Subgenomic RNA and Limited Cross-Reactive Neutralising Antibodies Point to Potential Improvements in SARS-CoV-2 Clinical Handling
Abstract
1. Introduction
2. Results
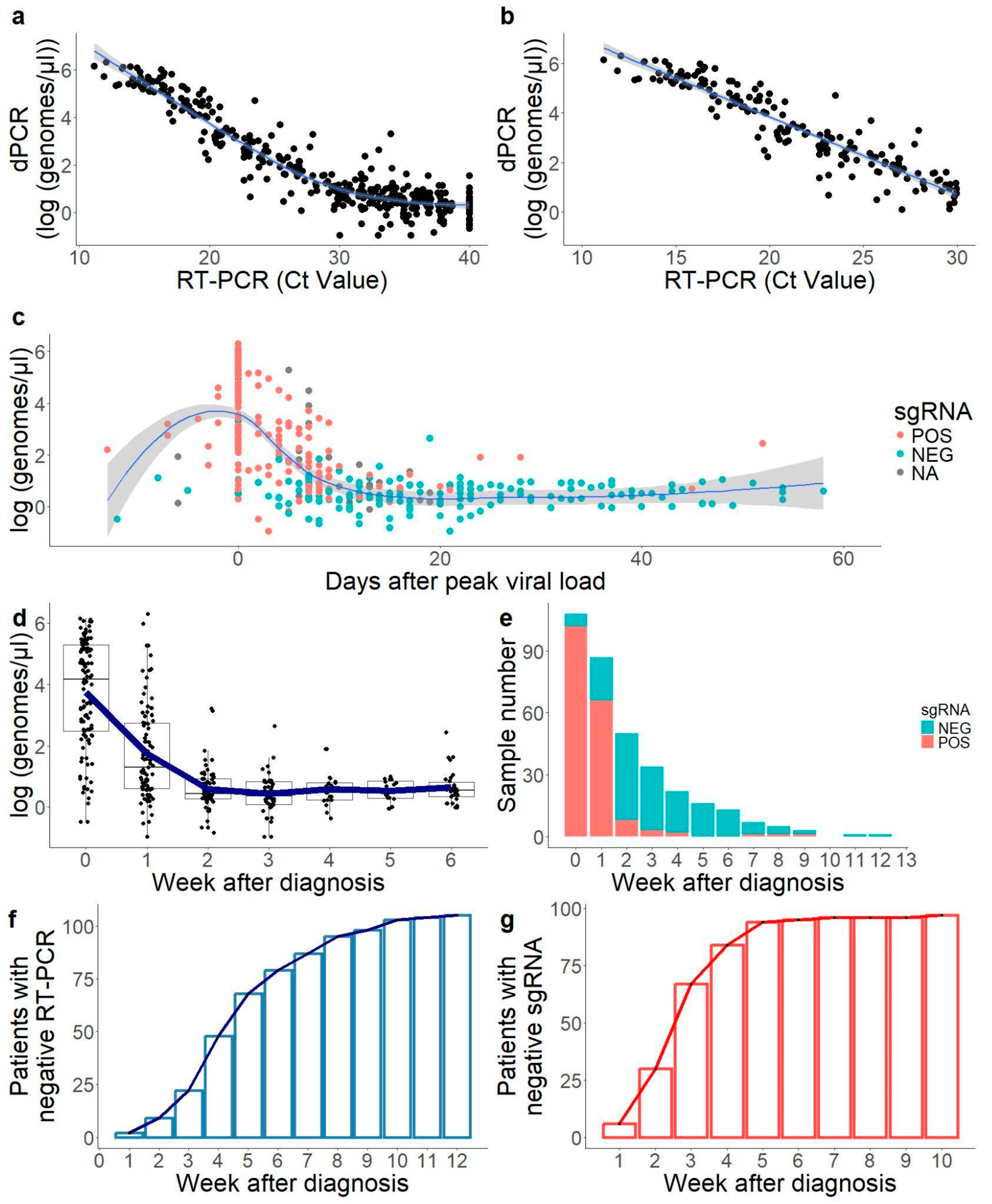
2.1. Correlation Between RT-PCR Results and dPCR Was Solid Below Cycle Threshold (Ct) 30
2.2. Convalescent Period with Low Viral Load Only Detected by Genomic RNA RT-PCR
2.3. sgRNA Was Only Detectable in the First Weeks of Infection in Most Samples
2.4. Age Had a Moderate Impact in Peak Viral Load
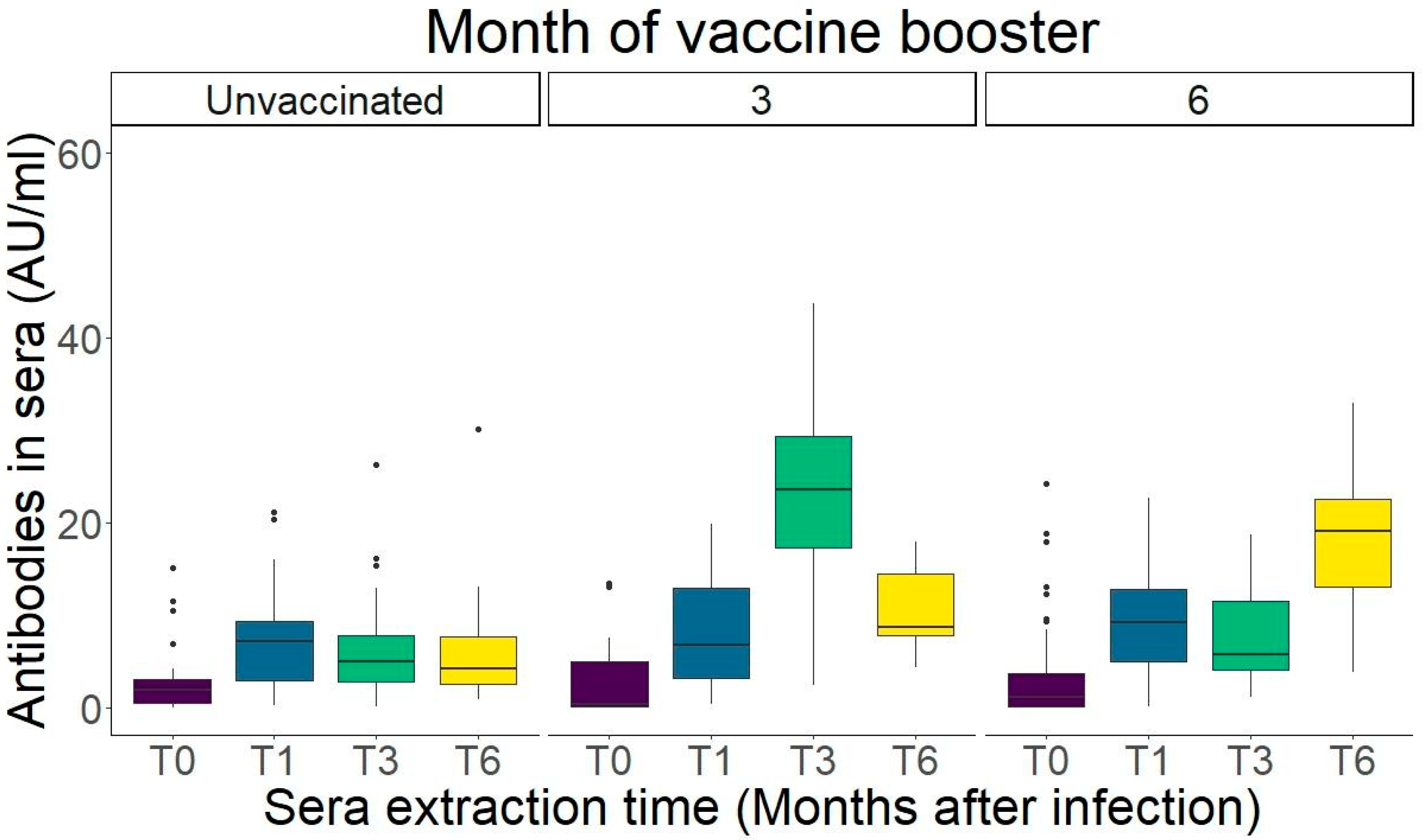
2.5. IgG Levels Peak One Month After Infection
2.6. Stronger Neutralisation Capacity Against the Variant of Infection
2.7. Live Virus Neutralisation Assay Showed a Lower Omicron Neutralisation Capacity for Sera Generated Against Pre-Omicron Variants of Concern (VOCs)
3. Discussion
4. Materials and Methods
4.1. Patient Selection and Sample Collection
4.2. Inclusion Criteria
4.3. Viral RNA Quantification
4.3.1. RT-PCR
4.3.2. Digital PCR (dPCR)
4.4. SARS-CoV-2 Next-Generation Sequencing (NGS)
4.5. Measurement of Antibodies in Sera
4.6. Pseudovirus Neutralisation Assay
4.7. Live Virus Neutralisation Assay
4.8. Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tsang, N.N.Y.; So, H.C.; Ng, K.Y.; Cowling, B.J.; Leung, G.M.; Ip, D.K.M. Diagnostic performance of different sampling approaches for SARS-CoV-2 RT-PCR testing: A systematic review and meta-analysis. Lancet Infect. Dis. 2021, 21, 1233–1245. [Google Scholar] [CrossRef] [PubMed]
- Richard-Greenblatt, M.; Ziegler, M.J.; Bromberg, V.; Huang, E.; Abdallah, H.; Tolomeo, P.; Lautenbach, E.; Glaser, L.; Kell, B.J. Quantifying the Impact of Nasopharyngeal Specimen Quality on Severe Acute Respiratory Syndrome Coronavirus 2 Test Performance. Open Forum Infect. Dis. 2021, 8, ofab235. [Google Scholar] [CrossRef]
- Puhach, O.; Meyer, B.; Eckerle, I. SARS-CoV-2 viral load and shedding kinetics. Nat. Rev. Microbiol. 2023, 21, 147–161. [Google Scholar] [CrossRef]
