Characterization of Gut Microbiota in Patients with Active Spreading Vitiligo Based on Whole-Genome Shotgun Sequencing
Abstract
1. Introduction
2. Results
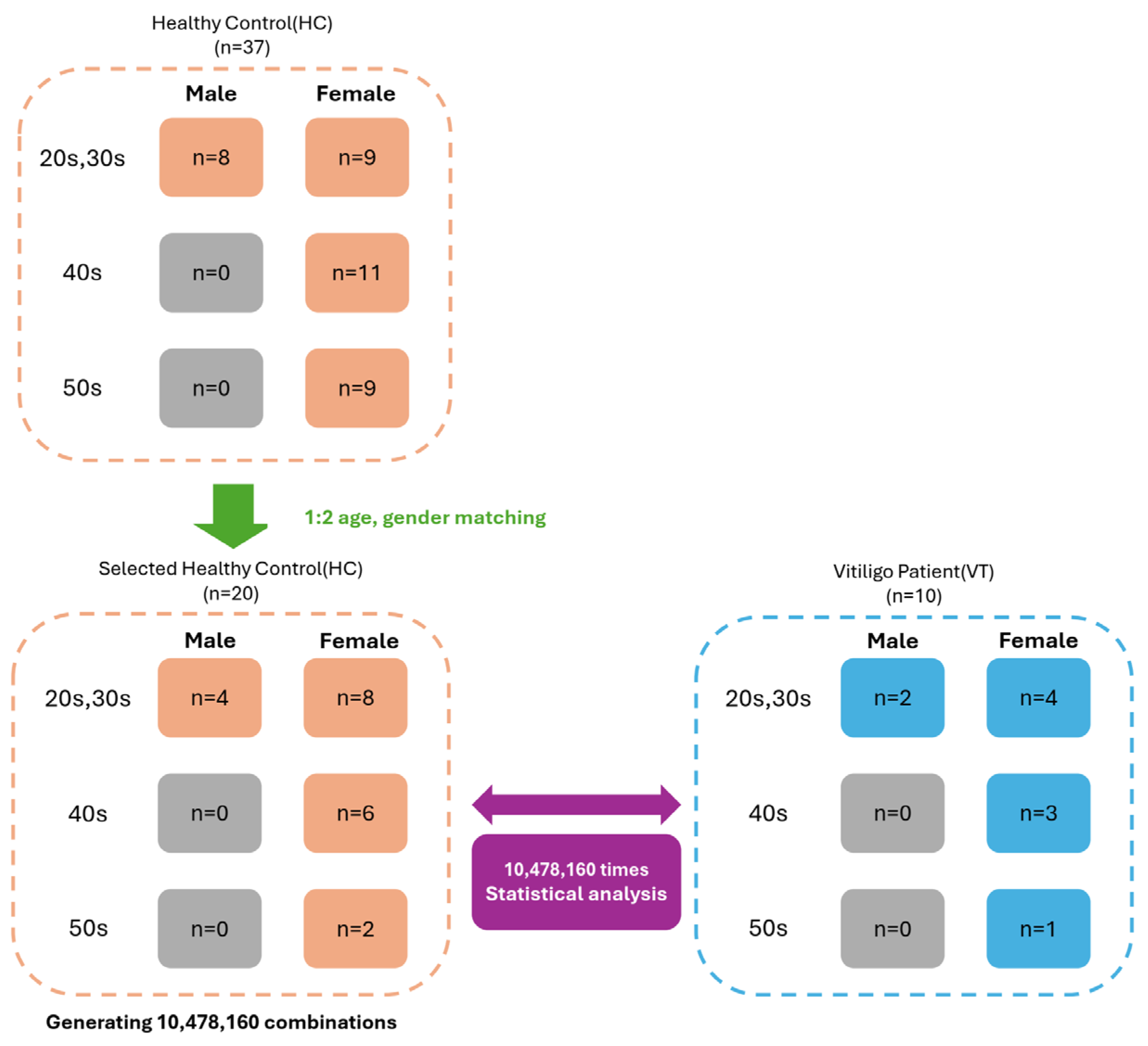
2.1. Clinical Characteristics
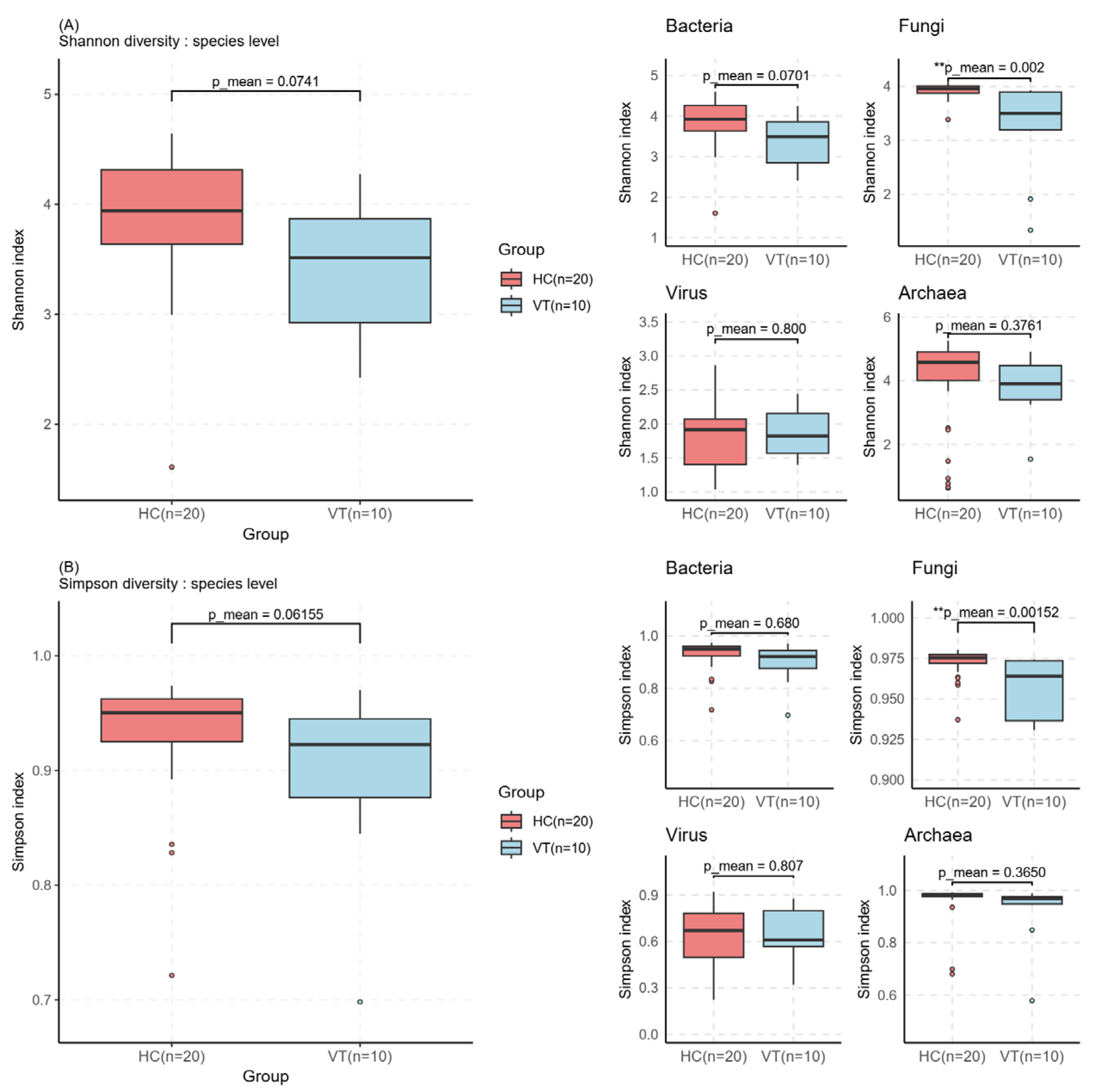
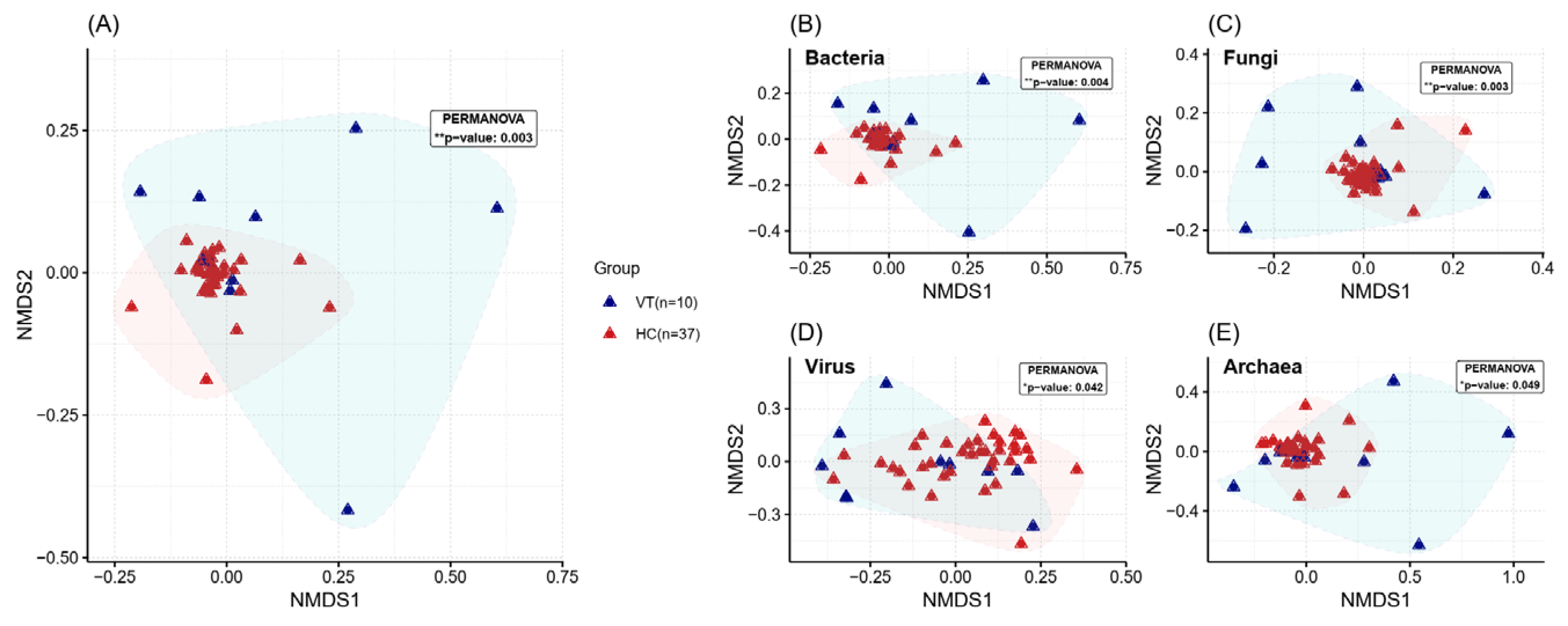
2.2. Alpha and Beta Diversity Assessment
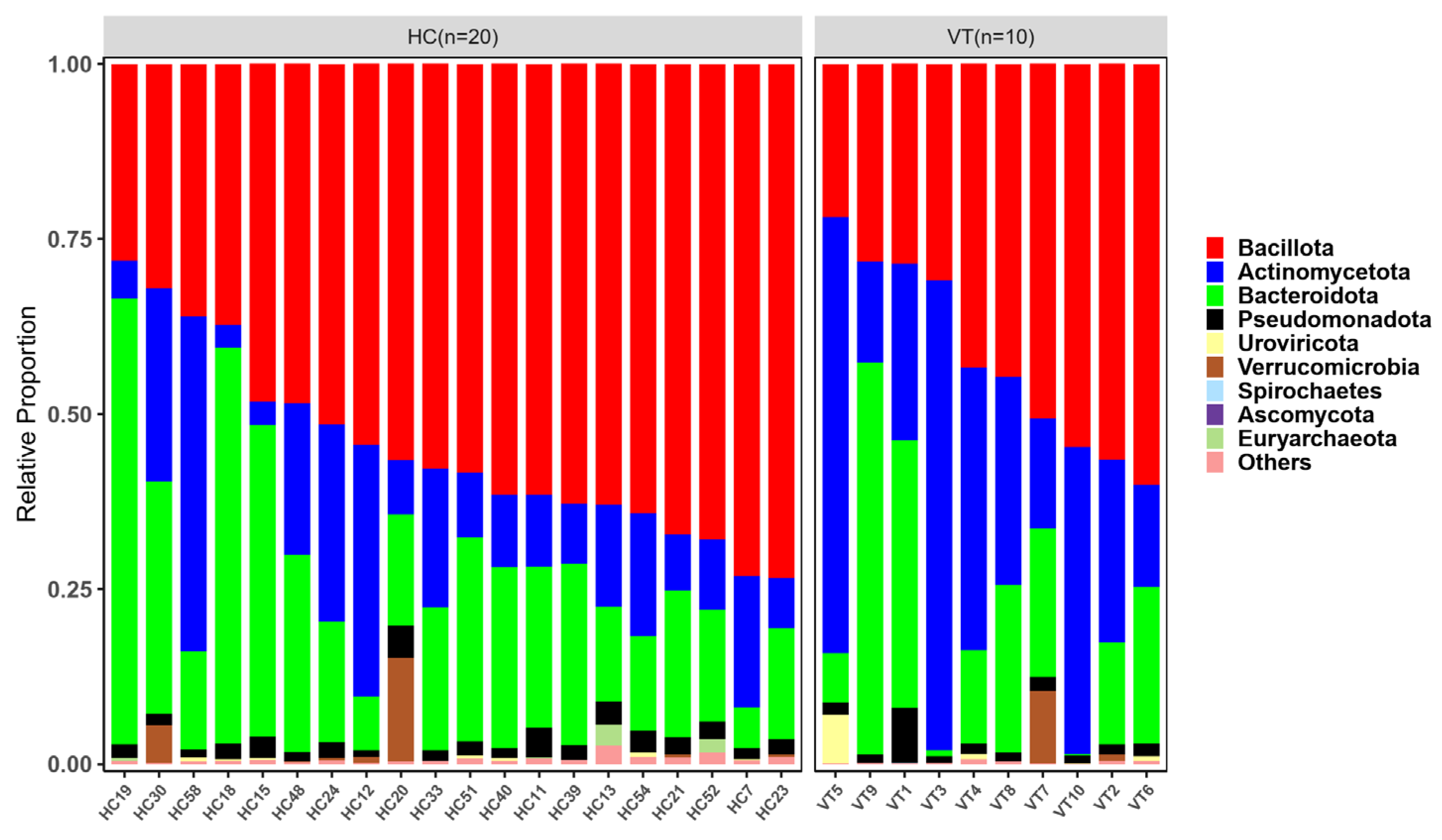
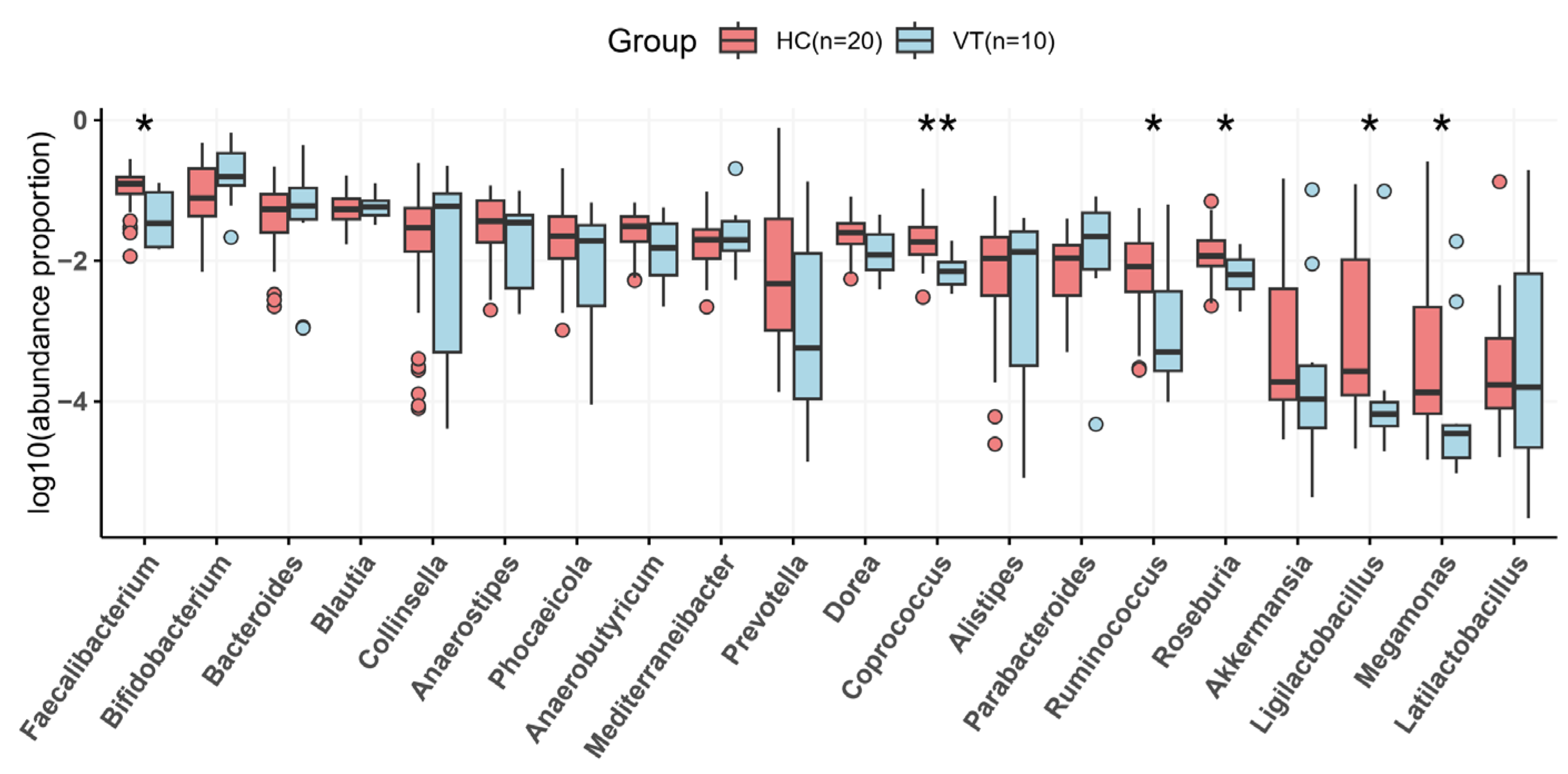
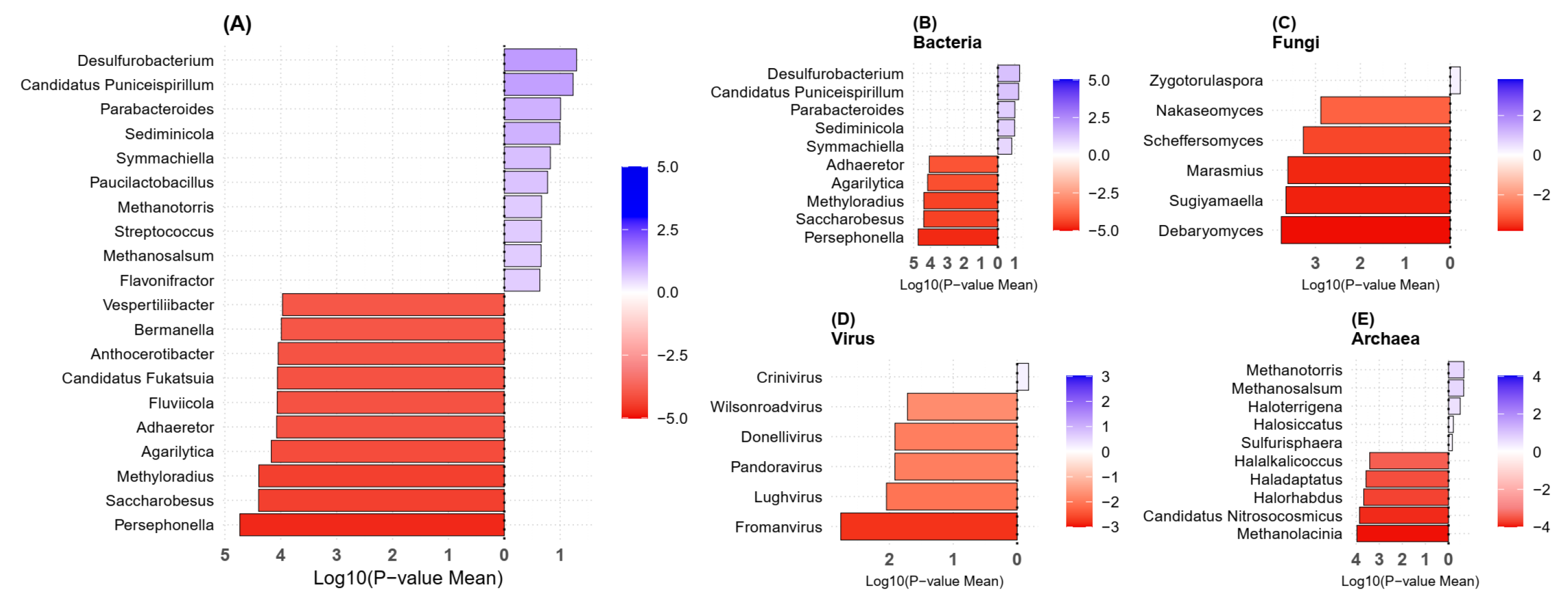
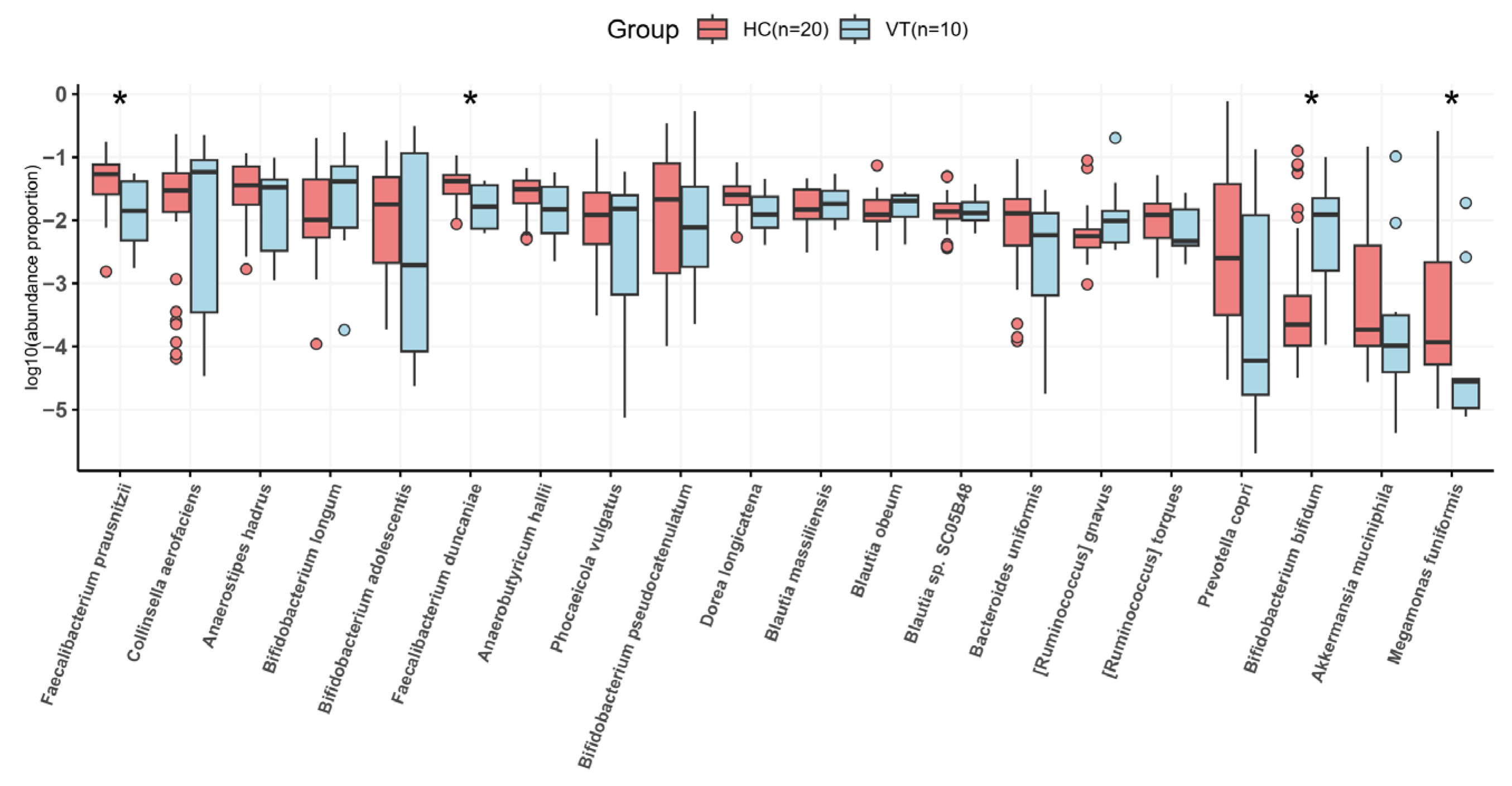
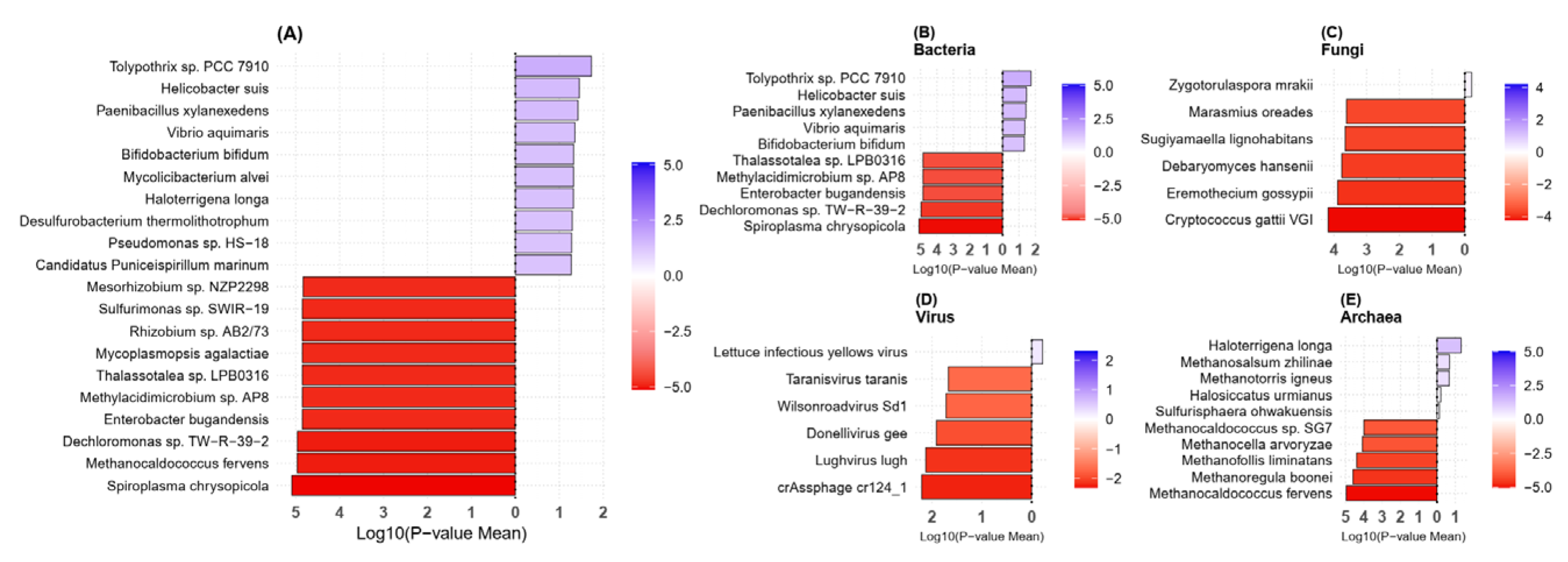
2.3. Taxonomy Analysis
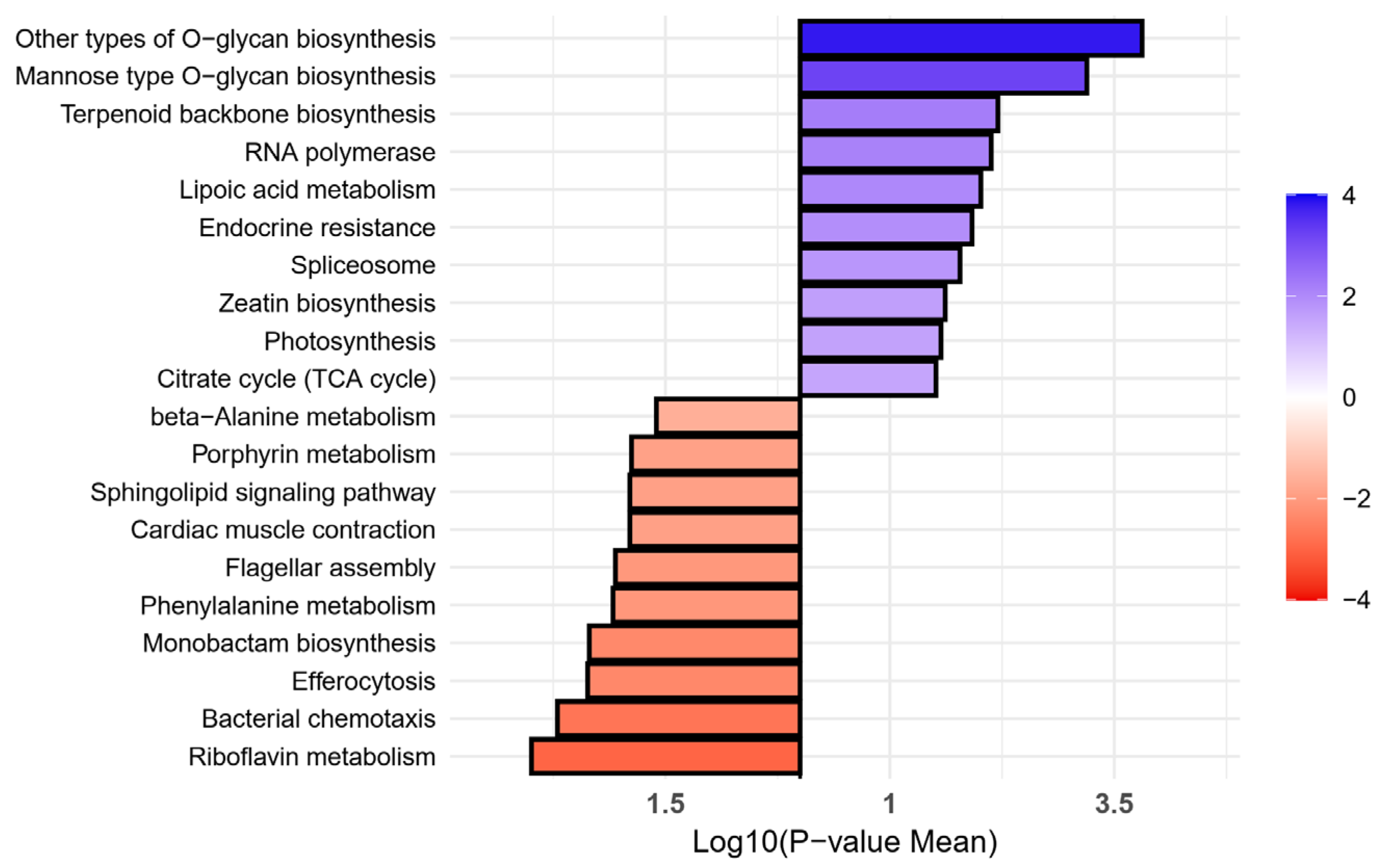
2.4. Metabolic Pathway Enrichment Analysis
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Demographic Matching for Comparative Analysis
4.3. Shotgun Sequencing Data Analysis
4.4. Taxonomic Classification
4.5. Calculation of Diversity Indices
4.6. Estimation of Metabolic Pathway Enrichment
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Author and Year | Country | Methodology | Sample Size | Alpha Diversity Compared to Controls | Beta Diversity | Main Findings | Pathway Analysis | Metabolome Data |
|---|---|---|---|---|---|---|---|---|
| Bzioueche et al., 2021 [8] | France | 16S rRNA | 10 vitiligo, 10 controls | Reduced | Significantly altered microbial composition | Reduced Bacteroides | Not available | Not available |
| Kumar et al., 2024 [53] | India | 16S rRNA | 22 vitiligo, 10 controls | Reduced | Significantly altered microbial composition | Reduced SCFA-producing taxa; increased gut mucosal degradation genes | Not available | SCFA reduction |
| Ni et al., 2020 [21] | China | 16S rRNA | 30 vitiligo, 30 controls | Reduced | Significantly altered microbial composition | Decreased Bacteroidetes to Firmicutes ratio; increased Corynebacterium and Psychrobacter | Not available | 23 altered metabolites, including taurochenodeoxycholate |
| Wu et al., 2023 [54] | China | 16S rRNA | 32 vitiligo, 27 controls | Not available | Significantly altered microbial composition | Increased Firmicutes-to-Bacteroidota ratio in the young adult vitiligo patients; increased Megamonas, Bifidobacterium and Psychrobacter | Paraxanthine, caffeine, and xanthosine from caffeine metabolism pathway downregulated in young vitiligo | 1,7-dimethyl uric acid reduction |
