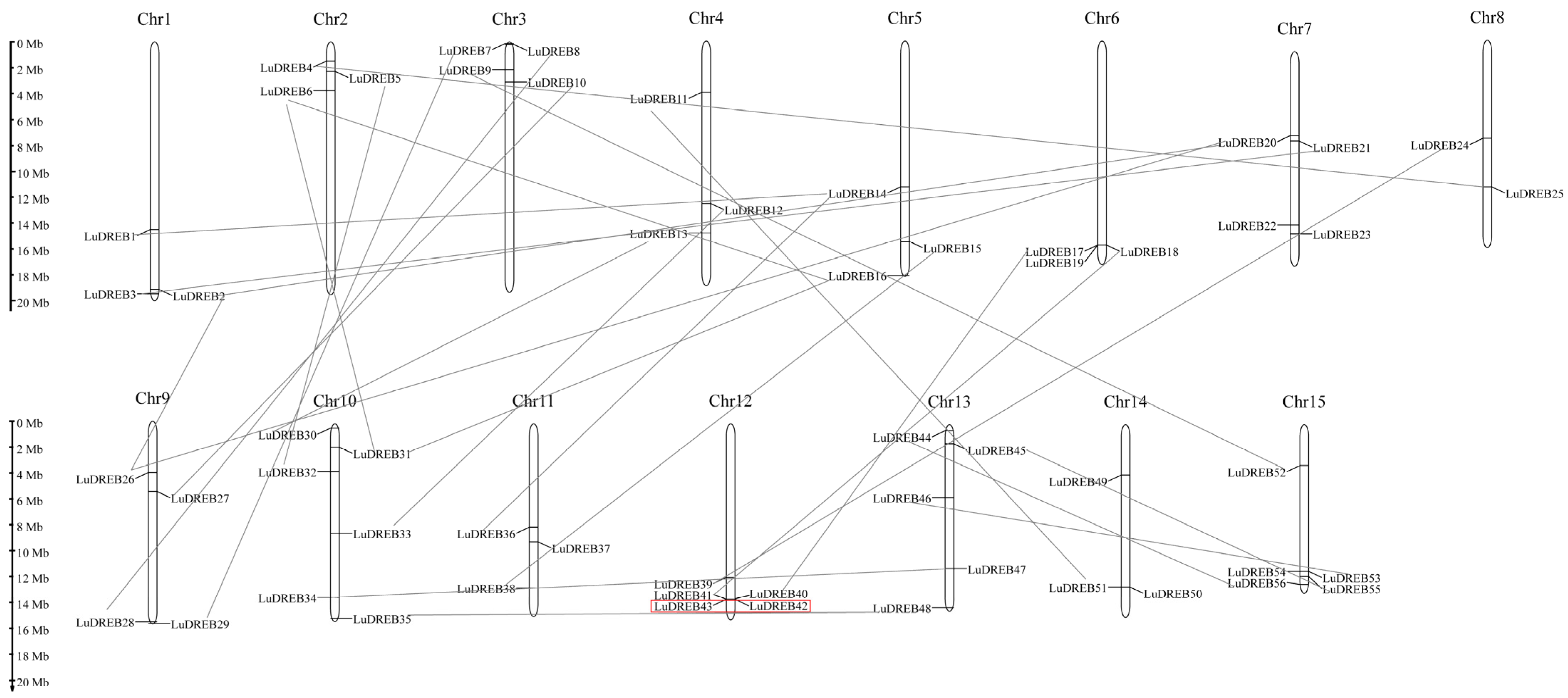
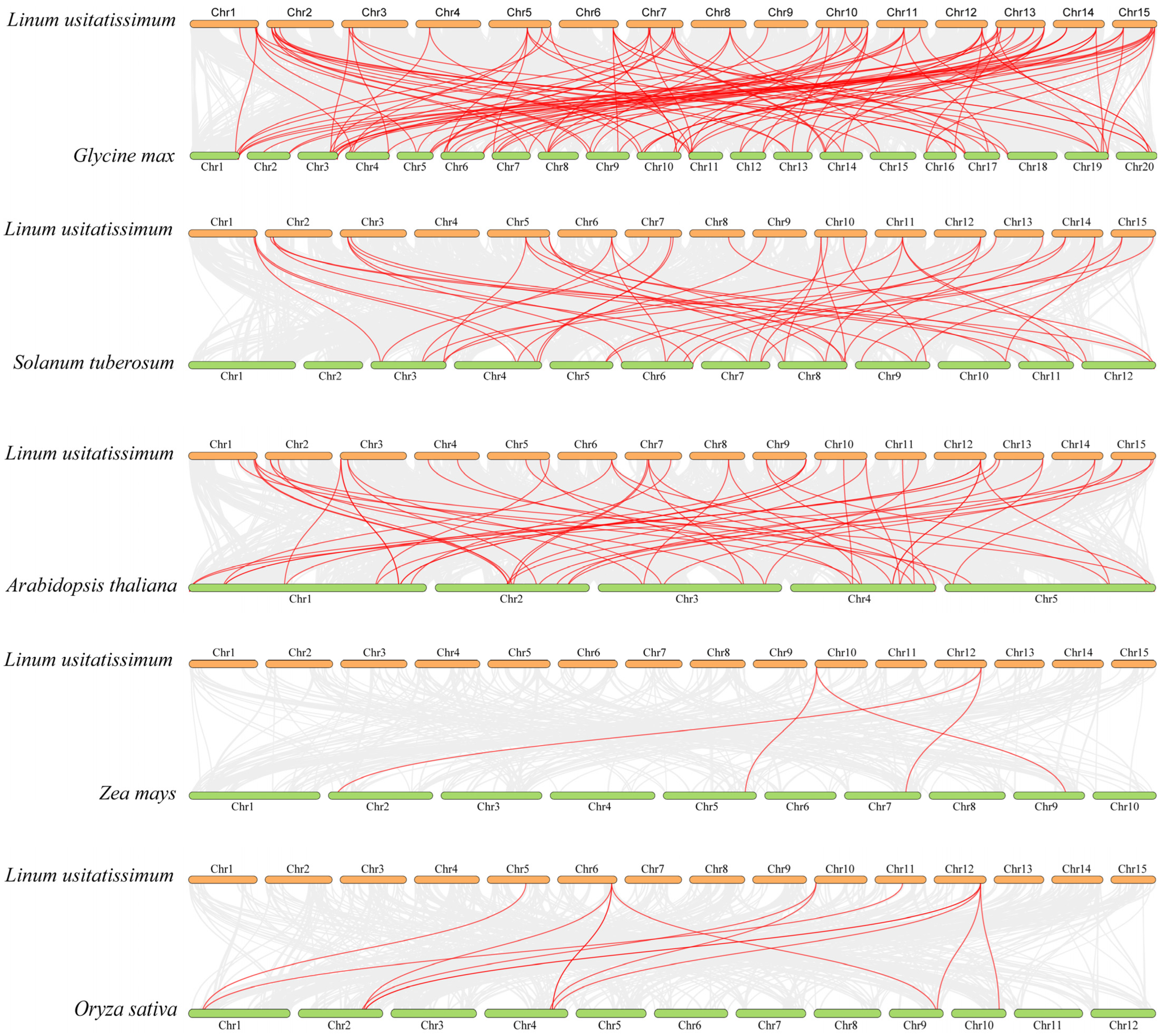
2.4. Chromosomal Location and Synteny Analysis
2.5. Cis-Acting Elements on LuDREB Promoters
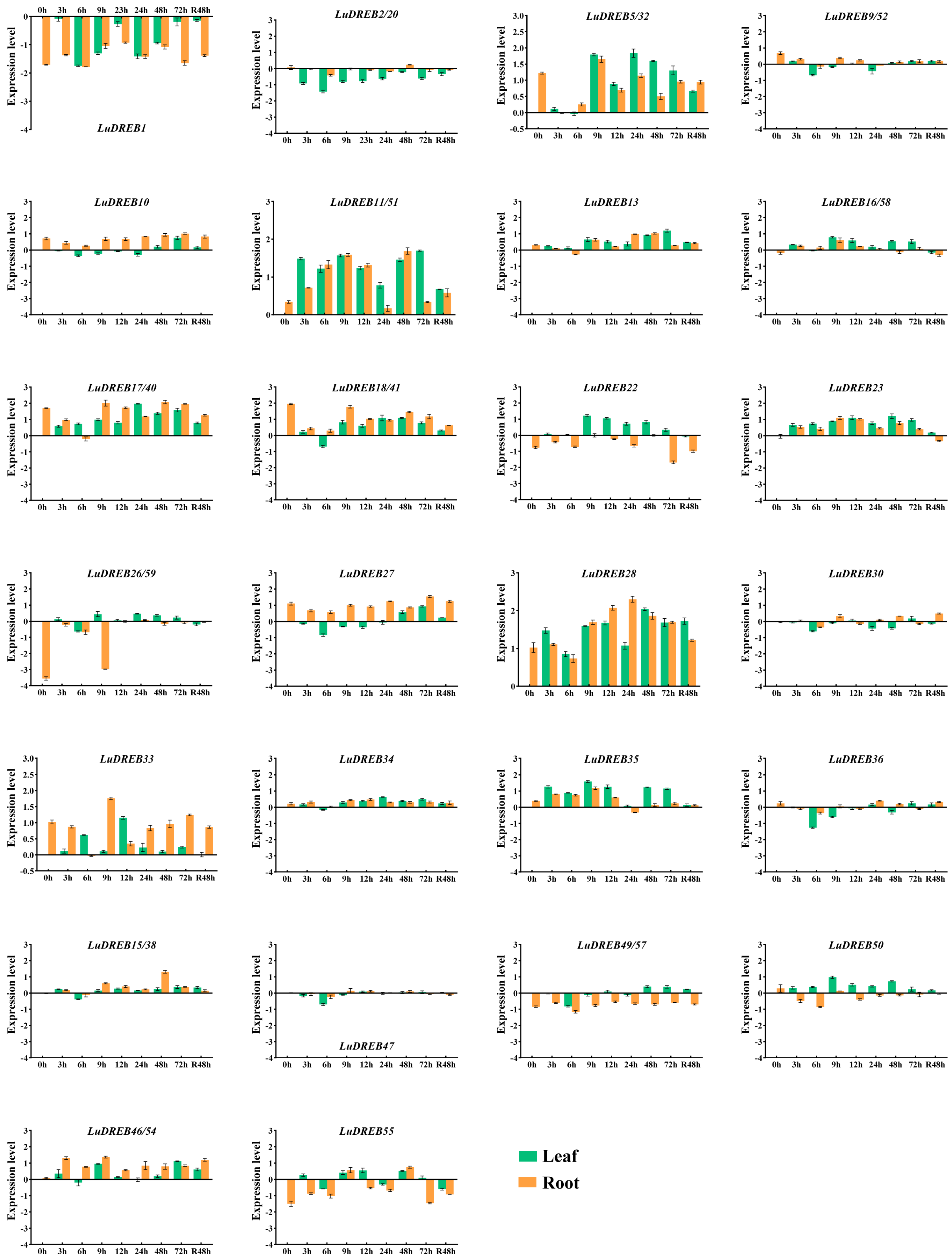
2.6. Expression Analysis of LuDREB Genes in Different Tissues of Flax Under Drought Stress
3. Discussion
4. Materials and Methods
4.1. Plant Growth and Drought Treatment
4.2. Identification of DREB Genes
4.3. Chromosomal Mapping and Collinearity Analysis
4.4. Gene Structure and Promoter Analysis
4.5. Phylogenetic Analysis
4.6. Gene Expression Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Peck, S.; Mittler, R. Plant signaling in biotic and abiotic stress. J. Exp. Bot. 2020, 71, 1649–1651. [Google Scholar] [CrossRef] [PubMed]
- Zandalinas, S.I.; Fichman, Y.; Devireddy, A.R.; Sengupta, S.; Azad, R.K.; Mittler, R. Systemic signaling during abiotic stress combination in plants. Proc. Natl. Acad. Sci. USA 2020, 117, 13810–13820. [Google Scholar] [CrossRef]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 2005, 10, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Goyal, R.K.; Tulpan, D.; Chomistek, N.; Fundora, D.G.P.; West, C.; Ellis, B.E.; Frick, M.; Laroche, A.; Foroud, N.A. Analysis of MAPK and MAPKK gene families in wheat and related Triticeae species. BMC Genom. 2018, 19, 178. [Google Scholar] [CrossRef]
- Wan, Y.Q.; Mao, M.Z.; Wan, D.L.; Yang, Q.; Yang, F.Y.; Mandlaa; Li, G.J.; Wang, R.G. Identification of the WRKY gene family and functional analysis of two genes in Caragana intermedia. BMC Plant Biol. 2018, 18, 31. [Google Scholar] [CrossRef]
- Wani, S.H.; Anand, S.; Singh, B.; Bohra, A.; Joshi, R. WRKY transcription factors and plant defense responses: Latest discoveries and future prospects. Plant Cell Rep. 2021, 40, 1071–1085. [Google Scholar] [CrossRef]
- Li, C.N.; Ng, C.K.Y.; Fan, L.M. MYB transcription factors, active players in abiotic stress signaling. Environ. Exp. Bot. 2015, 114, 80–91. [Google Scholar] [CrossRef]
- Phukan, U.J.; Jeena, G.S.; Tripathi, V.; Shukla, R.K. Regulation of Apetala2/Ethylene Response Factors in Plants. Front. Plant Sci. 2017, 8, 150. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, Y.; Liu, Q.; Dubouzet, J.G.; Abe, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 2002, 290, 998–1009. [Google Scholar] [CrossRef]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef]
- Ali, N.; Hadi, F. CBF/DREB transcription factor genes play role in cadmium tolerance and phytoaccumulation in Ricinus communis under molybdenum treatments. Chemosphere 2018, 208, 425–432. [Google Scholar] [CrossRef]
- Kudo, M.; Kidokoro, S.; Yoshida, T.; Mizoi, J.; Todaka, D.; Fernie, A.R.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Double overexpression of DREB and PIF transcription factors improves drought stress tolerance and cell elongation in transgenic plants. Plant Biotechnol. J. 2017, 15, 458–471. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.S.; Chen, M.; Li, L.C.; Ma, Y.Z. Functions and Application of the AP2/ERF Transcription Factor Family in Crop Improvement. J. Integr. Plant Biol. 2011, 53, 570–585. [Google Scholar] [CrossRef]