- Cevik, M.; Tate, M.; Lloyd, O.; Maraolo, A.E.; Schafers, J.; Ho, A. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: A systematic review and meta-analysis. Lancet Microbe 2021, 2, e13–e22. [Google Scholar] [CrossRef]
- Mishra, B.; Ranjan, J.; Purushotham, P.; Saha, S.; Payal, P.; Kar, P.; Das, S.; Deshmukh, V. High proportion of low cyclethreshold value as an early indicator of COVID-19 surge. J. Med. Virol. 2022, 94, 240–245. [Google Scholar] [CrossRef] [PubMed]
- Jefferson, T.; Spencer, E.A.; Brassey, J.; Heneghan, C. Viral Cultures for Coronavirus Disease 2019 Infectivity Assessment: A Systematic Review. Clin. Infect. Dis. 2021, 73, e3884–e3899. [Google Scholar] [CrossRef]
- Wölfel, R.; Corman, V.M.; Guggemos, W.; Seilmaier, M.; Zange, S.; Müller, M.A.; Niemeyer, D.; Jones, T.C.; Vollmar, P.; Rothe, C.; et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020, 581, 465–469. [Google Scholar] [CrossRef] [PubMed]
- van Kampen, J.J.A.; van de Vijver, D.A.M.C.; Fraaij, P.L.A.; Haagmans, B.L.; Lamers, M.M.; Okba, N.; van der Akker, J.P.C.; Endeman, H.; Gommers, D.A.M.P.J.; Cornelissen, J.J.; et al. Duration and key determinants of infectious virus shedding in hospitalized patients with coronavirus disease-2019 (COVID-19). Nat. Commun. 2021, 12, 267. [Google Scholar] [CrossRef]
- Owusu, D.; Pomeroy, M.A.; Lewis, N.M.; Wadhwa, A.; Yousaf, A.R.; Whitaker, B.; Dietrich, E.; Hall, A.J.; Chu, V.; Thornburg, N.; et al. Persistent SARS-CoV-2 RNA Shedding Without Evidence of Infectiousness: A Cohort Study of Individuals With COVID-19. J. Infect. Dis. 2021, 224, 1362–1371. [Google Scholar] [CrossRef]
- Zheng, S.; Fan, J.; Yu, F.; Feng, B.; Lou, B.; Zou, Q.; Xie, G.; Lin, S.; Wang, R.; Yang, X.; et al. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: Retrospective cohort study. BMJ 2020, 369, m1443. [Google Scholar] [CrossRef]