| Luan et al., 2023 [7] | China | Shotgun | 25 vitiligo, 25 healthy control | Reduced | No significant difference | Phylum: increased Bacillota; decreased Bacteroidota species: reduced Staphylococcus thermophiles; increased Bacteroides fragilis | NOD-like receptor signaling pathway enriched in vitiligo | Cysteine degradation down-regulated, and galactose degradation up-regulated |
| Our study | South Korea | Shotgun | 10 active spreading vitiligo, 20 controls | Reduced | Significantly altered microbial composition | Phylum: Actinomycetota and Bacteroidota dominance in vitiligo Species: Feacalibacterium prausnizii, Feacalibacterium duncanieae, and Megamonas funiformis were reduced and Bifidobacterium bifidum was enriched | O-glycan biosynthesis enriched in vitiligo | Not available |
References
- Ezzedine, K.; Eleftheriadou, V.; Whitton, M.; van Geel, N. Vitiligo. Lancet 2015, 386, 74–84. [Google Scholar]
- Ezzedine, K.; Soliman, A.M.; Li, C.; Camp, H.S.; Pandya, A.G. Comorbidity Burden Among Patients with Vitiligo in the United States: A Large-Scale Retrospective Claims Database Analysis. Dermatol. Ther. 2023, 13, 2265–2277. [Google Scholar]
- Spritz, R.A.; Santorico, S.A. The Genetic Basis of Vitiligo. J. Investig. Dermatol. 2021, 141, 265–273. [Google Scholar] [PubMed]
- Bhutta, N.K.; Xu, X.; Jian, C.; Wang, Y.; Liu, Y.; Sun, J.; Han, B.; Wu, S.; Javeed, A. Gut microbiota mediated T cells regulation and autoimmune diseases. Front. Microbiol. 2024, 15, 1477187. [Google Scholar]
- Mousa, W.K.; Chehadeh, F.; Husband, S. Microbial dysbiosis in the gut drives systemic autoimmune diseases. Front. Immunol. 2022, 13, 906258. [Google Scholar]
- Oliveira, G.L.V.d. The Gut Microbiome in Autoimmune Diseases. In Microbiome and Metabolome in Diagnosis, Therapy, and Other Strategic Applications; Academic Press: Cambridge, MA, USA, 2019. [Google Scholar]