- Ain-Ali, Q.-U.; Mushtaq, N.; Amir, R.; Gul, A.; Tahir, M.; Munir, F. Genome-wide promoter analysis, homology modeling and protein interaction network of Dehydration Responsive Element Binding (DREB) gene family in Solanum tuberosum. PLoS ONE 2021, 16, e0261215. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhang, L.T.; Nie, L.B.; Zheng, Y.S.; Zhu, S.D.; Hou, J.F.; Li, R.J.; Chen, G.H.; Tang, X.Y.; Wang, C.G.; et al. Genome-wide analysis of the DREB family genes and functional identification of the involvement of BrDREB2B in abiotic stress in wucai (Brassica campestris L.). Bmc Genom. 2022, 23, 598. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.L.; Hu, L.L.; Wang, L.X.; Wang, S.H.; Cheng, X.Z. Genome-wide identification and expression profiles of AP2/ERF transcription factor family in mung bean (Vigna radiata L.). J. Appl. Genet. 2022, 63, 223–236. [Google Scholar] [CrossRef]
- Wei, T.; Deng, K.J.; Liu, D.Q.; Gao, Y.H.; Liu, Y.; Yang, M.L.; Zhang, L.P.; Zheng, X.L.; Wang, C.G.; Song, W.Q.; et al. Ectopic Expression of DREB Transcription Factor, AtDREB1A, Confers Tolerance to Drought in Transgenic Salvia miltiorrhiza. Plant Cell Physiol. 2016, 57, 1593–1609. [Google Scholar] [CrossRef]
- Wei, T.; Deng, K.J.; Gao, Y.H.; Liu, Y.; Yang, M.L.; Zhang, L.P.; Zheng, X.L.; Wang, C.G.; Song, W.Q.; Chen, C.B.; et al. Arabidopsis DREB1B in transgenic Salvia miltiorrhiza increased tolerance to drought stress without stunting growth. Plant Physiol. Biochem. 2016, 104, 17–28. [Google Scholar] [CrossRef]
- Zhou, Y.B.; Chen, M.; Guo, J.K.; Wang, Y.X.; Min, D.H.; Jiang, Q.Y.; Ji, H.T.; Huang, C.Y.; Wei, W.; Xu, H.J.; et al. Overexpression of soybean DREB1 enhances drought stress tolerance of transgenic wheat in the field. J. Exp. Bot. 2020, 71, 1842–1857. [Google Scholar] [CrossRef]
- Moon, S.J.; Min, M.K.; Kim, J.A.; Kim, D.Y.; Yoon, I.S.; Kwon, T.R.; Byun, M.O.; Kim, B.G. Ectopic Expression of OsDREB1G, a Member of the OsDREB1 Subfamily, Confers Cold Stress Tolerance in Rice. Front. Plant Sci. 2019, 10, 297. [Google Scholar] [CrossRef]
- Wen, W.W.; Xie, Z.N.; Yu, G.H.; Zhao, C.L.; Zhang, J.; Huang, L.K.; Xu, B.; Huang, B.R. Switchgrass PvDREB1C plays opposite roles in plant cold and salt tolerance in transgenic tobacco. Hereditas 2017, 155, 15. [Google Scholar] [CrossRef]
- Challam, C.; Ghosh, T.; Rai, M.; Tyagi, W. Allele mining across DREB1A and DREB1B in diverse rice genotypes suggest a highly conserved pathway inducible by low temperature. J. Genet. 2015, 94, 231–238. [Google Scholar] [CrossRef]
- Sakuma, Y.; Maruyama, K.; Qin, F.; Osakabe, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc. Natl. Acad. Sci. USA 2006, 103, 18822–18827. [Google Scholar] [CrossRef] [PubMed]
