Berberine Derivative Compound 13 as a Potent Promoter of Osteoblast Differentiation via Akt and PKC Signaling Pathways
Abstract
1. Introduction
2. Results
2.1. Structure–Activity Relationship of Berberine Derivatives Evaluated Through ALP Staining Assay
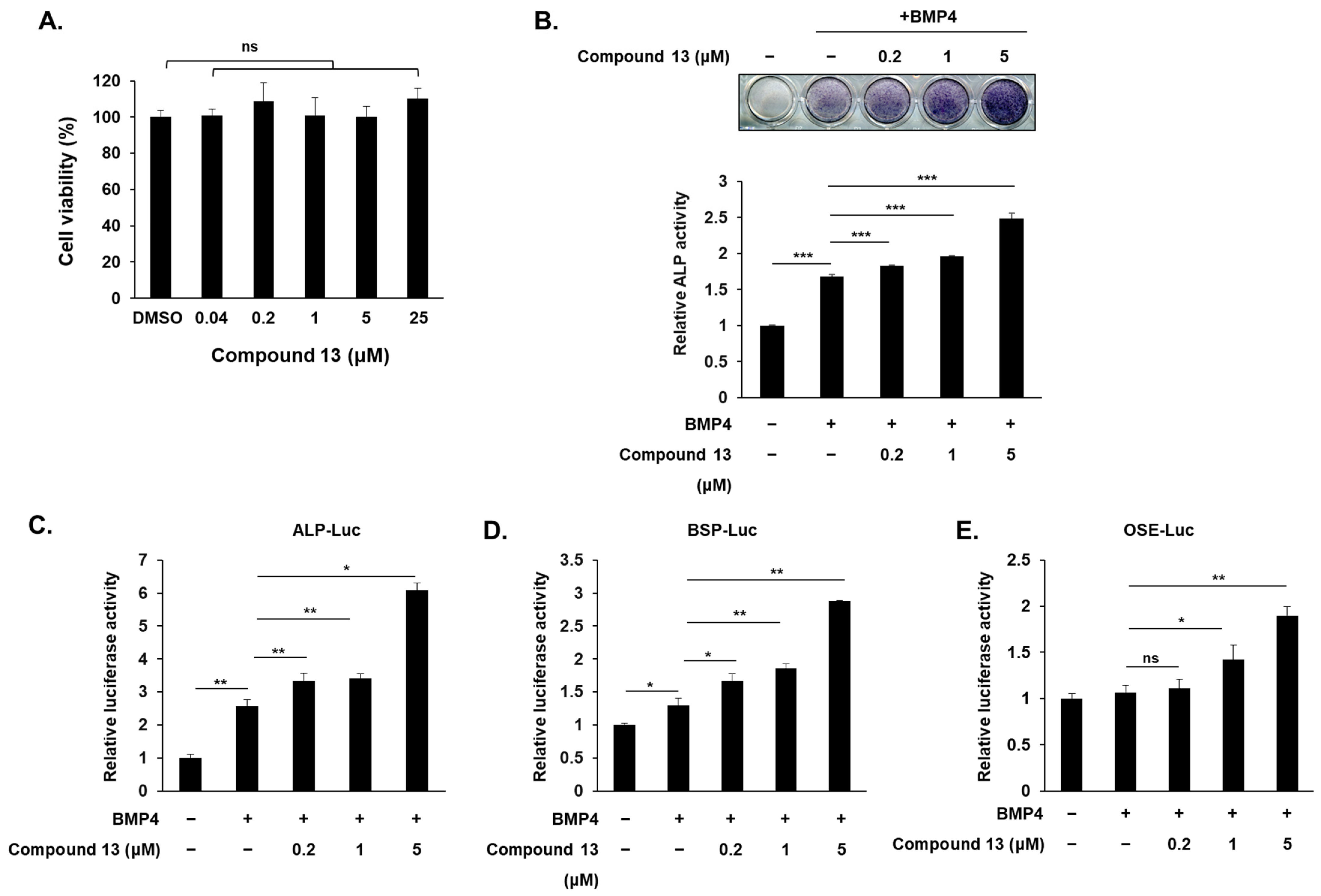
2.2. Compound Promotes Osteoblast Differentiation in C2C12 Cell
2.3. Compound Increases Osteogenic Gene Expression at mRNA and Protein Levels
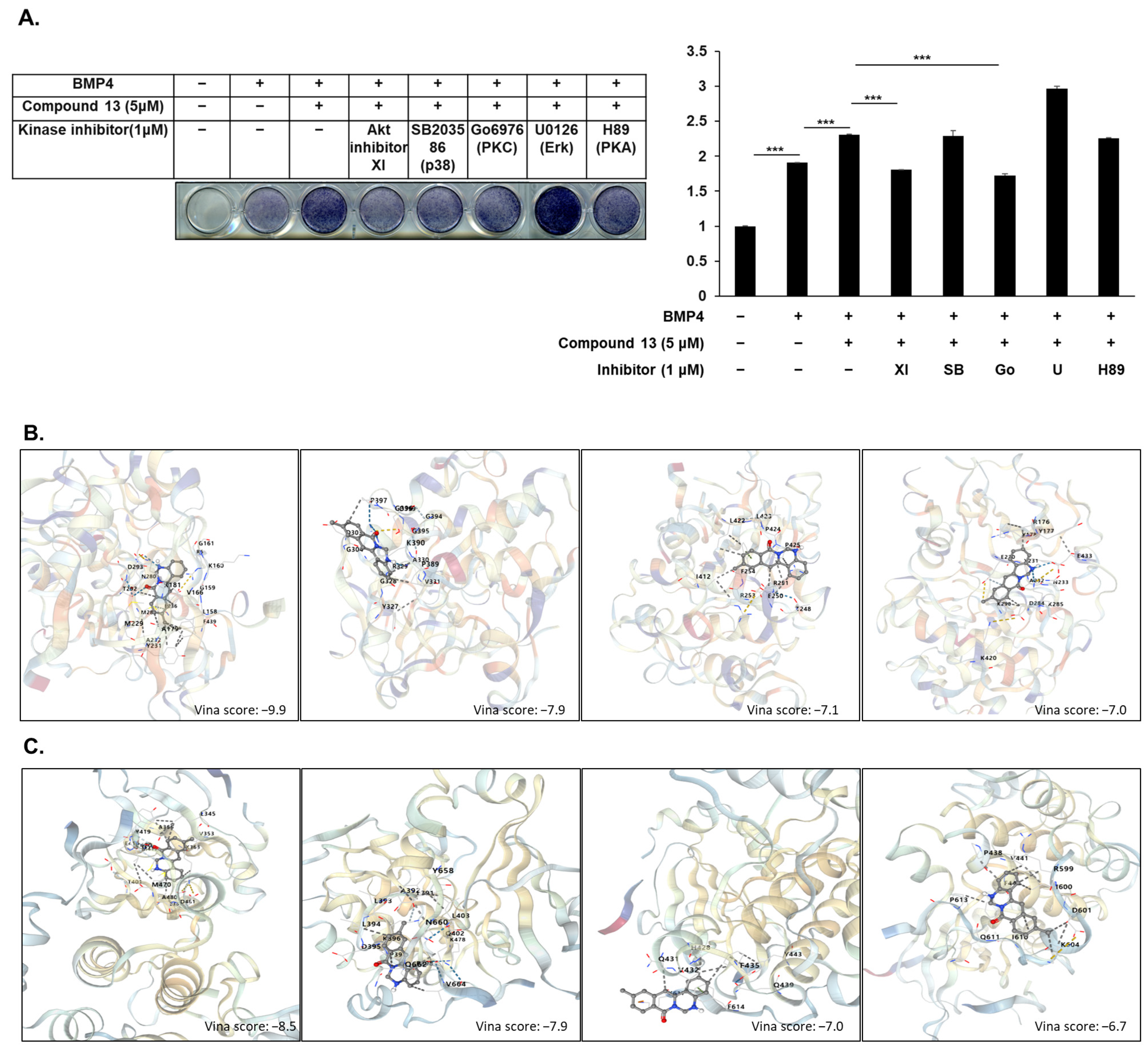
2.4. Akt and PKC Signaling Involved in Compound -Regulated Osteoblast Differentiation
3. Discussion
4. Materials and Methods
4.1. Reagent and Antibody
4.2. Cell Culture and Differentiation
4.3. MTT Assay
4.4. Alkaline Phosphatase (ALP) Staining
4.5. Luciferase Assay and Transfection
4.6. Reverse Transcription Followed by Quantitative Polymerase Chain Reaction (RT-qPCR)
4.7. Immunoblotting
4.8. Molecular Docking
4.9. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Danielson, L.; Zamulko, A. Osteoporosis: A Review. South Dak. Med. 2015, 68, 503-5–507-9. [Google Scholar]
- Rachner, T.D.; Khosla, S.; Hofbauer, L.C. Osteoporosis: Now and the future. Lancet 2011, 377, 1276–1287. [Google Scholar] [CrossRef] [PubMed]
- Manolagas, S.C.; Jilka, R.L.; Epstein, F.H. Bone Marrow, Cytokines, and Bone Remodeling—Emerging Insights into the Pathophysiology of Osteoporosis. N. Engl. J. Med. 1995, 332, 305–311. [Google Scholar] [CrossRef] [PubMed]
- Riggs, B.L.; Melton, L.J. The prevention and treatment of osteoporosis. N. Engl. J. Med. 1992, 327, 620–627. [Google Scholar]
