m6A-Modified GATA2 Enhances Odontogenic Differentiation in Stem Cells from the Apical Papilla
Abstract
1. Introduction
2. Results
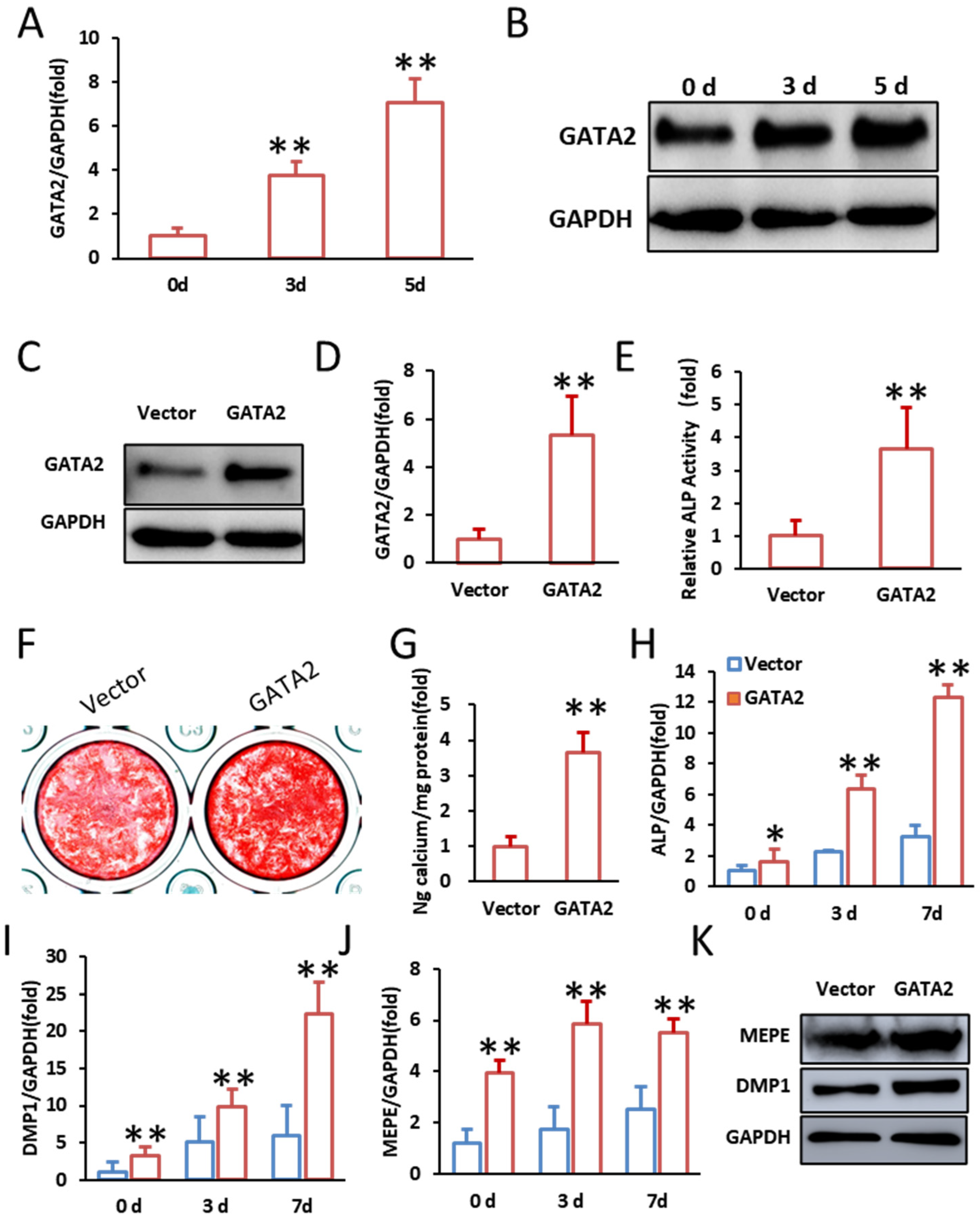
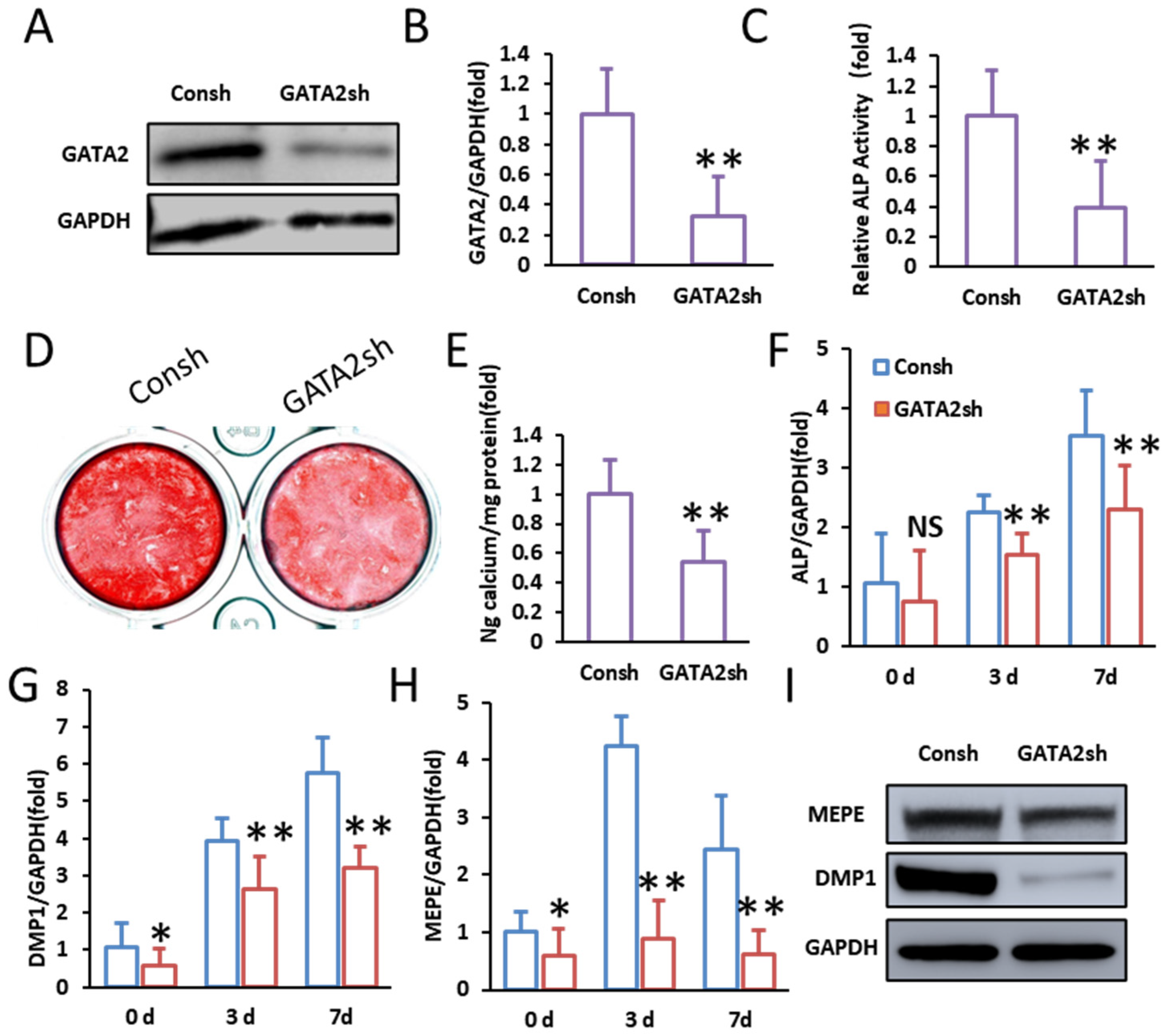
2.1. GATA2 Enhances the Odontogenic Differentiation of SCAPs
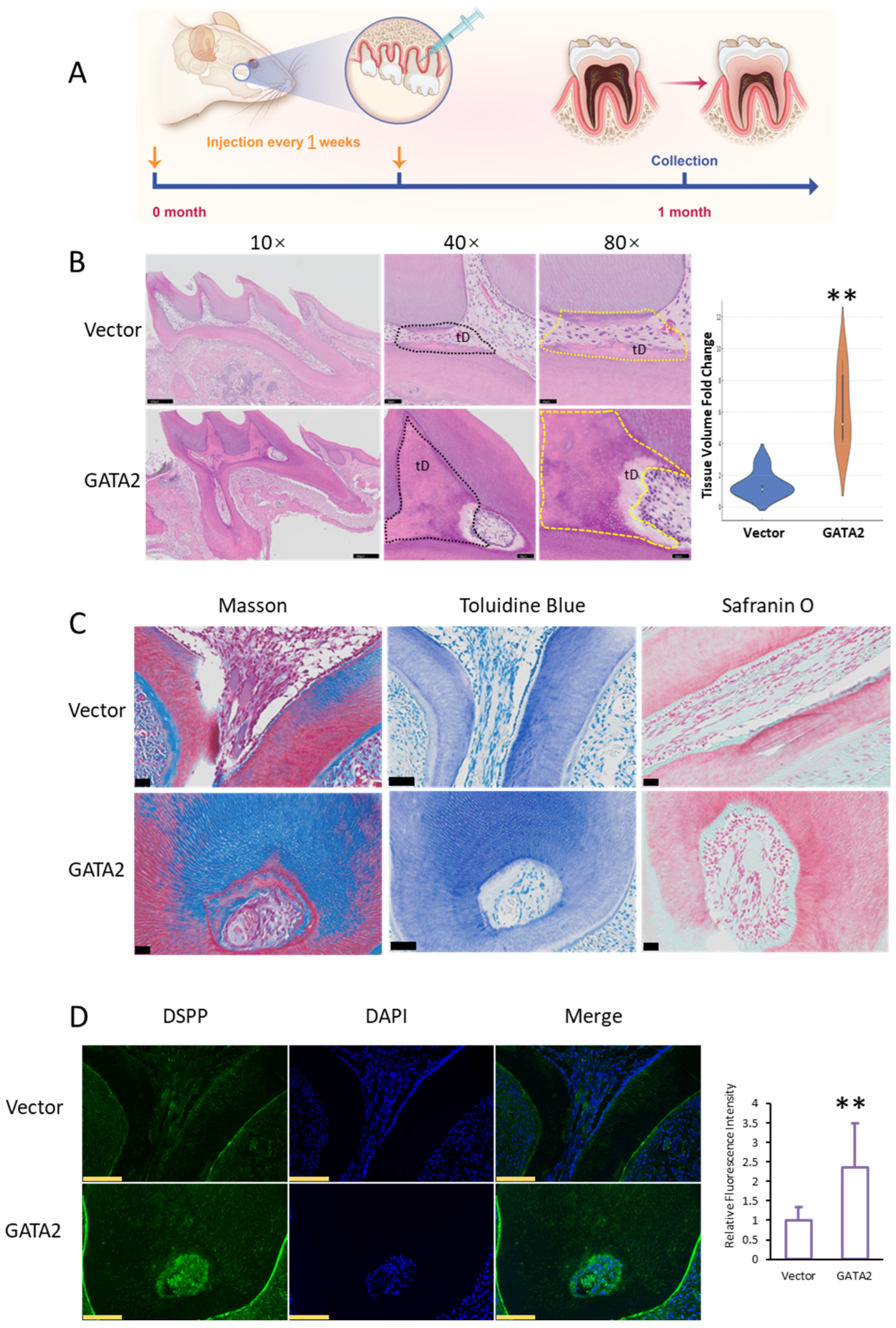
2.2. GATA2 Promotes Dentin Formation In Vivo
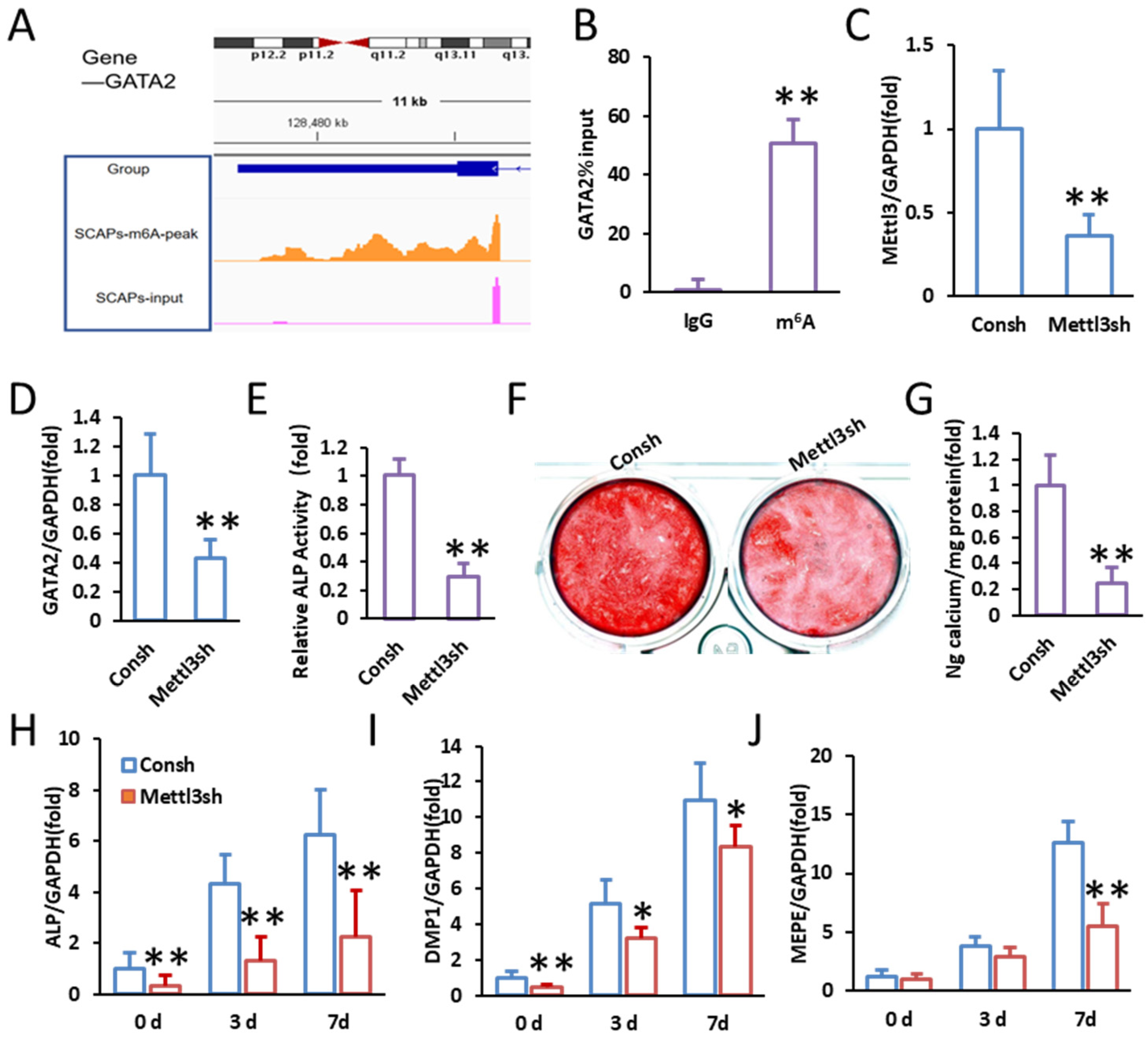
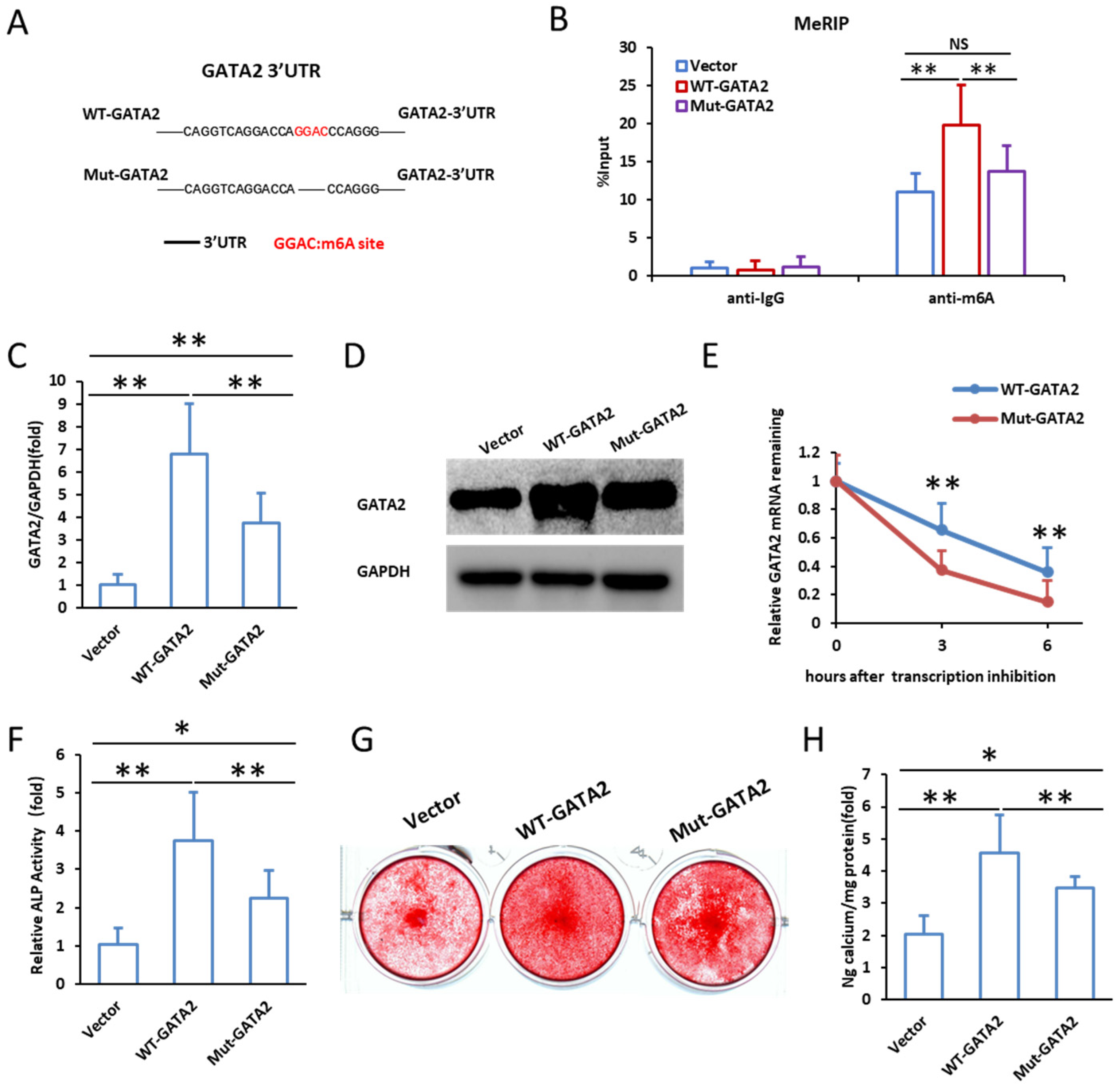
2.3. Identification of m6A Modification in GATA2
2.4. Mutation of m6A Modification Sites Reduces GATA2 Stability and Impairs Odontogenic Differentiation
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Cell Culture and Odontogenic Differentiation Induction
4.3. ALP Activity Assay
4.4. Alizarin Red S Staining
4.5. Lentiviral Vector Construction and Transduction
4.6. RNA Extraction and Quantitative Real-Time PCR (qPCR)
4.7. Western Blot Analysis
4.8. RNA Methylation and m6A Site Mutations
4.9. RNA Stability Assay
4.10. Methylated RNA Immunoprecipitation Quantitative PCR (MeRIP-qPCR)
4.11. In Vivo Dentin Formation Assay
4.12. Immunofluorescence Staining
4.13. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hashemi-Beni, B.; Khoroushi, M.; Foroughi, M.R.; Karbasi, S.; Khademi, A.A. Tissue engineering: Dentin–pulp complex regeneration approaches (A review). Tissue Cell 2017, 49, 552–564. [Google Scholar] [CrossRef]