- van Kasteren, P.B.; van der Veer, B.; van den Brink, S.; Wijsman, L.; de Jonge, J.; van den Brandt, A.; Molenkamp, R.; Reusken, C.B.E.M.; Meijer, A. Comparison of seven commercial RT-PCR diagnostic kits for COVID-19. J. Clin. Virol. 2020, 128, 104412. [Google Scholar] [CrossRef]
- Long, S. SARS-CoV-2 Subgenomic RNAs: Characterization, Utility, and Perspectives. Viruses 2021, 13, 1923. [Google Scholar] [CrossRef] [PubMed]
- Suo, T.; Liu, X.; Feng, J.; Guo, M.; Hu, W.; Guo, D.; Ullah, H.; Yang, Y.; Zhang, Q.; Wang, X.; et al. ddPCR: A more accurate tool for SARS-CoV-2 detection in low viral load specimens. Emerg. Microbes Infect. 2020, 9, 1259–1268. [Google Scholar] [CrossRef] [PubMed]
- Vasudevan, H.N.; Xu, P.; Servellita, V.; Miller, S.; Liu, L.; Gopez, A.; Chiu, C.Y.; Abate, A.R. Digital droplet PCR accurately quantifies SARS-CoV-2 viral load from crude lysate without nucleic acid purification. Sci. Rep. 2021, 11, 780. [Google Scholar] [CrossRef] [PubMed]
- Addetia, A.; Crawford, K.H.D.; Dingens, A.; Zhu, H.; Roychoudhury, P.; Huang, M.-L.; Jerome, K.; Bloom, J.; Greninger, A.L. Neutralizing Antibodies Correlate with Protection from SARS-CoV-2 in Humans during a Fishery Vessel Outbreak with a High Attack Rate. J. Clin. Microbiol. 2020, 58, e02107–e02120. [Google Scholar] [CrossRef]
- Houlihan, C.F.; Beale, R. The complexities of SARS-CoV-2 serology. Lancet Infect. Dis. 2020, 20, 1350–1351. [Google Scholar] [CrossRef]
- Weidner, L.; Gänsdorfer, S.; Unterweger, S.; Weseslindtner, L.; Drexler, C.; Farcet, M.; Witt, V.; Schistal, E.; Schlenke, P.; Kreil, T.R. Quantification of SARS-CoV-2 antibodies with eight commercially available immunoassays. J. Clin. Virol. 2020, 129, 104540. [Google Scholar] [CrossRef]
- Khoury, D.S.; Schlub, T.E.; Cromer, D.; Steain, M.; Fong, Y.; Gilbert, P.B.; Subbarao, K.; Triccas, J.A.; Kent, S.J.; Davenport, M.P. Correlates of Protection, Thresholds of Protection, and Immunobridging among Persons with SARS-CoV-2 Infection. Emerg. Infect. Dis. 2023, 29, 381–388. [Google Scholar] [CrossRef]