- Caetano-Lopes, J.; Canhão, H.; Fonseca, J.E. Osteoblasts and bone formation. Acta Reumatol. Port. 2007, 32, 103–110. [Google Scholar]
- Siddhanti, S.R.; Quarles, L.D. Molecular to pharmacologic control of osteoblast proliferation and differentiation. J. Cell. Biochem. 1994, 55, 310–320. [Google Scholar] [CrossRef]
- Lin, G.L.; Hankenson, K.D. Integration of BMP, Wnt, and notch signaling pathways in osteoblast differentiation. J. Cell. Biochem. 2011, 112, 3491–3501. [Google Scholar] [CrossRef]
- Huang, W.; Yang, S.; Shao, J.; Li, Y.P. Signaling and transcriptional regulation in osteoblast commitment and differentiation. Front. Biosci. 2007, 12, 3068–3092. [Google Scholar] [CrossRef]
- Blair, H.C.; Zaidi, M.; Huang, C.L.; Sun, L. The developmental basis of skeletal cell differentiation and the molecular basis of major skeletal defects. Biol. Rev. Camb. Philos. Soc. 2008, 83, 401–415. [Google Scholar] [CrossRef]
- Karner, C.M.; Lee, S.-Y.; Long, F. Bmp Induces Osteoblast Differentiation through both Smad4 and mTORC1 Signaling. Mol. Cell. Biol. 2017, 37, e00253-16. [Google Scholar] [CrossRef]
- Komori, T. Regulation of osteoblast differentiation by transcription factors. J. Cell. Biochem. 2006, 99, 1233–1239. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, L.M.G.; Lombardi, P.; Tillhon, M.; Scovassi, A.I. Berberine, an epiphany against cancer. Molecules 2014, 19, 12349–12367. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, Y.; Du, X.; Ma, H.; Yao, J. The Anti-Cancer Mechanisms of Berberine: A Review. Cancer Manag. Res. 2020, 12, 695–702. [Google Scholar] [CrossRef]