- Zhao, J.; Zhou, Y.H.; Zhao, Y.Q.; Gao, Z.R.; Ouyang, Z.Y.; Ye, Q.; Liu, Q.; Chen, Y.; Tan, L.; Zhang, S.H.; et al. Oral cavity-derived stem cells and preclinical models of jaw-bone defects for bone tissue engineering. Stem Cell Res. Ther. 2023, 14, 39. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Miteva, M.; Mihaylova, Z.; Mitev, V.; Aleksiev, E.; Stanimirov, P.; Praskova, M.; Dimitrova, V.S.; Vasileva, A.; Vasileva, A.; Vasileva, A.; et al. A Review of Stem Cell Attributes Derived from the Oral Cavity. Int. Dent. J. 2024, 74, 1129–1141. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Gao, Y.; He, J. Stem Cells from the Apical Papilla (SCAPs): Past, Present, Prospects, and Challenges. Biomedicines 2023, 11, 2047. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, X.; Wu, Z.; Jia, S.; Wan, M. Epigenetic regulation of dental-derived stem cells and their application in pulp and periodontal regeneration. PeerJ 2023, 11, e14550. [Google Scholar] [CrossRef]
- Schwechheimer, C.; Schröder, P.M.; Blaby-Haas, C.E. Plant GATA Factors: Their Biology, Phylogeny, and Phylogenomics. Annu. Rev. Plant Biol. 2022, 73, 123–148. [Google Scholar] [CrossRef] [PubMed]
- Patient, R.K.; McGhee, J.D. The GATA family (vertebrates and invertebrates). Curr. Opin. Genet. Dev. 2002, 12, 416–422. [Google Scholar] [CrossRef]
- Gao, J.; Chen, Y.H.; Peterson, L.C. GATA family transcriptional factors: Emerging suspects in hematologic disorders. Exp. Hematol. Oncol. 2015, 4, 28. [Google Scholar] [CrossRef]