- Chi, W.-Y.; Li, Y.-D.; Huang, H.-C.; Chan, T.E.H.; Chow, S.-Y.; Su, J.-H.; Ferral, L.; Hung, C.; Wu, T.C. COVID-19 vaccine update: Vaccine effectiveness, SARS-CoV-2 variants, boosters, adverse effects, and immune correlates of protection. J. Biomed. Sci. 2022, 29, 82. [Google Scholar] [CrossRef]
- Cabrera-Alvargonzalez, J.-J.; Davina-Nunez, C.; Rey-Cao, S.; Rodriguez Calviño, L.; Silva-Bea, S.; Gonzalez-Alonso, E.; Carballo-Fernandez, R.; Lameiro Vilariño, C.; Cortizo-Vidal, S.; Valiño-Prieto, S.; et al. Comparative analysis of eleven SARS-CoV-2 immunoassays and neutralisation data: Time to enhance standardisation and correlation of protection. Infect. Dis. 2024, 56, 1067–1079. [Google Scholar] [CrossRef]
- Lukaszuk, K.; Kiewisz, J.; Rozanska, K.; Dabrowska, M.; Podolak, A.; Jakiel, G.; Woclawek-Potocka, I.; Lukaszuk, A.; Rabalski, L. Usefulness of IVD Kits for the Assessment of SARS-CoV-2 Antibodies to Evaluate the Humoral Response to Vaccination. Vaccines 2021, 9, 840. [Google Scholar] [CrossRef] [PubMed]
- Dépéry, L.; Bally, I.; Amen, A.; Némoz, B.; Buisson, M.; Grossi, L.; Truffot, A.; Germi, R.; Guilligay, D.; Veloso, M.; et al. Anti-SARS-CoV-2 serology based on ancestral RBD antigens does not correlate with the presence of neutralizing antibodies against Omicron variants. Microbiol. Spectr. 2024, 13, e01568-e24. [Google Scholar] [CrossRef]
- Marking, U.; Bladh, O.; Aguilera, K.; Yang, Y.; Greilert Norin, N.; Blom, K.; Hober, S.; Klingström, J.; Haverhall, S.; Aberg, M.; et al. Humoral immune responses to the monovalent XBB.1.5-adapted BNT162b2 mRNA booster in Sweden. Lancet Infect. Dis. 2024, 24, e80–e81. [Google Scholar] [CrossRef] [PubMed]
- FDA. Updated COVID-19 Vaccines for Use in the United States Beginning in Fall 2024. FDA 2024. Available online: https://www.fda.gov/vaccines-blood-biologics/updated-covid-19-vaccines-use-united-states-beginning-fall-2024 (accessed on 19 March 2025).