- Kuo, C.-L.; Chi, C.-W.; Liu, T.-Y. The anti-inflammatory potential of berberine in vitro and in vivo. Cancer Lett. 2003, 203, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.W.; Suh, J.H.; Kim, H.-N.; Kim, A.Y.; Park, S.Y.; Shin, C.S.; Choi, J.-Y.; Kim, J.B. Berberine promotes osteoblast differentiation by Runx2 activation with p38 MAPK. J. Bone Miner. Res. 2008, 23, 1227–1237. [Google Scholar] [CrossRef]
- Tao, K.; Xiao, D.; Weng, J.; Xiong, A.; Kang, B.; Zeng, H. Berberine promotes bone marrow-derived mesenchymal stem cells osteogenic differentiation via canonical Wnt/β-catenin signaling pathway. Toxicol. Lett. 2016, 240, 68–80. [Google Scholar] [CrossRef]
- Han, Y.; Kim, M.J.; Lee, K.Y. Berberine derivative, Q8, stimulates osteogenic differentiation. Biochem. Biophys. Res. Commun. 2018, 504, 340–345. [Google Scholar]
- Peng, Y.; Kang, Q.; Cheng, H.; Li, X.; Sun, M.H.; Jiang, W.; Luu, H.H.; Park, J.Y.; Haydon, R.C.; He, T. Transcriptional characterization of bone morphogenetic proteins (BMPs)-mediated osteogenic signaling. J. Cell. Biochem. 2003, 90, 1149–1165. [Google Scholar] [CrossRef]
- Sun, Y.; Xun, K.; Wang, Y.; Chen, X. A systematic review of the anticancer properties of berberine, a natural product from Chinese herbs. Anti-Cancer Drugs 2009, 20, 757–769. [Google Scholar] [CrossRef]
- Zhou, J.; Zhou, S.; Tang, J.; Zhang, K.; Guang, L.; Huang, Y.; Xu, Y.; Ying, Y.; Zhang, L.; Li, D. Protective effect of berberine on beta cells in streptozotocin- and high-carbohydrate/high-fat diet-induced diabetic rats. Eur. J. Pharmacol. 2009, 606, 262–268. [Google Scholar] [CrossRef]