- Katsumura, K.R.; Bresnick, E.H. GATA Factor Mechanisms Group. The GATA factor revolution in hematology. Blood 2017, 129, 2092–2102. [Google Scholar] [CrossRef]
- Tremblay, M.; Sanchez-Ferras, O.; Bouchard, M. GATA transcription factors in development and disease. Development 2018, 145, dev164384. [Google Scholar] [CrossRef]
- Crispino, J.D.; Horwitz, M.S. GATA factor mutations in hematologic disease. Blood 2017, 129, 2103–2110. [Google Scholar] [CrossRef]
- Leubolt, G.; Redondo Monte, E.; Greif, P.A. GATA2 mutations in myeloid malignancies: Two zinc fingers in many pies. IUBMB Life 2020, 72, 151–158. [Google Scholar] [CrossRef]
- Peters, I.J.A.; de Pater, E.; Zhang, W. The role of GATA2 in adult hematopoiesis and cell fate determination. Front. Cell Dev. Biol. 2023, 11, 1250827. [Google Scholar] [CrossRef] [PubMed]
- Aktar, A.; Heit, B. Role of the pioneer transcription factor GATA2 in health and disease. J. Mol. Med. 2023, 101, 1191–1208. [Google Scholar] [CrossRef] [PubMed]
- Bresnick, E.H.; Katsumura, K.R.; Lee, H.Y.; Johnson, K.D.; Perkins, A.S. Master regulatory GATA transcription factors: Mechanistic principles and emerging links to hematologic malignancies. Nucleic Acids Res. 2012, 40, 5819–5831. [Google Scholar] [CrossRef]
- Wlodarski, M.W.; Hirabayashi, S.; Pastor, V.; Starý, J.; Hasle, H.; Masetti, R.; Dworzak, M.; Schmugge, M.; van den Heuvel-Eibrink, M.; Ussowicz, M.; et al. Prevalence, clinical characteristics, and prognosis of GATA2-related myelodysplastic syndromes in children and adolescents. Blood 2016, 127, 1387–1397. [Google Scholar] [CrossRef] [PubMed]