- WHO. Statement on the Antigen Composition of COVID-19 Vaccines 2024. Available online: https://www.who.int/news/item/26-04-2024-statement-on-the-antigen-composition-of-covid-19-vaccines (accessed on 13 December 2024).
- Mlcochova, P.; Kemp, S.A.; Dhar, M.S.; Papa, G.; Meng, B.; Ferreira, I.A.T.M.; Datir, R.; Collier, D.A.; Albecka, A.; Singh, S.; et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature 2021, 599, 114–119. [Google Scholar] [CrossRef] [PubMed]
- Singanallur, N.B.; van Vuren, P.J.; McAuley, A.J.; Bruce, M.P.; Kuiper, M.J.; Gwini, S.M.; Ridell, S.; Goldie, S.; Drew, T.W.; Blasdell, K.R.; et al. At Least Three Doses of Leading Vaccines Essential for Neutralisation of SARS-CoV-2 Omicron Variant. Front. Immunol. 2022, 13, 883612. [Google Scholar] [CrossRef]
- Collier, A.-R.Y.; Miller, J.; Hachmann, N.P.; McMahan, K.; Liu, J.; Bondzie, E.A.; Gallup, L.; Rowe, M.; Schonberg, E.; Thai, S.; et al. Immunogenicity of BA.5 Bivalent mRNA Vaccine Boosters. N. Engl. J. Med. 2023, 388, 565–567. [Google Scholar] [CrossRef]
- Scutari, R.; Renica, S.; Cento, V.; Nava, A.; Sammartino, J.C.; Ferrari, A.; Pani, A.; Merli, M.; Fanti, D.; Vismara, C.; et al. Quantitative SARS-CoV-2 subgenomic RNA as a surrogate marker for viral infectivity: Comparison between culture isolation and direct sgRNA quantification. PLoS ONE 2023, 18, e0291120. [Google Scholar] [CrossRef]
- Kissler, S.M.; Fauver, J.R.; Mack, C.; Olesen, S.W.; Tai, C.; Shiue, K.Y.; Kalinich, C.C.; Jednak, S.; Ott, I.M.; Vogels, C.B.F.; et al. Viral dynamics of acute SARS-CoV-2 infection and applications to diagnostic and public health strategies. PLoS Biol. 2021, 19, e3001333. [Google Scholar] [CrossRef]
- Romero-Alvarez, D.; Garzon-Chavez, D.; Espinosa, F.; Ligña, E.; Teran, E.; Mora, F.; Espin, E.; Alban, C.; Galarza, J.M.; Reyes, J. Cycle Threshold Values in the Context of Multiple RT-PCR Testing for SARS-CoV-2. Risk Manag. Healthc. Policy 2021, 14, 1311–1317. [Google Scholar] [CrossRef]
- Hindson, C.M.; Chevillet, J.R.; Briggs, H.A.; Gallichotte, E.N.; Ruf, I.K.; Hindson, B.J.; Vesella, R.L.; Tewari, M. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat. Methods 2013, 10, 1003–1005. [Google Scholar] [CrossRef]
- Ahmed, W.; Simpson, S.L.; Bertsch, P.M.; Bibby, K.; Bivins, A.; Blackall, L.L.; Bofill-Mas, S.; Bosch, A.; Brandao, J.; Choi, P.M.; et al. Minimizing errors in RT-PCR detection and quantification of SARS-CoV-2 RNA for wastewater surveillance. Sci. Total Environ. 2022, 805, 149877. [Google Scholar] [CrossRef] [PubMed]
- de la Cruz Barron, M.; Kneis, D.; Geissler, M.; Dumke, R.; Dalpke, A.; Berendonk, T.U. Evaluating the sensitivity of droplet digital PCR for the quantification of SARS-CoV-2 in wastewater. Front. Public Health 2023, 11, 1271594. [Google Scholar] [CrossRef]
- Yang, Y.; Feng, X.; Pan, Y.; Wang, X.; Peng, T.; Niu, C.; Wang, Q.; Zou, Q.; Dong, L.; Dai, X.; et al. A culture-free method for rapidly and accurately quantifying active SARS-CoV-2. Anal. Bioanal. Chem. 2023, 415, 5745–5753. [Google Scholar] [CrossRef]
- dMIQE Group; Huggett, J.F. The Digital MIQE Guidelines Update: Minimum Information for Publication of Quantitative Digital PCR Experiments for 2020. Clin. Chem. 2020, 66, 1012–1029. [Google Scholar] [CrossRef]