- Kong, W.; Wei, J.; Abidi, P.; Lin, M.; Inaba, S.; Li, C.; Wang, Y.; Wang, Z.; Si, S.; Pan, H.; et al. Berberine is a novel cholesterol-lowering drug working through a unique mechanism distinct from statins. Nat. Med. 2004, 10, 1344–1351. [Google Scholar] [CrossRef]
- Zou, K.; Li, Z.; Zhang, Y.; Zhang, H.-Y.; Li, B.; Zhu, W.-L.; Shi, J.-Y.; Jia, Q.; Li, Y.-M. Advances in the study of berberine and its derivatives: A focus on anti-inflammatory and anti-tumor effects in the digestive system. Acta Pharmacol. Sin. 2016, 38, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Jin, Y.; Lee, S.H.; Khadka, D.B.; Cho, W.-J.; Lee, K.Y. Berberine bioisostere Q8 compound stimulates osteoblast differentiation and function in vitro. Pharmacol. Res. 2017, 119, 463–475. [Google Scholar] [CrossRef]
- Kim, H.-J.; Kim, J.-H.; Bae, S.-C.; Choi, J.-Y.; Ryoo, H.-M. The protein kinase C pathway plays a central role in the fibroblast growth factor-stimulated expression and transactivation activity of runx2. J. Biol. Chem. 2003, 278, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Miraoui, H.; Miraoui, H.; Oudina, K.; Petite, H.; Tanimoto, Y.; Moriyama, K.; Marie, P.J. Fibroblast growth factor receptor 2 promotes osteogenic differentiation in mesenchymal cells via ERK1/2 and protein kinase C signaling. J. Biol. Chem. 2009, 284, 4897–4904. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.; Rotwein, P. Akt promotes BMP2-mediated osteoblast differentiation and bone development. J. Cell Sci. 2009, 122 Pt 5, 716–726. [Google Scholar] [CrossRef]