- Chandola, U.; Das, R.; Panda, B. Role of the N6-methyladenosine RNA mark in gene regulation and its implications on development and disease. Brief. Funct. Genom. 2015, 14, 169–179. [Google Scholar] [CrossRef]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Salmon-Divon, M.; Amariglio, N.; Rechavi, G. Transcriptome-wide mapping of N(6)-methyladenosine by m(6)A-seq based on immunocapturing and massively parallel sequencing. Nat. Protoc. 2013, 8, 176–189. [Google Scholar] [CrossRef]
- Maity, A.; Das, B. N6-methyladenosine modification in mRNA: Machinery, function and implications for health and diseases. FEBS J. 2016, 283, 1607–1630. [Google Scholar] [CrossRef]
- Jiang, X.; Liu, B.; Nie, Z.; Duan, L.; Xiong, Q.; Jin, Z.; Yang, C.; Chen, Y. The role of m6A modification in the biological functions and diseases. Signal Transduct. Target. Ther. 2021, 6, 74. [Google Scholar] [CrossRef]
- Geula, S.; Moshitch-Moshkovitz, S.; Dominissini, D.; Mansour, A.A.; Kol, N.; Salmon-Divon, M.; Hershkovitz, V.; Peer, E.; Mor, N.; Manor, Y.S. Stem cells m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 2015, 347, 1002–1006. [Google Scholar] [CrossRef]
- Wang, X.; Feng, J.; Xue, Y.; Guan, Z.; Zhang, D.; Liu, Z.; Gong, Z.; Wang, Q.; Huang, J.; Tang, C.; et al. Structural basis of N 6 adenosine methylation by the METTL3–METTL14 complex. Nature 2016, 534, 575. [Google Scholar] [CrossRef] [PubMed]
