Identification and Characterization of Histone Modification Gene Families and Their Expression Patterns During Pod and Seed Development in Peanut
Abstract
1. Introduction
2. Results and Discussion
2.1. Identification and Chromosomes Location of Histone Modification Genes in Peanut
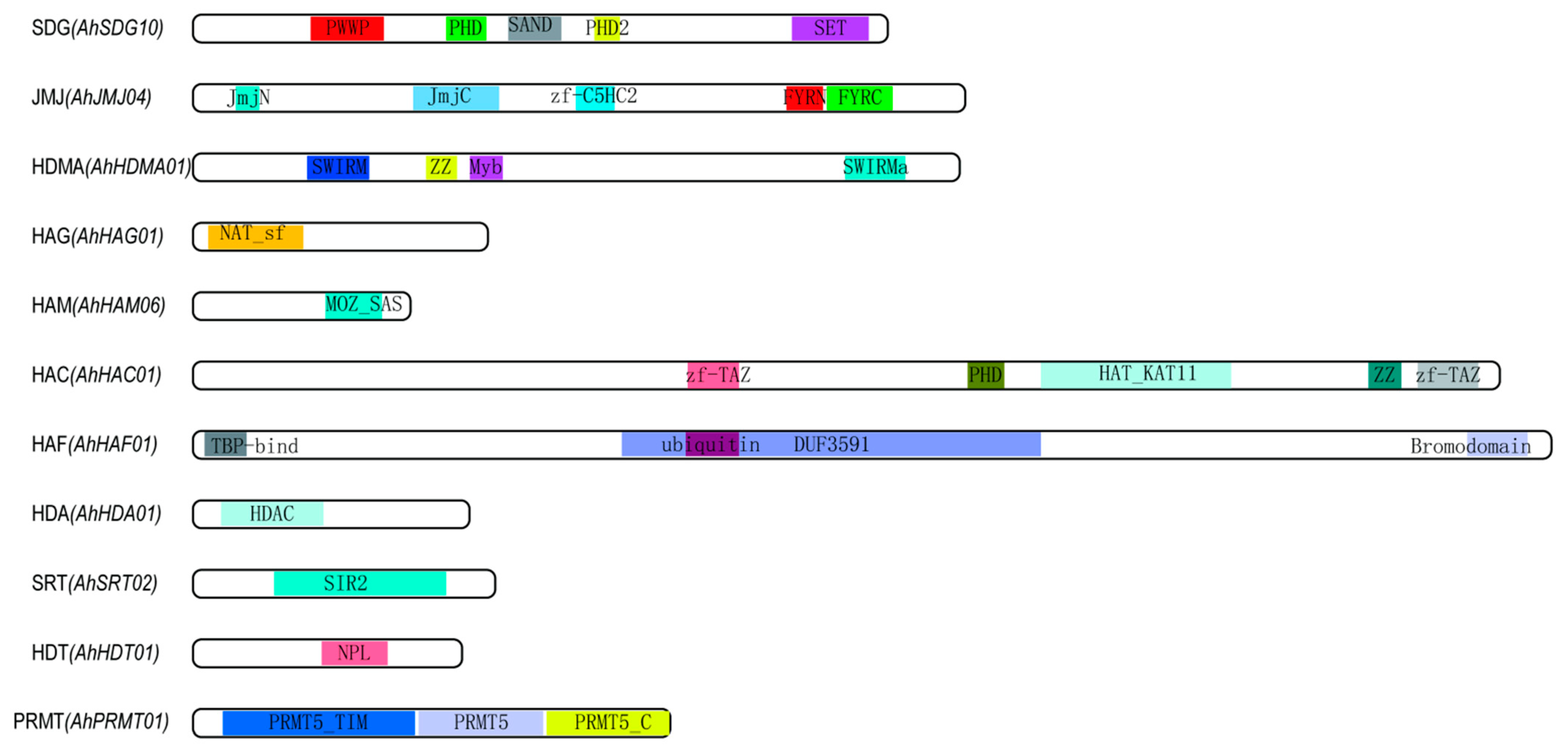
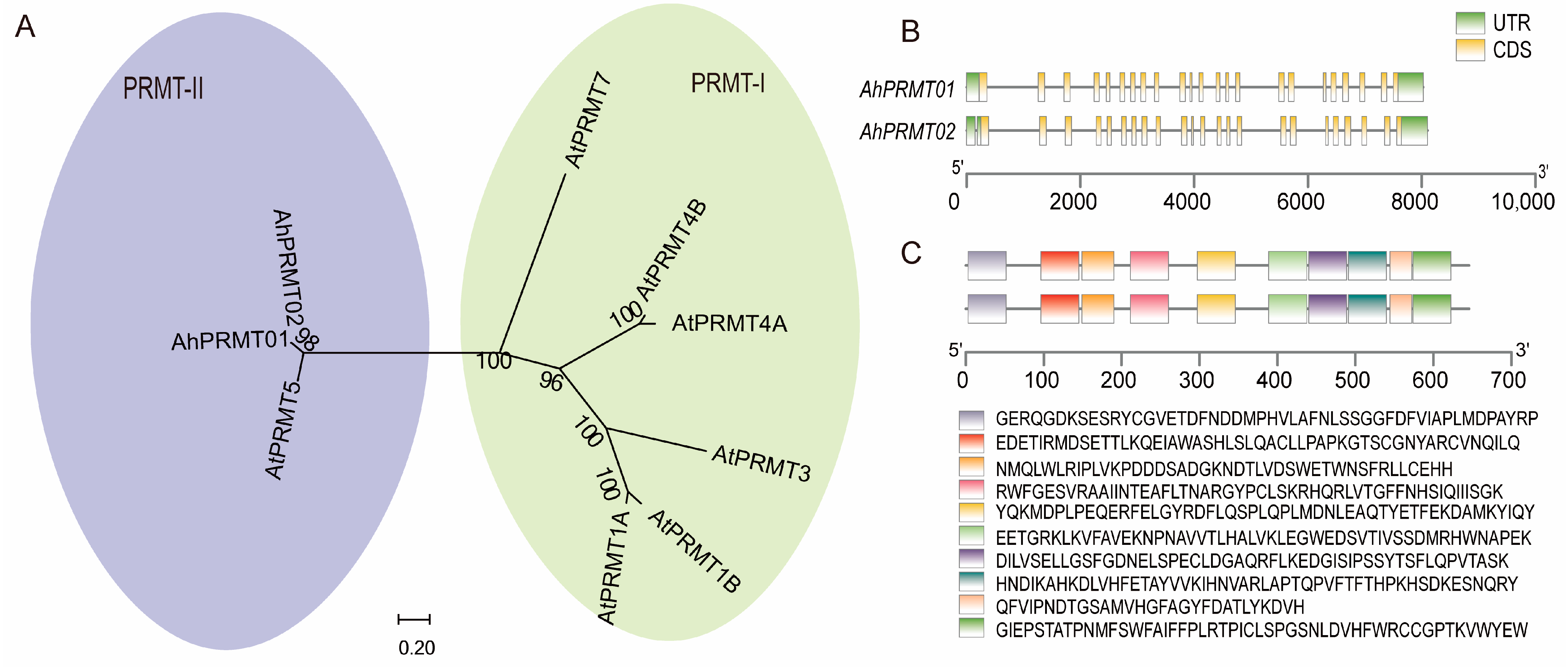
2.2. Gene Structure, Conserved Motif, and Phylogenetic Analysis of Histone Modification Genes
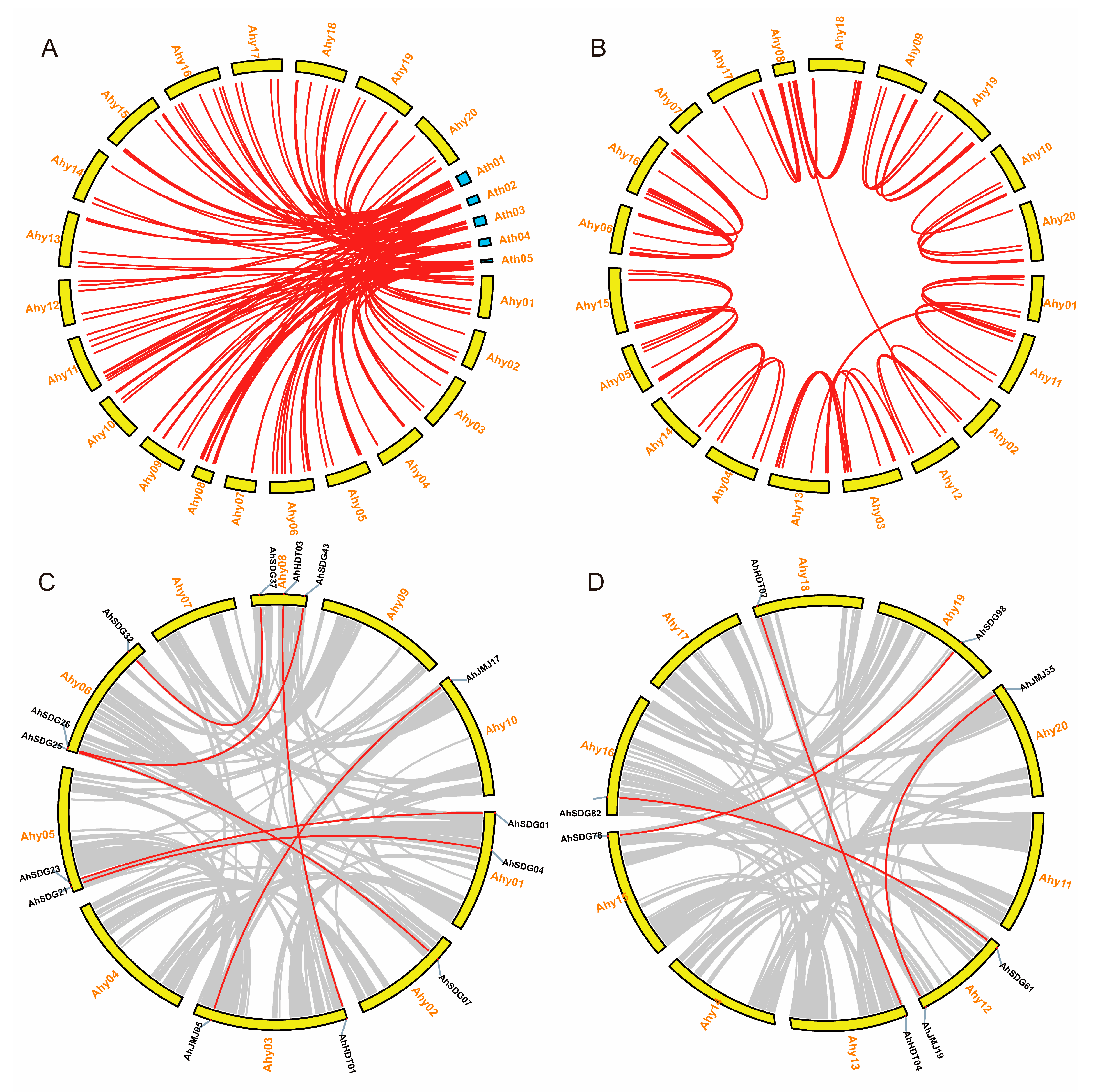
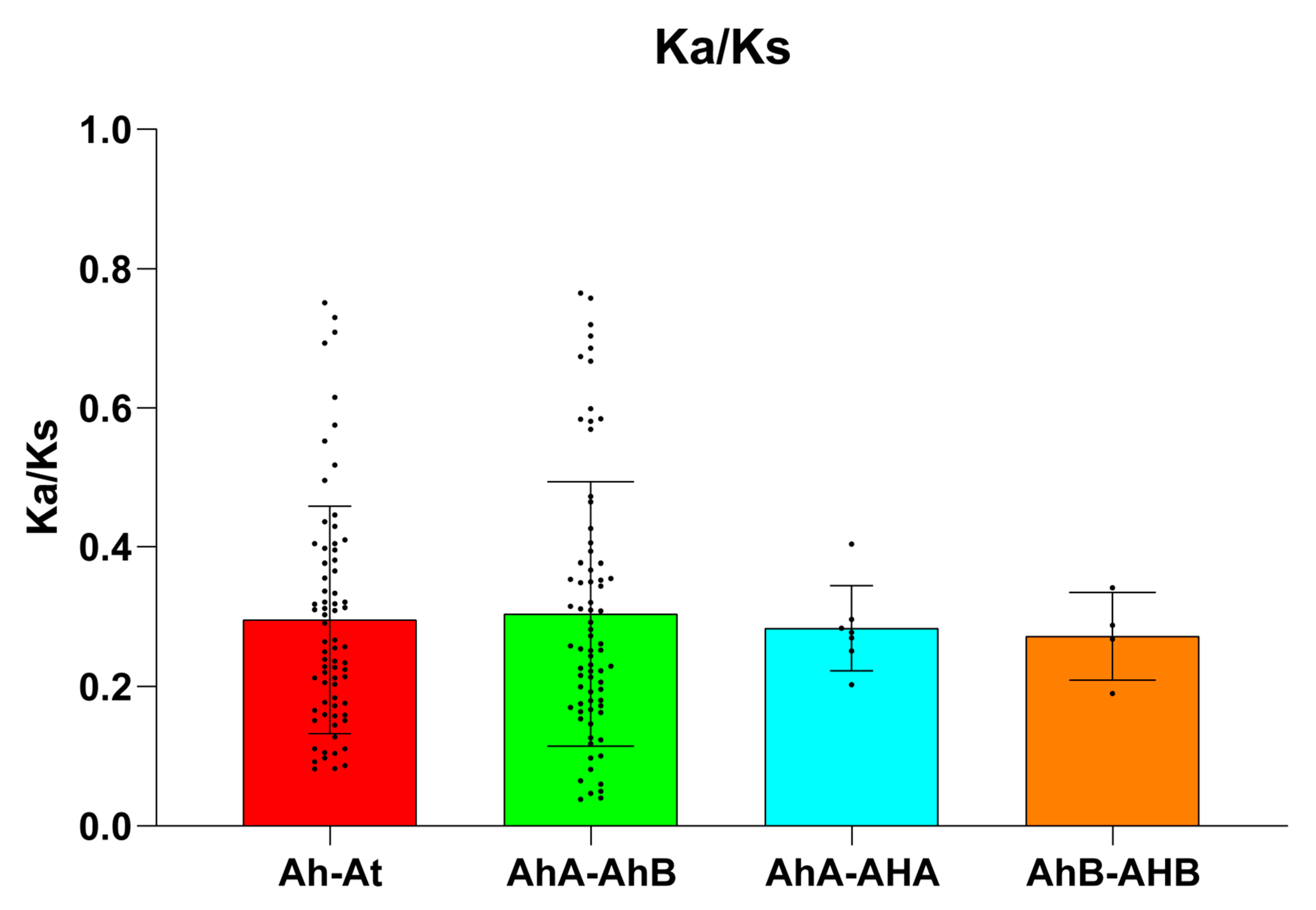
2.3. Synteny and Duplication of Histone Modification Genes
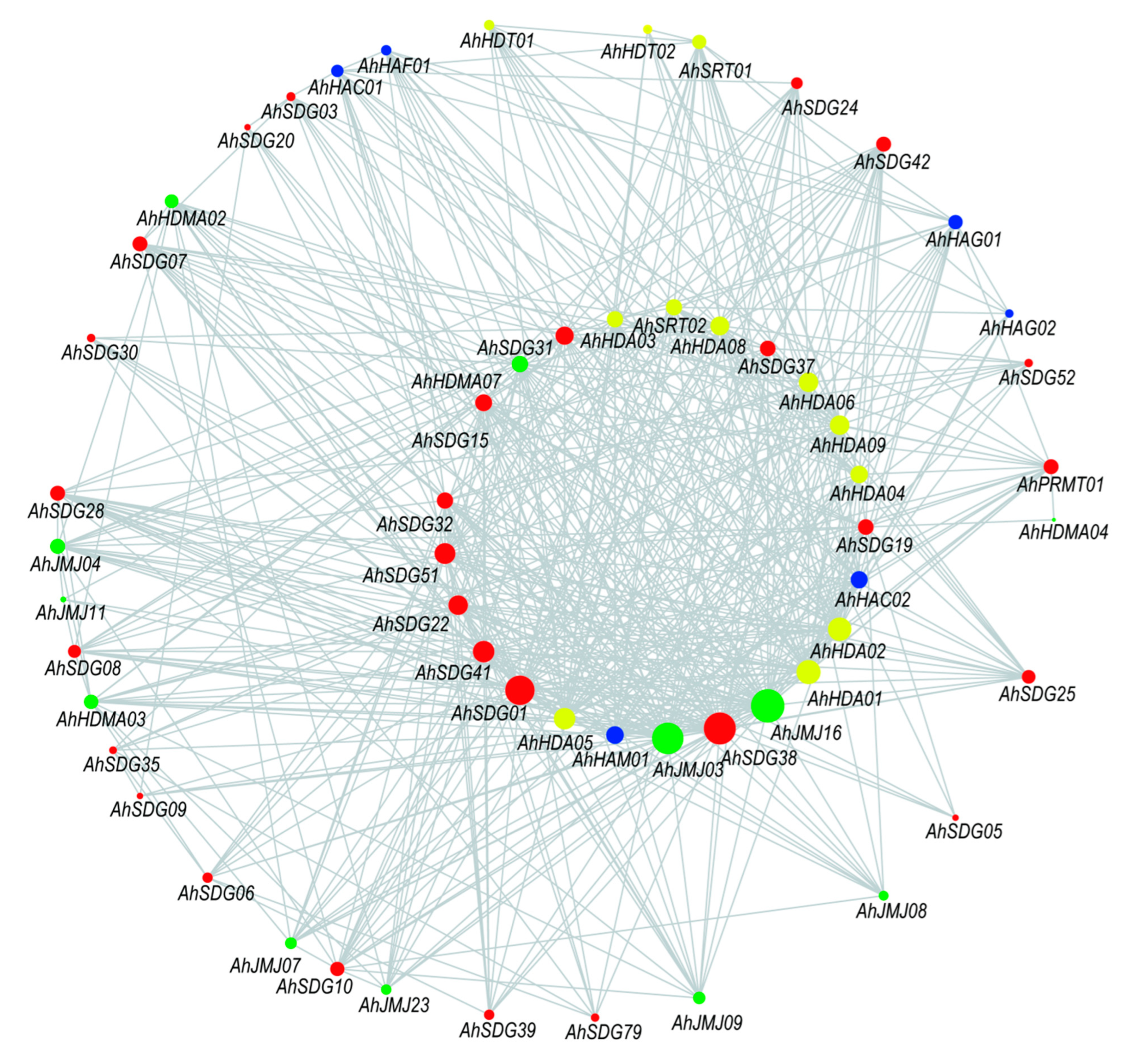
2.4. Potential Interaction of Peanut Histone Modification Genes
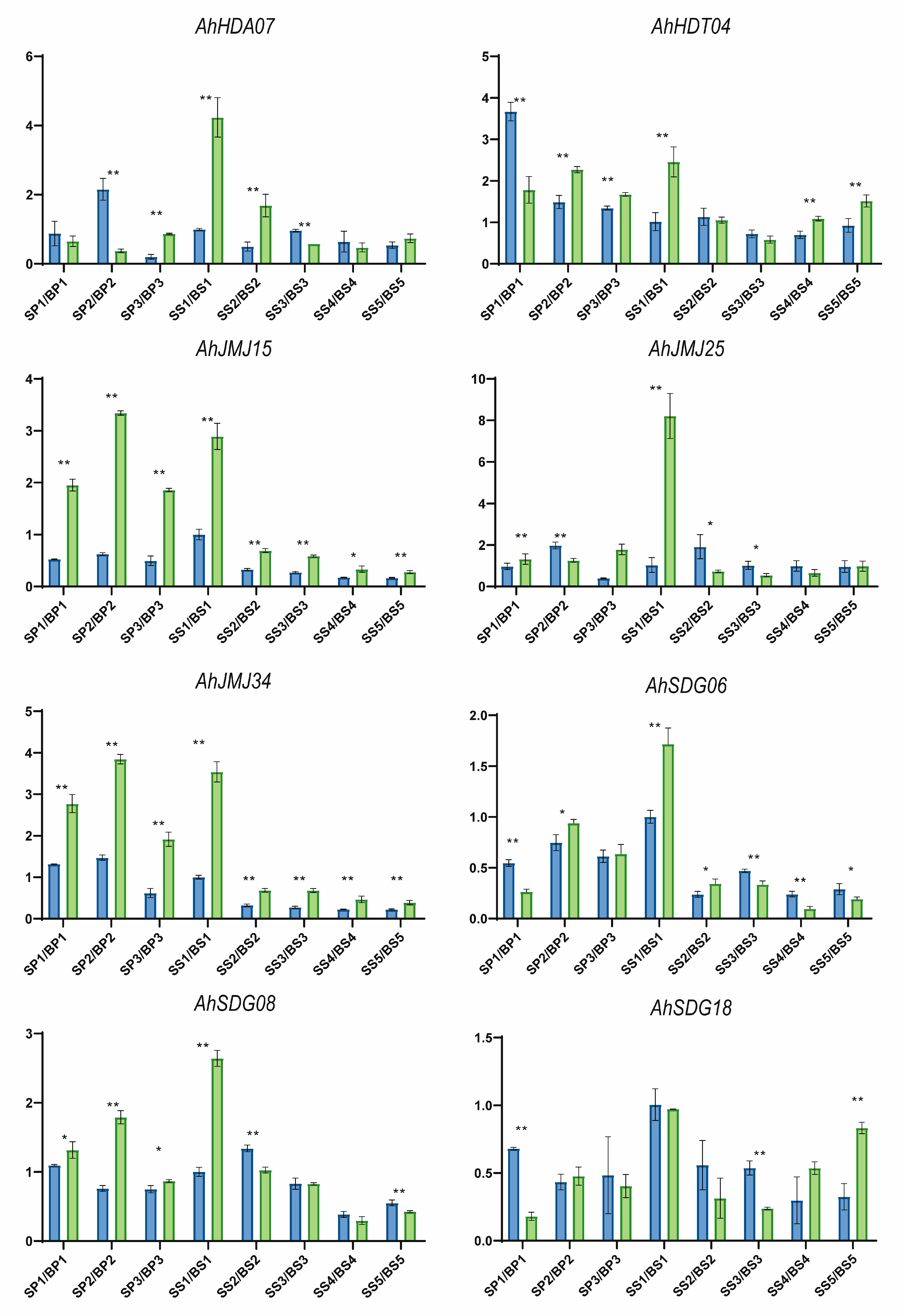
2.5. Identification of Histone Modification Genes Involved in Peanut Pod and Seed Development
3. Materials and Methods
3.1. Identification and Characterization of Histone Modification Genes in Peanut
3.2. Phylogenetic Analysis of Histone Modification Genes
3.3. Gene Structure and Motif Analysis of Peanut Histone Modification Genes
3.4. Synteny Analysis of Histone Modification Genes
3.5. Interaction Analysis of Peanut Histone Modification Genes
3.6. Expression Profiling of Histone Modification Genes During Peanut Seed and Pod Development
3.7. Total RNA Extraction and Quantitative Real-Time PCR Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yashar, W.M.; Kong, G.; VanCampen, J.; Curtiss, B.M.; Coleman, D.J.; Carbone, L.; Yardimci, G.G.; Maxson, J.E.; Braun, T.P. GoPeaks: Histone modification peak calling for CUT&Tag. Genome Biol. 2022, 23, 144. [Google Scholar]