- Mitchell, S.L.; Loeffelholz, M.J. Considerations regarding Interpretation of Positive SARS-CoV-2 Molecular Results with Late Cycle Threshold Values. J. Clin. Microbiol. 2022, 60, e00501–e00522. [Google Scholar] [CrossRef]
- La Scola, B.; Le Bideau, M.; Andreani, J.; Hoang, V.T.; Grimaldier, C.; Colson, P.; Gautret, P.; Raoult, D. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 1059–1061. [Google Scholar] [CrossRef]
- Bruce, E.A.; Mills, M.G.; Sampoleo, R.; Perchetti, G.A.; Huang, M.; Despres, H.W.; Schmidt, M.M.; Roychoudhury, P.; Shirley, D.J.; Jerome, K.R.; et al. Predicting infectivity: Comparing four PCR-based assays to detect culturable SARS-CoV-2 in clinical samples. EMBO Mol. Med. 2022, 14, e15290. [Google Scholar] [CrossRef]
- Santos Bravo, M.; Berengua, C.; Marín, P.; Esteban, M.; Rodriguez, C.; del Cuerpo, M.; Miro, E.; Cuesta, G.; Mosquera, M.; Sanchez-Palomino, S.; et al. Viral Culture Confirmed SARS-CoV-2 Subgenomic RNA Value as a Good Surrogate Marker of Infectivity. J. Clin. Microbiol. 2022, 60, e01609–e01621. [Google Scholar] [CrossRef] [PubMed]
- Alexandersen, S.; Chamings, A.; Bhatta, T.R. SARS-CoV-2 genomic and subgenomic RNAs in diagnostic samples are not an indicator of active replication. Nat. Commun. 2020, 11, 6059. [Google Scholar] [CrossRef]
- Verma, R.; Kim, E.; Martínez-Colón, G.J.; Jagannathan, P.; Rustagi, A.; Parsonnet, J.; Bonilla, H.; Khosla, C.; Holubar, M.; Subramanian, A.; et al. SARS-CoV-2 Subgenomic RNA Kinetics in Longitudinal Clinical Samples. Open Forum Infect. Dis. 2021, 8, ofab310. [Google Scholar] [CrossRef]
- Jones, T.C.; Biele, G.; Mühlemann, B.; Veith, T.; Schneider, J.; Beheim-Schwarzbach, J.; Bleicker, T.; Tesch, J.; Schmidt, M.L.; Sander, L.E.; et al. Estimating infectiousness throughout SARS-CoV-2 infection course. Science 2021, 373, eabi5273. [Google Scholar] [CrossRef] [PubMed]
- Mazzoni, A.; Lauria, N.D.; Maggi, L.; Salvati, L.; Vanni, A.; Capone, M.; Lamacchia, G.; Mantengoli, E.; Spinicci, M.; Zammarchi, L.; et al. First-dose mRNA vaccination is sufficient to reactivate immunological memory to SARS-CoV-2 in subjects who have recovered from COVID-19. J. Clin. Investig. 2021, 131, e149150. [Google Scholar] [CrossRef]
- Tarkowski, M.; de Jager, W.; Schiuma, M.; Covizzi, A.; Lai, A.; Gabrieli, A.; Corbellino, M.; Bergna, A.; Della Ventura, C.; Galli, M.; et al. Anti-SARS-CoV-2 Immunoglobulin Isotypes, and Neutralization Activity Against Viral Variants, According to BNT162b2-Vaccination and Infection History. Front. Immunol. 2021, 12, 793191. [Google Scholar] [CrossRef]
- Bates, T.A.; Leier, H.C.; McBride, S.K.; Schoen, D.; Lyski, Z.L.; Lee, D.X.; Messer, W.B.; Curlin, M.E.; Tafesse, F.G. An extended interval between vaccination and infection enhances hybrid immunity against SARS-CoV-2 variants. JCI Insight 2023, 8, e165265. [Google Scholar] [CrossRef]
- Erice, A.; Varillas-Delgado, D.; Caballero, C. Decline of antibody titres 3 months after two doses of BNT162b2 in non-immunocompromised adults. Clin. Microbiol. Infect. 2022, 28, 139-e1. [Google Scholar] [CrossRef]