- Batista, P.J.; Molinie, B.; Wang, J.; Qu, K.; Zhang, J.; Li, L.; Bouley, D.M.; Lujan, E.; Haddad, B.; Daneshvar, K.; et al. m6A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 2014, 15, 707–719. [Google Scholar] [CrossRef]
- Yin, J.; Ding, F.; Cheng, Z.; Ge, X.; Li, Y.; Zeng, A.; Zhang, J.; Yan, W.; Shi, Z.; Qian, X.; et al. METTL3-mediated m6A modification of LINC00839 maintains glioma stem cells and radiation resistance by activating Wnt/β-catenin signaling. Cell. Death. Dis. 2023, 14, 417. [Google Scholar] [CrossRef]
- Lu, Y.; Liu, Z.; Zhang, Y.; Wu, X.; Bian, W.; Shan, S.; Yang, D.; Ren, T. METTL3-mediated m6A RNA methylation induces the differentiation of lung resident mesenchymal stem cells into myofibroblasts via the miR-21/PTEN pathway. Respir. Res. 2023, 24, 300. [Google Scholar] [CrossRef] [PubMed]
- Oerum, S.; Meynier, V.; Catala, M.; Tisné, C. A comprehensive review of m6A/m6Am RNA methyltransferase structures. Nucleic Acids Res. 2021, 49, 7239–7255. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Li, G.; Yang, H.; Zhang, C.; Cao, Y.; Wang, N.; Ge, L.; Fan, Z. METTL3 Promotes Osteo/Odontogenic Differentiation of Stem Cells by Inhibiting miR-196b-5p Maturation. Stem Cells Int. 2023, 2023, 8992284. [Google Scholar] [CrossRef]