- Qin, F.; Kakimoto, M.; Sakuma, Y.; Maruyama, K.; Osakabe, Y.; Tran, L.S.P.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Regulation and functional analysis of ZmDREB2A in response to drought and heat stresses in Zea mays L. Plant J. 2007, 50, 54–69. [Google Scholar] [CrossRef]
- Gupta, K.; Agarwal, P.K.; Reddy, M.K.; Jha, B. SbDREB2A, an A-2 type DREB transcription factor from extreme halophyte Salicornia brachiata confers abiotic stress tolerance in Escherichia coli. Plant Cell Rep. 2010, 29, 1131–1137. [Google Scholar] [CrossRef]
- Gumi, A.M.; Guha, P.K.; Mazumder, A.; Jayaswal, P.; Mondal, T.K. Characterization of OglDREB2A gene from African rice (Oryza glaberrima), comparative analysis and its transcriptional regulation under salinity stress. 3 Biotech 2018, 8, 91. [Google Scholar] [CrossRef]
- Chen, H.L.; Liu, L.P.; Wang, L.X.; Wang, S.H.; Cheng, X.Z. VrDREB2A, a DREB-binding transcription factor from Vigna radiata, increased drought and high-salt tolerance in transgenic Arabidopsis thaliana. J. Plant Res. 2016, 129, 263–273. [Google Scholar] [CrossRef] [PubMed]
- Niu, X.; Luo, T.L.; Zhao, H.Y.; Su, Y.L.; Ji, W.Q.; Li, H.F. Identification of wheat DREB genes and functional characterization of TaDREB3 in response to abiotic stresses. Gene 2020, 740, 144514. [Google Scholar] [CrossRef]
- Guo, B.J.; Wei, Y.F.; Xu, R.B.; Lin, S.; Luan, H.Y.; Lv, C.; Zhang, X.Z.; Song, X.Y.; Xu, R.G. Genome-Wide Analysis of APETALA2/Ethylene-Responsive Factor (AP2/ERF) Gene Family in Barley (Hordeum vulgare L.). PloS ONE 2016, 11, e0161322. [Google Scholar] [CrossRef]
- Nakashima, K.; Shinwari, Z.K.; Sakuma, Y.; Seki, M.; Miura, S.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Organization and expression of two Arabidopsis DREB2 genes encoding DRE-binding proteins involved in dehydration- and high-salinity-responsive gene expression. Plant Mol. Biol. 2000, 42, 657–665. [Google Scholar] [CrossRef]
- Li, Z.; Wang, G.; Liu, X.H.; Wang, Z.C.; Zhang, M.Q.; Zhang, J.S. Genome-wide identification and expression profiling of DREB genes in Saccharum spontaneum. BMC Genom. 2021, 22, 456. [Google Scholar] [CrossRef]
- Wei, S.B.; Li, X.; Lu, Z.F.; Zhang, H.; Ye, X.Y.; Zhou, Y.J.; Li, J.; Yan, Y.Y.; Pei, H.C.; Duan, F.Y.; et al. A transcriptional regulator that boosts grain yields and shortens the growth duration of rice. Science 2022, 377, eabi8455. [Google Scholar] [CrossRef]
- Zohary, D. Monophyletic vs. polyphyletic origin of the crops on which agriculture was founded in the Near East. Genet. Resour. Crop Evol. 1999, 46, 133–142. [Google Scholar] [CrossRef]
- Smykal, P.; Bacová-Kerteszová, N.; Kalendar, R.; Corander, J.; Schulman, A.H.; Pavelek, M. Genetic diversity of cultivated flax (Linum usitatissimum L.) germplasm assessed by retrotransposon-based markers. Theor. Appl. Genet. 2011, 122, 1385–1397. [Google Scholar] [CrossRef] [PubMed]