- Luan, M.; Niu, M.; Yang, P.; Han, D.; Zhang, Y.; Li, W.; He, Q.; Zhao, Y.; Mao, B.; Chen, J.; et al. Metagenomic sequencing reveals altered gut microbial compositions and gene functions in patients with non-segmental vitiligo. BMC Microbiol. 2023, 23, 265. [Google Scholar]
- Bzioueche, H.; Simonyté Sjödin, K.; West, C.E.; Khemis, A.; Rocchi, S.; Passeron, T.; Tulic, M.K. Analysis of Matched Skin and Gut Microbiome of Patients with Vitiligo Reveals Deep Skin Dysbiosis: Link with Mitochondrial and Immune Changes. J. Investig. Dermatol. 2021, 141, 2280–2290. [Google Scholar]
- Ganju, P.; Nagpal, S.; Mohammed, M.H.; Nishal Kumar, P.; Pandey, R.; Natarajan, V.T.; Mande, S.S.; Gokhale, R.S. Microbial community profiling shows dysbiosis in the lesional skin of Vitiligo subjects. Sci. Rep. 2016, 6, 18761. [Google Scholar]
- Dellacecca, E.R.; Cosgrove, C.; Mukhatayev, Z.; Akhtar, S.; Engelhard, V.H.; Rademaker, A.W.; Knight, K.L.; Le Poole, I.C. Antibiotics Drive Microbial Imbalance and Vitiligo Development in Mice. J. Investig. Dermatol. 2020, 140, 676–687.e676. [Google Scholar]
- Peralta-Pedrero, M.L.; Morales-Sánchez, M.A.; Cruz-Peralta, A.; Guerrero-Oliva, G. [Translated article] Quantification of the Progression Rate in Vitiligo and Prognostic Implications. Actas Dermosifiliogr. 2024, 115, T505–T507. [Google Scholar]
- Bars-Cortina, D.; Ramon, E.; Rius-Sansalvador, B.; Guinó, E.; Garcia-Serrano, A.; Mach, N.; Khannous-Lleiffe, O.; Saus, E.; Gabaldón, T.; Ibáñez-Sanz, G.; et al. Comparison between 16S rRNA and shotgun sequencing in colorectal cancer, advanced colorectal lesions, and healthy human gut microbiota. BMC Genom. 2024, 25, 730. [Google Scholar]
- Moreno-Gallego, J.L.; Chou, S.-P.; Rienzi, S.C.D.; Goodrich, J.; Spector, T.D.; Bell, J.T.; Youngblut, N.D.; Hewson, I.; Reyes, A.; Reyes, A.; et al. Virome Diversity Correlates with Intestinal Microbiome Diversity in Adult Monozygotic Twins. Cell Host Microbe 2019, 25, 261–272.e265. [Google Scholar] [PubMed]
- Reyes, A.; Haynes, M.; Hanson, N.C.; Angly, F.E.; Heath, A.C.; Rohwer, F.L.; Gordon, J.I. Viruses in the fecal microbiota of monozygotic twins and their mothers. Nature 2010, 466, 334–338. [Google Scholar] [PubMed]