- Lopez-Hernandez, L.; Toolan-Kerr, P.; Bannister, A.J.; Millan-Zambrano, G. Dynamic histone modification patterns coordinating DNA processes. Mol. Cell 2025, 85, 225–237. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Huang, Y.; Hao, Y.; Song, X.; Zhu, T.; Liu, C.; Li, C. The HISTONE ACETYLTRANSFERASE 1 interacts with CONSTANS to promote flowering in Arabidopsis. J. Genet. Genom. 2025; in press. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, W.; Luan, S.; Xiang, X.; Guo, M.; Zhang, Y.; Li, G.; Li, X. Profiling plant histone modification at single-cell resolution using snCUT&Tag. Plant Biotechnol. J. 2022, 20, 420–422. [Google Scholar] [CrossRef]
- Lopez, L.; Perrella, G.; Calderini, O.; Porceddu, A.; Panara, F. Genome-wide identification of histone modification gene families in the model legume Medicago truncatula and their expression analysis in nodules. Plants 2022, 11, 322. [Google Scholar] [CrossRef]
- Xu, J.D.; Xu, H.D.; Liu, Y.L.; Wang, X.; Xu, Q.; Deng, X.X. Genome-wide identification of sweet orange (Citrus sinensis) histone modification gene families and their expression analysis during the fruit development and fruit-blue mold infection process. Front. Plant Sci. 2015, 6, 607. [Google Scholar] [CrossRef]
- Fan, S.; Wang, J.; Lei, C.; Gao, C.; Yang, Y.; Li, Y.M.; An, N.; Zhang, D.; Han, M.Y. Identification and characterization of histone modification gene family reveal their critical responses to flower induction in apple. BMC Plant Biol. 2018, 18, 173. [Google Scholar] [CrossRef]