- Xie, D.W.; Dai, Z.G.; Yang, Z.M.; Tang, Q.; Sun, J.; Yang, X.; Song, X.X.; Lu, Y.; Zhao, D.B.; Zhang, L.G.; et al. Genomic variations and association study of agronomic traits in flax. BMC Genom. 2018, 19, 512. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.P.; Qi, Y.N.; Wang, L.M.; Wang, L.L.; Yan, X.C.; Dang, Z.; Li, W.J.; Zhao, W.; Pei, X.W.; Li, X.M.; et al. Genomic Comparison and Population Diversity Analysis Provide Insights into the Domestication and Improvement of Flax. Iscience 2020, 23, 100967. [Google Scholar] [CrossRef]
- Yuan, H.M.; Guo, W.D.; Zhao, L.J.; Yu, Y.; Chen, S.; Tao, L.; Cheng, L.L.; Kang, Q.H.; Song, X.X.; Wu, J.Z.; et al. Genome-wide identification and expression analysis of the WRKY transcription factor family in flax (Linum usitatissimum L.). BMC Genom. 2021, 22, 375. [Google Scholar] [CrossRef]
- Tombuloglu, H. Genome-wide identification and expression analysis of R2R3, 3R-and 4R-MYB transcription factors during lignin biosynthesis in flax (Linum usitatissimum). Genomics 2020, 112, 782–795. [Google Scholar] [CrossRef]
- Li, Z.; Chi, H.; Liu, C.Y.; Zhang, T.B.; Han, L.D.; Li, L.; Pei, X.W.; Long, Y. Genome-wide identification and functional characterization of LEA genes during seed development process in linseed flax (Linum usitatissimum L.). BMC Plant Biol. 2021, 21, 193. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef]
- Nekrutenko, A.; Makova, K.D.; Li, W.H. The KA/KS ratio test for assessing the protein-coding potential of genomic regions: An empirical and simulation study. Genome Res. 2002, 12, 198–202. [Google Scholar] [CrossRef] [PubMed]
- Albert, V.A.; Barbazuk, W.B.; de Pamphilis, C.W.; Der, J.P.; Leebens-Mack, J.; Ma, H.; Palmer, J.D.; Rounsley, S.; Sankoff, D.; Schuster, S.C.; et al. The Amborella Genome and the Evolution of Flowering Plants. Science 2013, 342, 1241089. [Google Scholar] [CrossRef]
- Van De Peer, Y.; Mizrachi, E.; Marchal, K. The evolutionary significance of polyploidy. Nat. Rev. Genet. 2017, 18, 411–424. [Google Scholar] [CrossRef]
- Agarwal, P.K.; Agarwal, P.; Reddy, M.K.; Sopory, S.K. Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep. 2006, 25, 1263–1274. [Google Scholar] [CrossRef]
- Chen, Y.L.; Yang, J.L.; Wang, Z.C.; Zhang, H.Z.; Mao, X.L.; Li, C.H. Gene Structures, Classification, and Expression Models of the DREB Transcription Factor Subfamily in Populus trichocarpa. Sci. World J. 2013, 2013, 954640. [Google Scholar] [CrossRef]
- Hu, X.; Liang, J.X.; Wang, W.J.; Cai, C.Y.; Ye, S.W.; Wang, N.N.; Han, F.Y.; Wu, Y.X.; Zhu, Q. Comprehensive genome-wide analysis of the DREB gene family in Moso bamboo (Phyllostachys edulis): Evidence for the role of PeDREB28 in plant abiotic stress response. Plant J. 2023, 116, 1248–1270. [Google Scholar] [CrossRef] [PubMed]