- Chen, B.; Sun, L.; Zhang, X. Integration of microbiome and epigenome to decipher the pathogenesis of autoimmune diseases. J. Autoimmun. 2017, 83, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Matsuoka, K.; Sheikh, S.Z.; Russo, S.M.; Mishima, Y.; Collins, C.; DeZoeten, E.F.; Karp, C.L.; Ting, J.P.; Sartor, R.B. IL-10 regulates Il12b expression via histone deacetylation: Implications for intestinal macrophage homeostasis. J. Immunol. 2012, 189, 1792–1799. [Google Scholar] [PubMed]
- Brown, J.; Robusto, B.; Morel, L. Intestinal Dysbiosis and Tryptophan Metabolism in Autoimmunity. Front. Immunol. 2020, 11, 1741. [Google Scholar]
- Abdollahi-Roodsaz, S.; Joosten, L.A.; Koenders, M.I.; Devesa, I.; Roelofs, M.F.; Radstake, T.R.; Heuvelmans-Jacobs, M.; Akira, S.; Nicklin, M.J.; Ribeiro-Dias, F.; et al. Stimulation of TLR2 and TLR4 differentially skews the balance of T cells in a mouse model of arthritis. J. Clin. Investig. 2008, 118, 205–216. [Google Scholar]
- Maeda, Y.; Kurakawa, T.; Umemoto, E.; Motooka, D.; Ito, Y.; Gotoh, K.; Hirota, K.; Matsushita, M.; Furuta, Y.; Narazaki, M.; et al. Dysbiosis Contributes to Arthritis Development via Activation of Autoreactive T Cells in the Intestine. Arthritis Rheumatol. 2016, 68, 2646–2661. [Google Scholar]
- Rinaldi, M.; Perricone, R.; Blank, M.; Perricone, C.; Shoenfeld, Y. Anti-Saccharomyces cerevisiae autoantibodies in autoimmune diseases: From bread baking to autoimmunity. Clin. Rev. Allergy Immunol. 2013, 45, 152–161. [Google Scholar]
- Ni, Q.; Ye, Z.; Wang, Y.; Chen, J.; Zhang, W.; Ma, C.; Li, K.; Liu, Y.; Liu, L.; Han, Z.; et al. Gut Microbial Dysbiosis and Plasma Metabolic Profile in Individuals With Vitiligo. Front. Microbiol. 2020, 11, 592248. [Google Scholar]
- Wang, W.; Chen, L.; Zhou, R.; Wang, X.; Song, L.; Huang, S.; Wang, G.; Xia, B. Increased Proportions of Bifidobacterium and the Lactobacillus Group and Loss of Butyrate-Producing Bacteria in Inflammatory Bowel Disease. J. Clin. Microbiol. 2013, 52, 398–406. [Google Scholar] [CrossRef] [PubMed]