- Choi, Y.H.; Kim, Y.; Jeong, H.M.; Jin, Y.; Yeo, C.; Lee, K.Y. Akt enhances Runx2 protein stability by regulating Smurf2 function during osteoblast differentiation. FEBS J. 2014, 281, 3656–3666. [Google Scholar] [CrossRef]
- Choi, Y.H.; Jeong, H.M.; Jin, Y.-H.; Li, H.; Yeo, C.-Y.; Lee, K.-Y. Akt phosphorylates and regulates the osteogenic activity of Osterix. Biochem. Biophys. Res. Commun. 2011, 411, 637–641. [Google Scholar] [CrossRef]
- Jin, Y.; Han, Y.; Khadka, D.B.; Zhao, C.; Lee, K.Y.; Cho, W.-J. Discovery of Isoquinolinoquinazolinones as a Novel Class of Potent PPARγ Antagonists with Anti-adipogenic Effects. Sci. Rep. 2016, 6, 34661. [Google Scholar] [CrossRef]
- Katagiri, T.; Yamaguchi, A.; Komaki, M.; Abe, E.; Takahashi, N.; Ikeda, T.; Rosen, V.; Wozney, J.M.; Fujisawa-Sehara, A.; Suda, T. Bone morphogenetic protein-2 converts the differentiation pathway of C2C12 myoblasts into the osteoblast lineage. J. Cell Biol. 1994, 127 Pt 1, 1755–1766, Erratum in J. Cell Biol. 1995, 128, 713. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Sabokbar, A.; Millett, P.J.; Myer, B.; Rushton, N. A rapid, quantitative assay for measuring alkaline phosphatase activity in osteoblastic cells in vitro. Bone Miner. 1994, 27, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Ducy, P.; Karsenty, G. Two distinct osteoblast-specific cis-acting elements control expression of a mouse osteocalcin gene. Mol. Cell. Biol. 1995, 15, 1858–1869. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [PubMed]