- Mushtaq, N.; Munir, F.; Gul, A.; Amir, R.; Paracha, R.Z. Genome-wide analysis, identification, evolution and genomic organization of dehydration responsive element-binding (DREB) gene family in Solanum tuberosum. PeerJ 2021, 9, e11647. [Google Scholar] [CrossRef]
- Zhou, Y.X.; Zhou, W.; Liu, H.; Liu, P.; Li, Z.G. Genome-wide analysis of the soybean DREB gene family: Identification, genomic organization and expression profiles in response to drought stress. Plant Breed. 2020, 139, 1158–1167. [Google Scholar] [CrossRef]
- Salih, H.; Odongo, M.R.; Gong, W.F.; He, S.P.; Du, X.M. Genome-wide analysis of cotton C2H2-zinc finger transcription factor family and their expression analysis during fiber development. BMC Plant Biol. 2019, 19, 400. [Google Scholar] [CrossRef]
- Chung, B.Y.W.; Simons, C.; Firth, A.E.; Brown, C.M.; Hellens, R.P. Effect of 5′UTR introns on gene expression in Arabidopsis thaliana. BMC Genom. 2006, 7, 120. [Google Scholar] [CrossRef]
- Ren, X.Y.; Vorst, O.; Fiers, M.; Stiekema, W.J.; Nap, J.P. In plants, highly expressed genes are the least compact. Trends Genet. 2006, 22, 528–532. [Google Scholar] [CrossRef] [PubMed]
- Jeffares, D. Rapidly regulated genes are intron poor. Trends Genet. 2008, 24, 488. [Google Scholar] [CrossRef]
- Maqsood, H.; Munir, F.; Amir, R.; Gul, A. Genome-wide identification, comprehensive characterization of transcription factors, cis-regulatory elements, protein homology, and protein interaction network of DREB gene family in Solanum lycopersicum. Front. Plant Sci. 2022, 13, 1031679. [Google Scholar] [CrossRef]
- Wang, D.; Zeng, Y.Y.; Yang, X.X.; Nie, S.M. Characterization of DREB family genes in Lotus japonicus and LjDREB2B overexpression increased drought tolerance in transgenic Arabidopsis. BMC Plant Biol. 2024, 24, 497. [Google Scholar] [CrossRef]
- Ederli, L.; Bianchet, C.; Paolocci, F.; Alqurashi, M.; Gehring, C.; Pasqualini, S. Drought stress induces a biphasic NO accumulation in Arabidopsis thaliana. Plant Signal. Behav. 2019, 14, e1573098. [Google Scholar] [CrossRef] [PubMed]
- Shkolnik-Inbar, D.; Bar-Zvi, D. ABI4 Mediates Abscisic Acid and Cytokinin Inhibition of Lateral Root Formation by Reducing Polar Auxin Transport in Arabidopsis. Plant Cell 2010, 22, 3560–3573. [Google Scholar] [CrossRef]
- Liu, C.X.; Zhang, T.Z. Expansion and stress responses of the AP2/EREBP superfamily in cotton. BMC Genom. 2017, 18, 118. [Google Scholar] [CrossRef]
- Zhu, Y.; Wu, N.N.; Song, W.L.; Yin, G.J.; Qin, Y.J.; Yan, Y.M.; Hu, Y.K. Soybean (Glycine max) expansin gene superfamily origins: Segmental and tandem duplication events followed by divergent selection among subfamilies. BMC Plant Biol. 2014, 14, 93. [Google Scholar] [CrossRef]
- Ren, R.; Wang, H.F.; Guo, C.C.; Zhang, N.; Zeng, L.P.; Chen, Y.M.; Ma, H.; Qi, J. Widespread Whole Genome Duplications Contribute to Genome Complexity and Species Diversity in Angiosperms. Mol. Plant 2018, 11, 414–428. [Google Scholar] [CrossRef]