- Zhou, L.; Yarra, R.; Jin, L.; Yang, Y.; Cao, H.; Zhao, Z. Identification and expression analysis of histone modification gene (HM) family during somatic embryogenesis of oil palm. BMC Genom. 2022, 23, 11. [Google Scholar] [CrossRef]
- Panara, F.; Fasano, C.; Lopez, L.; Porceddu, A.; Facella, P.; Fantini, E.; Daddiego, L.; Perrella, G. Genome-wide identification and spatial expression analysis of histone modification gene families in the rubber dandelion Taraxacum kok-saghyz. Plants 2022, 11, 2077. [Google Scholar] [CrossRef]
- Borchetia, S.; Gogoi, M.; Rawal, H.C.; Patel, P.K.; Chakraborty, M.; Saikia, H.; Nishad, J.; Ilango, V.J.; Barooah, A.K.; Mondal, T.K. Genome-wide identification of histone modification (HM) gene family and their expression patterns under abiotic stress and different developmental stages of tea (Camellia assamica). J. Plant Growth Regul. 2023, 42, 2960–2982. [Google Scholar] [CrossRef]
- Wang, L.; Ahmad, B.; Liang, C.; Shi, X.; Sun, R.; Zhang, S.; Du, G. Bioinformatics and expression analysis of histone modification genes in grapevine predict their involvement in seed development, powdery mildew resistance, and hormonal signaling. BMC Plant Biol. 2020, 20, 412. [Google Scholar] [CrossRef] [PubMed]
- Hu, P.; Zhao, H.; Zhu, P.; Xiao, Y.; Miao, W.; Wang, Y.; Jin, H. Dual regulation of Arabidopsis AGO2 by arginine methylation. Nat. Commun. 2019, 10, 844. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Berr, A.; Chang, C.; Liu, C.; Shen, W.H.; Ruan, Y. Interplay of the histone methyltransferases SDG8 and SDG26 in the regulation of transcription and plant flowering and development. Biochim. Biophys. Acta 2016, 1859, 581–590. [Google Scholar] [CrossRef]