- Sai, A.; Shetty, G.B.; Shetty, P.; Nanjeshgowda, H.L. Influence of gut microbiota on autoimmunity: A narrative review. Brain Behav. Immun. Integr. 2024, 5, 100046. [Google Scholar] [CrossRef]
- Lopez-Siles, M.; Duncan, S.H.; Garcia-Gil, L.J.; Martinez-Medina, M. Faecalibacterium prausnitzii: From microbiology to diagnostics and prognostics. ISME J. 2017, 11, 841–852. [Google Scholar] [CrossRef] [PubMed]
- Cabezas-Cruz, A.; Bermúdez-Humarán, L.G. Exploring the relationship between Faecalibacterium duncaniae and Escherichia coli in inflammatory bowel disease (IBD): Insights and implications. Comput. Struct. Biotechnol. J. 2023, 23, 1–9. [Google Scholar] [CrossRef]
- Verstraeten, S.; Layec, S.; Auger, S.; Juste, C.; Henry, C.; Charif, S.; Jaszczyszyn, Y.; Sokol, H.; Beney, L.; Langella, P.; et al. Faecalibacterium duncaniae A2-165 regulates the expression of butyrate synthesis, ferrous iron uptake, and stress-response genes based on acetate consumption. Sci. Rep. 2024, 14, 987. [Google Scholar] [CrossRef] [PubMed]
- Chollet, L.; Heumel, S.; Deruyter, L.; Bouilloux, F.; Delval, L.; Robert, V.; Gevaert, M.-H.; Pichavant, M.; Sencio, V.; Robil, C.; et al. Faecalibacterium duncaniae as a novel next generation probiotic against influenza. Front. Immunol. 2024, 15, 1347676. [Google Scholar] [CrossRef]
- Shimizu, J.; Kubota, T.; Takada, E.; Takai, K.; Fujiwara, N.; Arimitsu, N.; Ueda, Y.; Wakisaka, S.; Suzuki, T.; Suzuki, N. Bifidobacteria Abundance-Featured Gut Microbiota Compositional Change in Patients with Behcet’s Disease. PLoS ONE 2016, 11, e0153746. [Google Scholar] [CrossRef]
- Nugumanova, G.; Ponomarev, E.D.; Askarova, S.; Fasler-Kan, E.; Barteneva, N.S. Freshwater Cyanobacterial Toxins, Cyanopeptides and Neurodegenerative Diseases. Toxins 2023, 15, 233. [Google Scholar] [CrossRef]
- Padra, M.; Adamczyk, B.; Flahou, B.; Erhardsson, M.; Chahal, G.; Smet, A.; Jin, C.; Thorell, A.; Ducatelle, R.; Haesebrouck, F.; et al. Helicobacter suis infection alters glycosylation and decreases the pathogen growth inhibiting effect and binding avidity of gastric mucins. Mucosal Immunol. 2019, 12, 784–794. [Google Scholar] [CrossRef]