- Towbin, H.; Staehelin, T.; Gordon, J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc. Natl. Acad. Sci. USA 1979, 76, 4350–4354. [Google Scholar] [CrossRef]
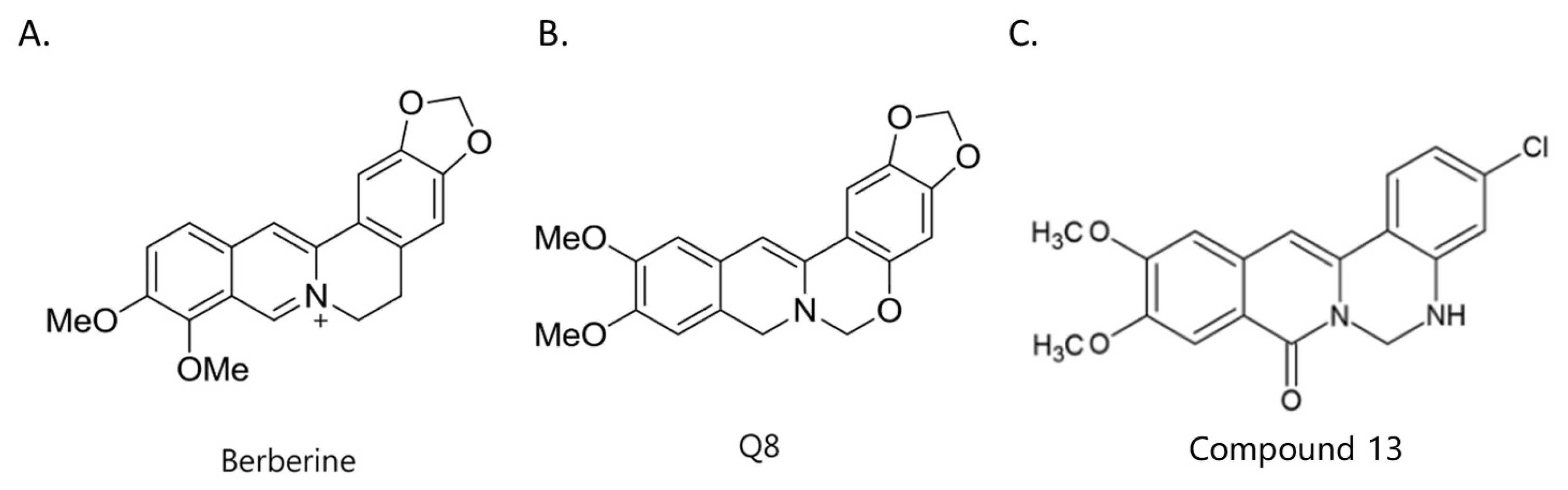
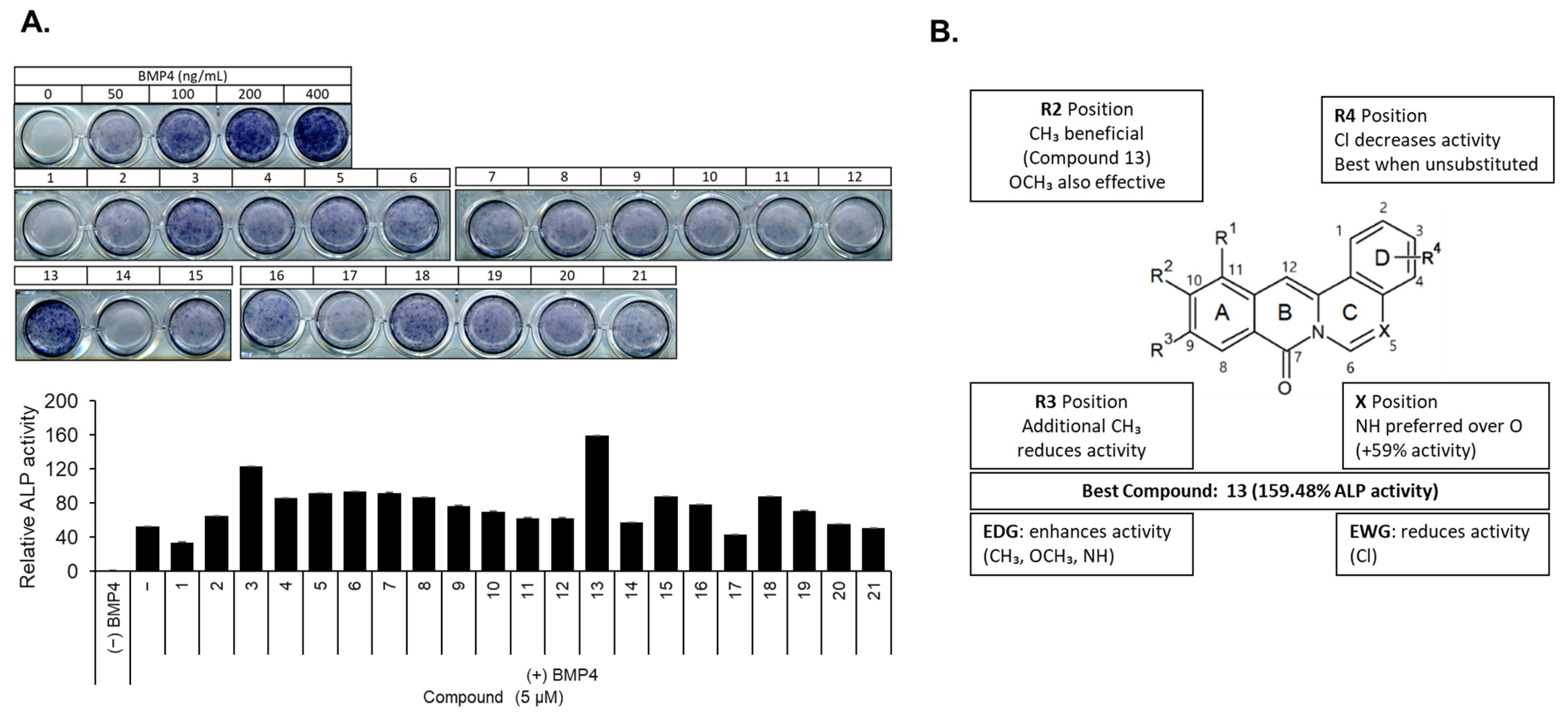

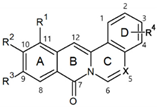 | |||||||
|---|---|---|---|---|---|---|---|
| No. | Structure | R1 | R2 | R3 | R4 | X | ALP Staining (%) |
| 1 | 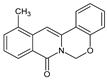 | CH3- | - | - | - | O | 33.80 |
| 2 | 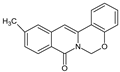 | - | CH3 | - | - | O | 64.91 |
| 3 | 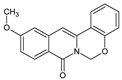 | - | OCH3 | - | - | O | 123.23 |
| 4 | 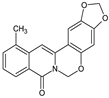 | CH3 | - | - | OCH2O | O | 85.83 |
| 5 | 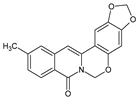 | - | CH3 | - | OCH2O | O | 91.39 |
| 6 | 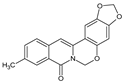 | - | - | CH3 | OCH2O | O | 93.70 |
| 7 | 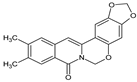 | - | CH3 | CH3 | OCH2O | O | 92.08 |
| 8 | 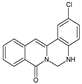 | - | - | - | Cl | NH | 86.66 |
| 9 | 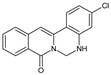 | - | - | - | Cl | NH | 76.60 |
| 10 | 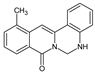 | CH3 | - | - | NH | 69.93 | |
| 11 | 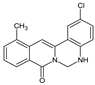 | CH3 | - | - | Cl | NH | 62.58 |
| 12 | 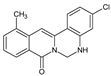 | CH3 | - | - | Cl | NH | 62.31 |
| 13 |  | - | CH3 | - | - | NH | 159.48 |
| 14 |  | - | CH3 | - | Cl | NH | 57.25 |