- Wang, Y.; Gu, D.; Deng, L.; He, C.; Zheng, F.; Liu, X. The Histone H3K27 demethylase REF6 is a positive regulator of light-initiated seed germination in Arabidopsis. Cells 2023, 12, 295. [Google Scholar] [CrossRef]
- Zhao, M.; Yang, S.; Liu, X.; Wu, K. Arabidopsis histone demethylases LDL1 and LDL2 control primary seed dormancy by regulating DELAY OF GERMINATION 1 and ABA signaling-related genes. Front. Plant Sci. 2015, 6, 159. [Google Scholar] [CrossRef]
- Luo, Y.; Shi, D.Q.; Jia, P.F.; Bao, Y.; Li, H.J.; Yang, W.C. Nucleolar histone deacetylases HDT1, HDT2, and HDT3 regulate plant reproductive development. J. Genet. Genom. 2022, 49, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Bai, X.; Cheng, N.; Xiao, J.; Li, X.; Xing, Y. Wide Grain 7 increases grain width by enhancing H3K4me3 enrichment in the OsMADS1 promoter in rice (Oryza sativa L.). Plant J. 2020, 102, 517–528. [Google Scholar] [CrossRef]
- He, C.; Bi, S.; Li, Y.; Song, C.; Zhang, H.; Xu, X.; Li, Q.; Saeed, S.; Chen, W.; Zhao, C.; et al. Dynamic atlas of histone modifications and gene regulatory networks in endosperm of bread wheat. Nat. Commun. 2024, 15, 9572. [Google Scholar] [CrossRef]
- Moreno-Pérez, A.J.; Santos-Pereira, J.M.; Martins-Noguerol, R.; DeAndrés-Gil, C.; Troncoso-Ponce, M.A.; Venegas-Calerón, M.; Sánchez, R.; Garcés, R.; Salas, J.J.; Tena, J.J.; et al. Genome-wide mapping of histone H3 lysine 4 trimethylation (H3K4me3) and its involvement in fatty acid biosynthesis in sunflower developing seeds. Plants 2021, 10, 706. [Google Scholar] [CrossRef]