- De Bruyne, E.; Flahou, B.; Chiers, K.; Meyns, T.; Kumar, S.; Vermoote, M.; Pasmans, F.; Millet, S.; Dewulf, J.; Haesebrouck, F.; et al. An experimental Helicobacter suis infection causes gastritis and reduced daily weight gain in pigs. Vet. Microbiol. 2012, 160, 449–454. [Google Scholar] [CrossRef]
- Ekim, B.; Calik, A.; Ceylan, A.; Saçaklı, P. Effects of Paenibacillus xylanexedens on growth performance, intestinal histomorphology, intestinal microflora, and immune response in broiler chickens challenged with Escherichia coli K88. Poult. Sci. 2020, 99, 214–223. [Google Scholar]
- Yamada, T.; Hino, S.; Iijima, H.; Genda, T.; Aoki, R.; Nagata, R.; Han, K.H.; Hirota, M.; Kinashi, Y.; Oguchi, H.; et al. Mucin O-glycans facilitate symbiosynthesis to maintain gut immune homeostasis. EBioMedicine 2019, 48, 513–525. [Google Scholar]
- Pinho, S.S.; Alves, I.; Gaifem, J.; Rabinovich, G.A. Immune regulatory networks coordinated by glycans and glycan-binding proteins in autoimmunity and infection. Cell. Mol. Immunol. 2023, 10, 1101–1113. [Google Scholar]
- Pagan, J.D.; Kitaoka, M.; Anthony, R.M. Engineered Sialylation of Pathogenic Antibodies In Vivo Attenuates Autoimmune Disease. Cell 2018, 172, 564–577.e13. [Google Scholar] [PubMed]
- Balasubramaniam, S.; Yaplito-Lee, J. In Riboflavin metabolism: Role in mitochondrial function. J. Transl. Genet. Genom. 2020, 4, 285–306. [Google Scholar]
- Henriques, B.J.; Olsen, R.K.; Bross, P.; Gomes, C.M. Emerging roles for riboflavin in functional rescue of mitochondrial β-oxidation flavoenzymes. Curr. Med. Chem. 2010, 17, 3842–3854. [Google Scholar] [PubMed]
- Sant’Anna-Silva, A.C.B.; Botton, T.; Rossi, A.; Dobner, J.; Bzioueche, H.; Thach, N.; Blot, L.; Pagnotta, S.; Kleszczynski, K.; Steinbrink, K.; et al. Vitiligo auto-immune response upon oxidative stress-related mitochondrial DNA release opens up new therapeutic strategies. Clin. Transl. Med. 2024, 14, e1810. [Google Scholar]
- Papaccio, F.; Bellei, B.; Ottaviani, M.; D’Arino, A.; Truglio, M.; Caputo, S.; Cigliana, G.; Sciuto, L.; Migliano, E.; Pacifico, A.; et al. A Possible Modulator of Vitiligo Metabolic Impairment: Rethinking a PPARγ Agonist. Cells 2022, 11, 3583. [Google Scholar] [CrossRef]