| 15 |  | - | CH3 | - | Cl | NH | 87.55 |
| 16 | 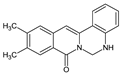 | - | CH3 | CH3 | - | NH | 78.40 |
| 17 | 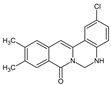 | - | CH3 | CH3 | Cl | NH | 42.86 |
| 18 |  | - | CH3 | CH3 | Cl | NH | 87.96 |
| 19 | 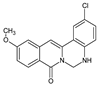 | - | OCH3 | - | Cl | NH | 71.12 |
| 20 | 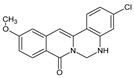 | - | OCH3 | - | Cl | NH | 55.44 |
| 21 |  | - | OCH3 | OCH3 | Cl | NH | 50.74 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piao, M.; Han, Y.H.; Lee, K.Y. Berberine Derivative Compound 13 as a Potent Promoter of Osteoblast Differentiation via Akt and PKC Signaling Pathways. Int. J. Mol. Sci. 2025, 26, 2984. https://doi.org/10.3390/ijms26072984
Piao M, Han YH, Lee KY. Berberine Derivative Compound 13 as a Potent Promoter of Osteoblast Differentiation via Akt and PKC Signaling Pathways. International Journal of Molecular Sciences. 2025; 26(7):2984. https://doi.org/10.3390/ijms26072984
Chicago/Turabian StylePiao, Meiyu, Youn Ho Han, and Kwang Youl Lee. 2025. "Berberine Derivative Compound 13 as a Potent Promoter of Osteoblast Differentiation via Akt and PKC Signaling Pathways" International Journal of Molecular Sciences 26, no. 7: 2984. https://doi.org/10.3390/ijms26072984
APA StylePiao, M., Han, Y. H., & Lee, K. Y. (2025). Berberine Derivative Compound 13 as a Potent Promoter of Osteoblast Differentiation via Akt and PKC Signaling Pathways. International Journal of Molecular Sciences, 26(7), 2984. https://doi.org/10.3390/ijms26072984