- Bertioli, D.; Jenkins, J.; Clevenger, J.; Dudchenko, O.; Gao, D.; Seijo, G.; Leal-Bertioli, S.C.M.; Ren, L.; Farmer, A.D.; Pandey, M.K.; et al. The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat. Genet. 2019, 51, 877–884. [Google Scholar] [CrossRef]
- Li, C.L.; Wu, K.Q.; Fu, G.H.; Li, Y.; Zhong, Y.J.; Lin, X.D.; Zhou, Y.; Tian, L.N.; Huang, S.Z. Regulation of oleosin expression in developing peanut (Arachis hypogaea L.) embryos through nucleosome loss and histone modifications. J. Exp. Bot. 2009, 60, 4371–4382. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Su, L.C.; Liu, S.; Liu, X.; Zhang, B.H.; Li, M.J.; Zeng, L.D.; Li, L. Transcriptome profiling reveals histone deacetylase 1 gene overexpression improves flavonoid, isoflavonoid, and phenylpropanoid metabolism in Arachis hypogaea hairy roots. PeerJ 2021, 9, e10976. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.H.; Su, L.C.; Hu, B.; Li, L. Expression of AhDREB1, an AP2/ERF transcription factor gene from peanut, is affected by histone acetylation and increases abscisic acid sensitivity and tolerance to osmotic Stress in Arabidopsis. Int. J. Mol. Sci. 2018, 19, 1441. [Google Scholar] [CrossRef]
- Cigliano, R.A.; Sanseverino, W.; Cremona, G.; Ercolano, M.R.; Conicella, C.; Consiglio, F.M. Genome-wide analysis of histone modifiers in tomato: Gaining an insight into their developmental roles. BMC Genom. 2013, 14, 57. [Google Scholar] [CrossRef] [PubMed]