- Qiao, X.; Li, Q.H.; Yin, H.; Qi, K.J.; Li, L.T.; Wang, R.Z.; Zhang, S.L.; Paterson, A.H. Gene duplication and evolution in recurring polyploidization-diploidization cycles in plants. Genome Biol. 2019, 20, 38. [Google Scholar] [CrossRef]
- Zhang, Y.; Xia, P.G. The DREB transcription factor, a biomacromolecule, responds to abiotic stress by regulating the expression of stress-related genes. Int. J. Biol. Macromol. 2023, 243, 125231. [Google Scholar] [CrossRef] [PubMed]
- Dubouzet, J.G.; Sakuma, Y.; Ito, Y.; Kasuga, M.; Dubouzet, E.G.; Miura, S.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J. 2003, 33, 751–763. [Google Scholar] [CrossRef]
- Ito, Y.; Katsura, K.; Maruyama, K.; Taji, T.; Kobayashi, M.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol. 2006, 47, 141–153. [Google Scholar] [CrossRef]
- Wang, Q.Y.; Guan, Y.C.; Wu, Y.R.; Chen, H.L.; Chen, F.; Chu, C.C. Overexpression of a rice OsDREB1F gene increases salt, drought, and low temperature tolerance in both Arabidopsis and rice. Plant Mol. Biol. 2008, 67, 589–602. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.Q.; Meng, X.P.; Zhang, Y.; Xia, M.; Wang, X.P. Over-expression of OsDREB genes lead to enhanced drought tolerance in rice. Biotechnol. Lett. 2008, 30, 2191–2198. [Google Scholar] [CrossRef] [PubMed]
- Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Et Biophys. Acta-Gene Regul. Mech. 2012, 1819, 86–96. [Google Scholar] [CrossRef]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 2006, 57, 781–803. [Google Scholar] [CrossRef]
- Qiu, W.J.; Li, H.Y.; Song, Y.; Ding, J.R.; Chen, S.X.; Ma, C.Q.; Yu, B. Genome-Wide Analysis of DREB Genes in Sugar Beet and Their Potential Functions in Response to Drought Stress. Sugar Tech 2024, 26, 1306–1322. [Google Scholar] [CrossRef]
- Rehman, S.; Mahmood, T. Functional role of DREB and ERF transcription factors: Regulating stress-responsive network in plants. Acta Physiol. Plant. 2015, 37, 178. [Google Scholar] [CrossRef]
- Liu, Q.; Kasuga, M.; Sakuma, Y.; Abe, H.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 1998, 10, 1391–1406. [Google Scholar] [CrossRef]
- Lata, C.; Prasad, M. Role of DREBs in regulation of abiotic stress responses in plants. J. Exp. Bot. 2011, 62, 4731–4748. [Google Scholar] [CrossRef]
- Nakashima, K.; Ito, Y.; Yamaguchi-Shinozaki, K. Transcriptional Regulatory Networks in Response to Abiotic Stresses in Arabidopsis and Grasses. Plant Physiol. 2009, 149, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, Y.; Maruyama, K.; Osakabe, Y.; Qin, F.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 2006, 18, 1292–1309. [Google Scholar] [CrossRef] [PubMed]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef]
- Qi, Y.N.; Wang, L.M.; Li, W.J.; Dang, Z.; Xie, Y.P.; Zhao, W.; Zhao, L.R.; Li, W.; Yang, C.X.; Xu, C.M.; et al. Genome-Wide Identification and Expression Analysis of Auxin Response Factor Gene Family in Linum usitatissimum. Int. J. Mol. Sci. 2023, 24, 11006. [Google Scholar] [CrossRef]