- Tian, Q.; Gao, S.; Li, S.; Wan, M.; Zhou, X.; Du, W.; Zhou, X.; Zheng, L.; Zhou, Y. Glutamine-αKG axis affects dentin regeneration and regulates osteo/odontogenic differentiation of mesenchymal adult stem cells via IGF2 m6A modification. Stem Cell Res. Ther. 2024, 15, 479. [Google Scholar] [CrossRef]
- Babb, R.C.; Chandrasekaran, D.; Zaugg, L.K.; Sharpe, P.T. A Mouse Model to Study Reparative Dentinogenesis. Methods Mol. Biol. 2019, 1922, 111–119. [Google Scholar] [CrossRef]
- Wang, S.; Tu, Y.; Yu, H.; Li, Z.; Feng, J.; Liu, S. Animal models and related techniques for dentin study. Odontology 2025, 113, 42–60. [Google Scholar] [CrossRef]
- Bakopoulou, A.; Leyhausen, G.; Volk, J.; Tsiftsoglou, A.; Garefis, P.; Koidis, P.; Geurtsen, W. Comparative analysis of in vitro osteo/odontogenic differentiation potential of human dental pulp stem cells (DPSCs) and stem cells from the apical papilla (SCAP). Arch. Oral Biol. 2011, 56, 709–721. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.S.; Xie, J.; Wang, D.; Kim, J.M.; Tai, P.W.; Gravallese, E.; Gao, G.; Shim, J.H. Bone-targeting AAV-mediated silencing of Schnurri-3 prevents bone loss in osteoporosis. Nat. Commun. 2019, 10, 2958. [Google Scholar] [CrossRef]
- Zhu, Z.X.; Liu, Y.; Wang, J.; Xie, Y.; Li, R.Y.; Ma, Q.; Tu, Q.; Melhem, N.A.; Couldwell, S.; El-Araby, R.E.; et al. A novel lncRNA-mediated epigenetic regulatory mechanism in periodontitis. Int. J. Biol. Sci. 2023, 19, 5187–5203. [Google Scholar] [CrossRef]
- Zaccara, S.; Ries, R.J.; Jaffrey, S.R. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 608–624. [Google Scholar] [CrossRef]