- Das, M.; Haberer, G.; Panda, A.; Das Laha, S.; Ghosh, T.C.; Schäffner, A.R. Expression pattern similarities support the prediction of orthologs retaining common functions after gene duplication events. Plant Physiol. 2016, 171, 2343–2357. [Google Scholar] [CrossRef]
- Zhao, Y.; Ma, R.; Xu, D.; Bi, H.; Xia, Z.; Peng, H. Genome-wide identification and analysis of the AP2 transcription factor gene family in wheat (Triticum aestivum L.). Front. Plant Sci. 2019, 10, 1286. [Google Scholar] [CrossRef]
- Lyons, E.; Pedersen, B.; Kane, J.; Alam, M.; Ming, R.; Tang, H.; Wang, X.; Bowers, J.; Paterson, A.; Lisch, D.; et al. Finding and comparing syntenic regions among Arabidopsis and the outgroups papaya, poplar, and grape: CoGe with rosids. Plant Physiol. 2008, 148, 1772–1781. [Google Scholar] [CrossRef]
- Morgan, C.C.; Loughran, N.B.; Walsh, T.A.; Harrison, A.J.; O’Connell, M.J. Positive selection neighboring functionally essential sites and disease-implicated regions of mammalian reproductive proteins. BMC Evol. Biol. 2010, 10, 39. [Google Scholar] [CrossRef]
- Mou, Y.; Liu, Y.; Tian, S.; Guo, Q.; Wang, C.; Wen, S. Genome-Wide identification and characterization of the OPR Gene family in wheat (Triticum aestivum L.). Int. J. Mol. Sci. 2019, 20, 1914. [Google Scholar] [CrossRef]
- Gao, S.; Li, L.; Han, X.; Liu, T.; Jin, P.; Cai, L.; Xu, M.; Zhang, T.; Zhang, F.; Chen, J.; et al. Genome-wide identification of the histone acetyltransferase gene family in Triticum aestivum. BMC Genom. 2021, 22, 49. [Google Scholar] [CrossRef]