- Ranjan, R.; Rani, A.; Metwally, A.; McGee, H.S.; Perkins, D.L. Analysis of the microbiome: Advantages of whole genome shotgun versus 16S amplicon sequencing. Biochem. Biophys. Res. Commun. 2016, 469, 967–977. [Google Scholar]
- Laudadio, I.; Fulci, V.; Palone, F.; Stronati, L.; Cucchiara, S.; Carissimi, C. Quantitative Assessment of Shotgun Metagenomics and 16S rDNA Amplicon Sequencing in the Study of Human Gut Microbiome. Omics 2018, 22, 248–254. [Google Scholar]
- Durazzi, F.; Sala, C.; Castellani, G.; Manfreda, G.; Remondini, D.; De Cesare, A. Comparison between 16S rRNA and shotgun sequencing data for the taxonomic characterization of the gut microbiota. Sci. Rep. 2021, 11, 3030. [Google Scholar]
- Rothschild, D.; Weissbrod, O.; Barkan, E.; Kurilshikov, A.; Korem, T.; Zeevi, D.; Costea, P.I.; Godneva, A.; Kalka, I.N.; Bar, N.; et al. Environment dominates over host genetics in shaping human gut microbiota. Nature 2018, 555, 210–215. [Google Scholar]
- Kim, J.; Park, K.Y.; Park, H.K.; Hwang, H.S.; Seo, M.R.; Kim, B.; Cho, Y.; Rho, M.; Pai, H. High fecal carriage of bla(CTX-M), bla(CMY-2), and plasmid-mediated quinolone resistance genes among healthy Korean people in a metagenomic analysis. Sci. Rep. 2021, 11, 5874. [Google Scholar]
- Joshi, N.; Fass, J. Sickle: A Sliding-Window, Adaptive, Quality-Based Trimming Tool for FastQ Files (Version 1.33) [Software]. 2011. Available online: https://github.com/najoshi/sickle (accessed on 19 March 2025).
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar]
- Wood, D.E.; Lu, J.; Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar]
- Wood, D.E.; Salzberg, S.L. Kraken: Ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014, 15, R46. [Google Scholar] [CrossRef]
- Whittaker, R.H. Evolution and measurement of species diversity. Taxon 1972, 21, 213–251. [Google Scholar]
- Legendre, P.; Gallagher, E.D. Ecologically meaningful transformations for ordination of species data. Oecologia 2001, 129, 271–280. [Google Scholar]