- Liu, S.X.; Wang, X.L.; Wang, H.W.; Xin, H.B.; Yang, X.H.; Yan, J.B.; Li, J.S.; Tran, L.S.P.; Shinozaki, K.; Yamaguchi-Shinozaki, K.; et al. Genome-Wide Analysis of ZmDREB Genes and Their Association with Natural Variation in Drought Tolerance at Seedling Stage of Zea mays L. PloS Genet. 2013, 9, e1003790. [Google Scholar] [CrossRef]
- Matsukura, S.; Mizoi, J.; Yoshida, T.; Todaka, D.; Ito, Y.; Maruyama, K.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Comprehensive analysis of rice DREB2-type genes that encode transcription factors involved in the expression of abiotic stress-responsive genes. Mol. Genet. Genom. 2010, 283, 185–196. [Google Scholar] [CrossRef]
- Mao, D.H.; Chen, C.Y. Colinearity and Similar Expression Pattern of Rice DREB1s Reveal Their Functional Conservation in the Cold-Responsive Pathway. PloS ONE 2012, 7, e47275. [Google Scholar] [CrossRef]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar]
- Chou, K.C.; Shen, H.B. Plant-mPLoc: A Top-Down Strategy to Augment the Power for Predicting Plant Protein Subcellular Localization. PloS ONE 2010, 5, e11335. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.Y.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.P.; Li, J.P.; Paterson, A.H. MCScanX-transposed: Detecting transposed gene duplications based on multiple colinearity scans. Bioinformatics 2013, 29, 1458–1460. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Lynch, M.; Conery, J.S. The evolutionary fate and consequences of duplicate genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.P.; Guo, A.Y.; Zhang, H.; Luo, J.C.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Huis, R.; Hawkins, S.; Neutelings, G. Selection of reference genes for quantitative gene expression normalization in flax (Linum usitatissimum L.). BMC Plant Biol. 2010, 10, 71. [Google Scholar] [CrossRef]
- Antonov, J.; Goldstein, D.R.; Oberli, A.; Baltzer, A.; Pirotta, M.; Fleischmann, A.; Altermatt, H.J.; Jaggi, R. Reliable gene expression measurements from degraded RNA by quantitative real-time PCR depend on short amplicons and a proper normalization. Lab. Investig. 2005, 85, 1040–1050. [Google Scholar] [CrossRef]


| Seq1 | Seq2 | Duplication Event | Ka | Ks | Ka/Ks | Divergence Time (Mya) |
|---|---|---|---|---|---|---|
| LuDREB1 | LuDREB14 | SD | 0.2477 | 1.3797 | 0.1795 | 113.0911 |
| LuDREB2 | LuDREB20 | SD | 0.0325 | 0.3025 | 0.1074 | 24.7955 |
| LuDREB2 | LuDREB26 | SD | 0.2824 | 1.3108 | 0.2154 | 107.4433 |
| LuDREB3 | LuDREB21 | SD | 0.0104 | 0.0783 | 0.1322 | 6.4207 |
| LuDREB4 | LuDREB25 | SD | 0.0161 | 0.11 | 0.1468 | 9.0186 |
| LuDREB6 | LuDREB16 | SD | 0.1906 | 1.069 | 0.1783 | 87.6266 |
| LuDREB7 | LuDREB29 | SD | 0.0164 | 0.2569 | 0.064 | 21.0547 |
| LuDREB8 | LuDREB28 | SD | 0.037 | 0.1822 | 0.2029 | 14.9325 |
| LuDREB10 | LuDREB27 | SD | 0.0529 | 0.117 | 0.4525 | 9.5896 |