- Zhuang, Y.; Wang, X.; Li, X.; Hu, J.; Fan, L.; Landis, J.B.; Cannon, S.B.; Grimwood, J.; Schmutz, J.; Jackson, S.A.; et al. Phylogenomics of the genus Glycine sheds light on polyploid evolution and life-strategy transition. Nat. Plants 2022, 8, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Guo, C.; Shan, H.; Kong, H. Divergence of duplicate genes in exon–intron structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1187–1192. [Google Scholar] [CrossRef]
- Si, W.; Hang, T.; Guo, M.; Chen, Z.; Liang, Q.; Gu, L.; Ding, T. Whole-genome and transposed duplication contributes to the expansion and diversification of TLC genes in maize. Int. J. Mol. Sci. 2019, 20, 5484. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef]
- Keyzor, C.; Mermaz, B.; Trigazis, E.; Jo, S.; Song, J. Histone demethylases ELF6 and JMJ13 antagonistically regulate self-fertility in Arabidopsis. Front. Plant Sci. 2021, 12, 640135. [Google Scholar] [CrossRef]
- Pan, J.; Zhang, H.; Zhan, Z.; Zhao, T.; Jiang, D. A REF6-dependent H3K27me3-depleted state facilitates gene activation during germination in Arabidopsis. J. Genet. Genom. 2023, 50, 178–191. [Google Scholar] [CrossRef]
- Zhou, Y.; Yang, P.; Zhang, F.; Luo, X.; Xie, J. Histone deacetylase HDA19 interacts with histone methyltransferase SUVH5 to regulate seed dormancy in Arabidopsis. Plant. Biol. 2020, 22, 1062–1071. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2018, 47, D427–D432. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Tang, H.; Wang, X.; Bowers, J.E.; Ming, R.; Alam, M.; Paterson, A.H. Unraveling ancient hexaploidy through multiply-aligned angiosperm gene maps. Genome Res. 2008, 18, 1944–1954. [Google Scholar] [CrossRef]