- Wan, W.; Ao, X.; Chen, Q.; Yu, Y.; Ao, L.; Xing, W.; Guo, W.; Wu, X.; Pu, C.; Hu, X.; et al. METTL3/IGF2BP3 axis inhibits tumor immune surveillance by upregulating N-methyladenosine modification of PD-L1 mRNA in breast cancer. Mol. Cancer 2022, 21, 60. [Google Scholar] [CrossRef] [PubMed]
- Flamand, M.N.; Tegowski, M.; Meyer, K.D. The Proteins of mRNA Modification: Writers, Readers, and Erasers. Annu. Rev. Biochem. 2023, 92, 145–173. [Google Scholar] [CrossRef]
- Wu, D.; Spencer, C.B.; Ortoga, L.; Zhang, H.; Miao, C. Histone lactylation-regulated METTL3 promotes ferroptosis via m6A-modification on ACSL4 in sepsis-associated lung injury. Redox Biol. 2024, 74, 103194. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, Y.; Xiao, H.; Liu, Z.; Liu, X.; Feng, Z.; Sheng, X.; Peng, B.; Ren, X.; Xu, L.; et al. METTL3-mediated m6A modification increases Hspa1a stability to inhibit osteoblast aging. Cell Death Discov. 2024, 10, 155. [Google Scholar] [CrossRef]
- Xu, M.; Li, B.; Huang, J.; Jia, R.; Guo, J. The N6-methyladenosine demethylase FTO is required for odontoblast differentiation in vitro and dentine formation in mice by promoting RUNX2 exon 5 inclusion through RBM4. Int. Endod. J. 2023, 56, 1534–1549. [Google Scholar] [CrossRef]
- Pan, Y.; Liu, Y.; Cui, D.; Yu, S.; Zhou, Y.; Zhou, X.; Du, W.; Zheng, L.; Wan, M. METTL3 enhances dentinogenesis differentiation of dental pulp stem cells via increasing GDF6 and STC1 mRNA stability. BMC Oral Health. 2023, 23, 209. [Google Scholar] [CrossRef]