- Coutant, F.; Touret, F.; Pin, J.-J.; Alonzo, M.; Baronti, C.; Munier, S.; Attia, M.; de Lamballerie, X.; Ferry, T.; Miossec, P. Neutralizing and enhancing monoclonal antibodies in SARS-CoV-2 convalescent patients: Lessons from early variant infection and impact on shaping emerging variants. Emerg. Microbes Infect. 2024, 13, 2307510. [Google Scholar] [CrossRef]
- Migueres, M.; Chapuy-Regaud, S.; Miédougé, M.; Jamme, T.; Lougarre, C.; Da Silva, I.; Pucelle, M.; Staes, L.; Porcheron, M.; Dimeglio, C. Current immunoassays and detection of antibodies elicited by Omicron SARS-CoV-2 infection. J. Med. Virol. 2023, 95, e28200. [Google Scholar] [CrossRef]
- Rössler, A.; Knabl, L.; Raschbichler, L.-M.; Peer, E.; Laer D von Borena, W.; Kimpel, J. Reduced sensitivity of antibody tests after omicron infection. Lancet Microbe 2023, 4, e10–e11. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Bretones, M.; Fouchier, R.A.M.; Koopmans, M.P.G.; van Nierop, G.P. Impact of antigenic evolution and original antigenic sin on SARS-CoV-2 immunity. J. Clin. Investig. 2023, 133, e162192. [Google Scholar] [CrossRef]
- Tang, L.; Zhang, R.; Cui, M.; Hong, P. Omicron-adapted vaccines might require longer follow-up to reveal true benefits. Lancet Microbe 2023, 4, e12. [Google Scholar] [CrossRef]
- Barros-Martins, J.; Hammerschmidt, S.I.; Morillas Ramos, G.; Cossmann, A.; Hetzel, L.; Odak, I.; Kohler, M.; Stankov, M.V.; Ritter, C.; Friedrichsen, M.; et al. Omicron infection-associated T- and B-cell immunity in antigen-naive and triple-COVID-19-vaccinated individuals. Front. Immunol. 2023, 14, 1166589. [Google Scholar] [CrossRef]
- Qu, P.; Faraone, J.N.; Evans, J.P.; Zheng, Y.-M.; Carlin, C.; Anghelina, M.; Stevens, P.; Fernandez, S.; Jones, D.; Panchal, A.R.; et al. Enhanced evasion of neutralizing antibody response by Omicron XBB.1.5, CH.1.1, and CA.3.1 variants. Cell Rep. 2023, 42, 112443. [Google Scholar] [CrossRef]
- Devasundaram, S.; Terpos, E.; Rosati, M.; Ntanasis-Stathopoulos, I.; Bear, J.; Burns, R.; Skourti, S.; Malandrakis, P.; Trougakos, I.P.; Dimopoulos, D.-A.; et al. XBB.1.5 neutralizing antibodies upon bivalent COVID-19 vaccination are similar to XBB but lower than BQ.1.1. Am. J. Hematol. 2023, 98, E123–E126. [Google Scholar] [CrossRef] [PubMed]
- Favresse, J.; Gillot, C.; Cabo, J.; David, C.; Dogné, J.-M.; Douxfils, J. Neutralizing antibody response to XBB.1.5, BA.2.86, FL.1.5.1, and JN.1 six months after the BNT162b2 bivalent booster. Int. J. Infect. Dis. 2024, 143, 107028. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Guo, Y.; Bowen, A.; Mellis, I.A.; Valdez, R.; Gherasim, C.; Gordon, A.; Liu, L.; Ho, D.D. XBB.1.5 monovalent mRNA vaccine booster elicits robust neutralizing antibodies against XBB subvariants and JN.1. Cell Host Microbe 2024, 32, 315–321.e3. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Edwards, R.J.; Manne, K.; Martinez, D.R.; Schäfer, A.; Alam, S.M.; Wiehe, K.; Lu, X.; Parks, R.; Sutherland, L.L.; et al. In vitro and in vivo functions of SARS-CoV-2 infection-enhancing and neutralizing antibodies. Cell 2021, 184, 4203–4219. [Google Scholar] [CrossRef]
- Bratcher-Bowman, N. On Titers for Hyperimmune Plasma (Letter of Authorisation 060320321). Food Drug Administration (FDA). 2021. Available online: https://img-cdn.tinkoffjournal.ru/-/ccp-eua-loatableamendment_6221.pdf (accessed on 19 March 2025).