- Abubucker, S.; Segata, N.; Goll, J.; Schubert, A.M.; Izard, J.; Cantarel, B.L.; Rodriguez-Mueller, B.; Zucker, J.; Thiagarajan, M.; Henrissat, B. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput. Biol. 2012, 8, e1002358. [Google Scholar] [CrossRef]
- Franzosa, E.A.; McIver, L.J.; Rahnavard, G.; Thompson, L.R.; Schirmer, M.; Weingart, G.; Lipson, K.S.; Knight, R.; Caporaso, J.G.; Segata, N. Species-level functional profiling of metagenomes and metatranscriptomes. Nat. Methods 2018, 15, 962–968. [Google Scholar]
- Kumar, S.; Mahajan, S.; Kale, D.; Chourasia, N.; Khan, A.; Asati, D.; Kotnis, A.; Sharma, V.K. Insights into the gut microbiome of vitiligo patients from India. BMC Microbiol. 2024, 24, 440. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Cheng, P.; Shao, T.; Li, Z.; Ji, Q.; Wang, L.; Yao, S.; Lu, B. Alterations of gut microbiota and gut metabolites in the young-adult vitiligo patients. J. Eur. Acad. Dermatol. Venereol. 2023, 37, e904–e907. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ju, H.J.; Song, W.H.; Shin, J.H.; Lee, J.H.; Bae, J.M.; Lee, Y.B.; Lee, M. Characterization of Gut Microbiota in Patients with Active Spreading Vitiligo Based on Whole-Genome Shotgun Sequencing. Int. J. Mol. Sci. 2025, 26, 2939. https://doi.org/10.3390/ijms26072939
Ju HJ, Song WH, Shin JH, Lee JH, Bae JM, Lee YB, Lee M. Characterization of Gut Microbiota in Patients with Active Spreading Vitiligo Based on Whole-Genome Shotgun Sequencing. International Journal of Molecular Sciences. 2025; 26(7):2939. https://doi.org/10.3390/ijms26072939
Chicago/Turabian StyleJu, Hyun Jeong, Woo Hyun Song, Ji Hae Shin, Ji Hae Lee, Jung Min Bae, Young Bok Lee, and Minho Lee. 2025. "Characterization of Gut Microbiota in Patients with Active Spreading Vitiligo Based on Whole-Genome Shotgun Sequencing" International Journal of Molecular Sciences 26, no. 7: 2939. https://doi.org/10.3390/ijms26072939
APA StyleJu, H. J., Song, W. H., Shin, J. H., Lee, J. H., Bae, J. M., Lee, Y. B., & Lee, M. (2025). Characterization of Gut Microbiota in Patients with Active Spreading Vitiligo Based on Whole-Genome Shotgun Sequencing. International Journal of Molecular Sciences, 26(7), 2939. https://doi.org/10.3390/ijms26072939