- Yang, H.; Wang, W.; Liu, H.; Zhang, C.; Cao, Y.; Long, L.; Han, X.; Wang, Y.; Yan, F.; Li, G.; et al. miR615-3p inhibited FBLN1 and osteogenic differentiation of umbilical cord mesenchymal stem cells by associated with YTHDF2 in a m6A-miRNA interaction manner. Cell Prolif. 2024, 57, e13607. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, H.; Yuan, F.; Song, J.; Huang, Y.; Shan, Z.; Fan, Z. m6A-Modified GATA2 Enhances Odontogenic Differentiation in Stem Cells from the Apical Papilla. Int. J. Mol. Sci. 2025, 26, 2920. https://doi.org/10.3390/ijms26072920
Yang H, Yuan F, Song J, Huang Y, Shan Z, Fan Z. m6A-Modified GATA2 Enhances Odontogenic Differentiation in Stem Cells from the Apical Papilla. International Journal of Molecular Sciences. 2025; 26(7):2920. https://doi.org/10.3390/ijms26072920
Chicago/Turabian StyleYang, Haoqing, Fengning Yuan, Jiaxin Song, Yishu Huang, Zhaochen Shan, and Zhipeng Fan. 2025. "m6A-Modified GATA2 Enhances Odontogenic Differentiation in Stem Cells from the Apical Papilla" International Journal of Molecular Sciences 26, no. 7: 2920. https://doi.org/10.3390/ijms26072920
APA StyleYang, H., Yuan, F., Song, J., Huang, Y., Shan, Z., & Fan, Z. (2025). m6A-Modified GATA2 Enhances Odontogenic Differentiation in Stem Cells from the Apical Papilla. International Journal of Molecular Sciences, 26(7), 2920. https://doi.org/10.3390/ijms26072920