| LuDREB16 | LuDREB58 | SD | 0.0305 | 0.2713 | 0.1125 | 22.2348 |
| LuDREB20 | LuDREB26 | SD | 0.2629 | 2.2378 | 0.1175 | 183.4266 |
| LuDREB26 | LuDREB59 | SD | 0.0231 | 0.0747 | 0.3096 | 6.1192 |
| LuDREB30 | LuDREB13 | SD | 0.0714 | 0.2016 | 0.3543 | 16.5232 |
| LuDREB31 | LuDREB6 | SD | 0.0291 | 0.1533 | 0.1898 | 12.5668 |
| LuDREB31 | LuDREB16 | SD | 0.1998 | 1.0254 | 0.1949 | 84.0454 |
| LuDREB32 | LuDREB5 | SD | 0.0338 | 0.1559 | 0.2169 | 12.7806 |
| LuDREB33 | LuDREB12 | SD | 0.0297 | 0.1248 | 0.2384 | 10.2259 |
| LuDREB34 | LuDREB47 | SD | 0.0321 | 0.0803 | 0.4 | 6.586 |
| LuDREB35 | LuDREB48 | SD | 0.0628 | 0.2461 | 0.2554 | 20.1684 |
| LuDREB36 | LuDREB14 | SD | 0.0149 | 0.1446 | 0.103 | 11.8563 |
| LuDREB38 | LuDREB15 | SD | 0.0178 | 0.1158 | 0.1541 | 9.4944 |
| LuDREB39 | LuDREB24 | SD | 0.0341 | 0.1839 | 0.1857 | 15.0722 |
| LuDREB40 | LuDREB17 | SD | 0.055 | 0.204 | 0.2697 | 16.7172 |
| LuDREB41 | LuDREB18 | SD | 0.0504 | 0.1612 | 0.3129 | 13.2142 |
| LuDREB44 | LuDREB56 | SD | 0.0759 | 0.2569 | 0.2954 | 21.0547 |
| LuDREB45 | LuDREB55 | SD | 0.0911 | 0.3122 | 0.2918 | 25.5866 |
| LuDREB46 | LuDREB53 | SD | 0.3364 | 1.838 | 0.183 | 150.657 |
| LuDREB49 | LuDREB57 | SD | 0.0059 | 0.1234 | 0.0478 | 10.1141 |
| LuDREB51 | LuDREB11 | SD | 0.0593 | 0.1328 | 0.4467 | 10.8847 |
| LuDREB52 | LuDREB9 | SD | 0.0141 | 0.1202 | 0.117 | 9.8513 |
| LuDREB42 | LuDREB43 | TD | 0.2188 | 0.9539 | 0.2294 | 78.1916 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Qi, Y.; Wang, L.; Xu, C.; Li, W.; Dang, Z.; Zhao, W.; Wang, P.; Xie, Y.; Niu, Y.; et al. Genome-Wide Identification and Expression Profiling of Dehydration-Responsive Element-Binding Family Genes in Flax (Linum usitatissimum L.). Int. J. Mol. Sci. 2025, 26, 3074. https://doi.org/10.3390/ijms26073074
Wang Y, Qi Y, Wang L, Xu C, Li W, Dang Z, Zhao W, Wang P, Xie Y, Niu Y, et al. Genome-Wide Identification and Expression Profiling of Dehydration-Responsive Element-Binding Family Genes in Flax (Linum usitatissimum L.). International Journal of Molecular Sciences. 2025; 26(7):3074. https://doi.org/10.3390/ijms26073074
Chicago/Turabian StyleWang, Yan, Yanni Qi, Limin Wang, Chenmeng Xu, Wenjuan Li, Zhao Dang, Wei Zhao, Ping Wang, Yaping Xie, Yamin Niu, and et al. 2025. "Genome-Wide Identification and Expression Profiling of Dehydration-Responsive Element-Binding Family Genes in Flax (Linum usitatissimum L.)" International Journal of Molecular Sciences 26, no. 7: 3074. https://doi.org/10.3390/ijms26073074
APA StyleWang, Y., Qi, Y., Wang, L., Xu, C., Li, W., Dang, Z., Zhao, W., Wang, P., Xie, Y., Niu, Y., Lu, N., Hu, Z., Liu, Z., & Zhang, J. (2025). Genome-Wide Identification and Expression Profiling of Dehydration-Responsive Element-Binding Family Genes in Flax (Linum usitatissimum L.). International Journal of Molecular Sciences, 26(7), 3074. https://doi.org/10.3390/ijms26073074