- Nie, J.; Li, Q.; Wu, J.; Zhao, C.; Hao, H.; Liu, H.; Zhang, L.; Nie, L.; Qin, Z.; Wang, M.; et al. Quantification of SARS-CoV-2 neutralizing antibody by a pseudotyped virus-based assay. Nat. Protoc. 2020, 15, 3699–3715. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2. WIREs Comput. Stat. 2011, 3, 180–185. [Google Scholar] [CrossRef]
- A Box and Whiskers Plot (In the Style of Tukey)—Geom_boxplot n.d. Available online: https://ggplot2.tidyverse.org/reference/geom_boxplot.html (accessed on 14 March 2025).


| Group 1 (n = 105) | Group 2 (n = 95) | |
|---|---|---|
| Age (mean ± sd) | 37.0 ± 9.46 | 38.0 ± 8.87 |
| Gender | ||
| Female | 61 (58.1%) | 58 (61.1%) |
| Male | 44 (41.9%) | 37 (38.9%) |
| SARS-CoV-2 variant | ||
| Alpha | 73 (69.5%) | 67 (70.5%) |
| Beta | 2 (1.90%) | 2 (2.11%) |
| Gamma | 5 (4.76%) | 4 (4.21%) |
| Delta | 10 (9.52%) | 9 (9.47%) |
| Mu | 6 (5.71%) | 4 (4.21%) |
| No sequence | 9 (8.57%) | 9 (9.47%) |
| Participants | Month 0 | Month 1 | Month 3 | Month 6 |
|---|---|---|---|---|
| Unvaccinated | 88 | 94 | 76 | 18 |
| Vaccinated | 0 | 0 | 19 | 58 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Davina-Nunez, C.; Perez-Castro, S.; Cabrera-Alvargonzalez, J.J.; Gonzalez-Alonso, E.; Silva-Bea, S.; Rodriguez-Perez, M.; Figueroa-Lamas, M.d.P.; Perez-Gonzalez, A.; del Campo, V.; Rojas, A.; et al. Subgenomic RNA and Limited Cross-Reactive Neutralising Antibodies Point to Potential Improvements in SARS-CoV-2 Clinical Handling. Int. J. Mol. Sci. 2025, 26, 2948. https://doi.org/10.3390/ijms26072948
Davina-Nunez C, Perez-Castro S, Cabrera-Alvargonzalez JJ, Gonzalez-Alonso E, Silva-Bea S, Rodriguez-Perez M, Figueroa-Lamas MdP, Perez-Gonzalez A, del Campo V, Rojas A, et al. Subgenomic RNA and Limited Cross-Reactive Neutralising Antibodies Point to Potential Improvements in SARS-CoV-2 Clinical Handling. International Journal of Molecular Sciences. 2025; 26(7):2948. https://doi.org/10.3390/ijms26072948
Chicago/Turabian StyleDavina-Nunez, Carlos, Sonia Perez-Castro, Jorge Julio Cabrera-Alvargonzalez, Elena Gonzalez-Alonso, Sergio Silva-Bea, Miriam Rodriguez-Perez, Maria del Pilar Figueroa-Lamas, Alexandre Perez-Gonzalez, Victor del Campo, Almudena Rojas, and et al. 2025. "Subgenomic RNA and Limited Cross-Reactive Neutralising Antibodies Point to Potential Improvements in SARS-CoV-2 Clinical Handling" International Journal of Molecular Sciences 26, no. 7: 2948. https://doi.org/10.3390/ijms26072948
APA StyleDavina-Nunez, C., Perez-Castro, S., Cabrera-Alvargonzalez, J. J., Gonzalez-Alonso, E., Silva-Bea, S., Rodriguez-Perez, M., Figueroa-Lamas, M. d. P., Perez-Gonzalez, A., del Campo, V., Rojas, A., Mendoza, J., & Regueiro-Garcia, B. (2025). Subgenomic RNA and Limited Cross-Reactive Neutralising Antibodies Point to Potential Improvements in SARS-CoV-2 Clinical Handling. International Journal of Molecular Sciences, 26(7), 2948. https://doi.org/10.3390/ijms26072948