- Tang, H.; Bowers, J.E.; Wang, X.; Ming, R.; Alam, M.; Paterson, A.H. Synteny and collinearity in plant genomes. Science 2008, 320, 486–488. [Google Scholar] [CrossRef]
- Tang, H.; Bowers, J.E.; Wang, X.; Paterson, A.H. Angiosperm genome comparisons reveal early polyploidy in the monocot lineage. Proc. Natl. Acad. Sci. USA 2010, 107, 472–477. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Bertioli, D.; Cannon, S.B.; Froenicke, L.; Huang, G.; Farmer, A.D.; Cannon, E.K.; Liu, X.; Gao, D.; Clevenger, J.; Dash, S.; et al. The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 2016, 48, 438–446. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Su, G.; Morris, J.H.; Demchak, B.; Bader, G.D. Biological network exploration with Cytoscape 3. Curr. Protoc. Bioinform. 2014, 47, 8.13.1–8.13.24. [Google Scholar] [CrossRef]
- Luo, H.; Guo, J.; Ren, X.; Chen, W.; Huang, L.; Zhou, X.; Chen, Y.; Liu, N.; Xiong, F.; Lei, Y.; et al. Chromosomes A07 and A05 associated with stable and major QTLs for pod weight and size in cultivated peanut (Arachis hypogaea L.). Theor. Appl. Genet. 2018, 131, 267–282. [Google Scholar] [CrossRef]
- Li, Y.M.; Zhang, D.; Xing, L.B.; Fan, S.; Ma, J.J.; Zhao, C.P.; Du, L.S.; Han, M.Y. A transcriptome analysis of two apple (Malus × domestica) cultivars with different flowering abilities reveals a gene network module associate with floral transitions. Sci. Hortic. 2018, 239, 269–281. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−△△CT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
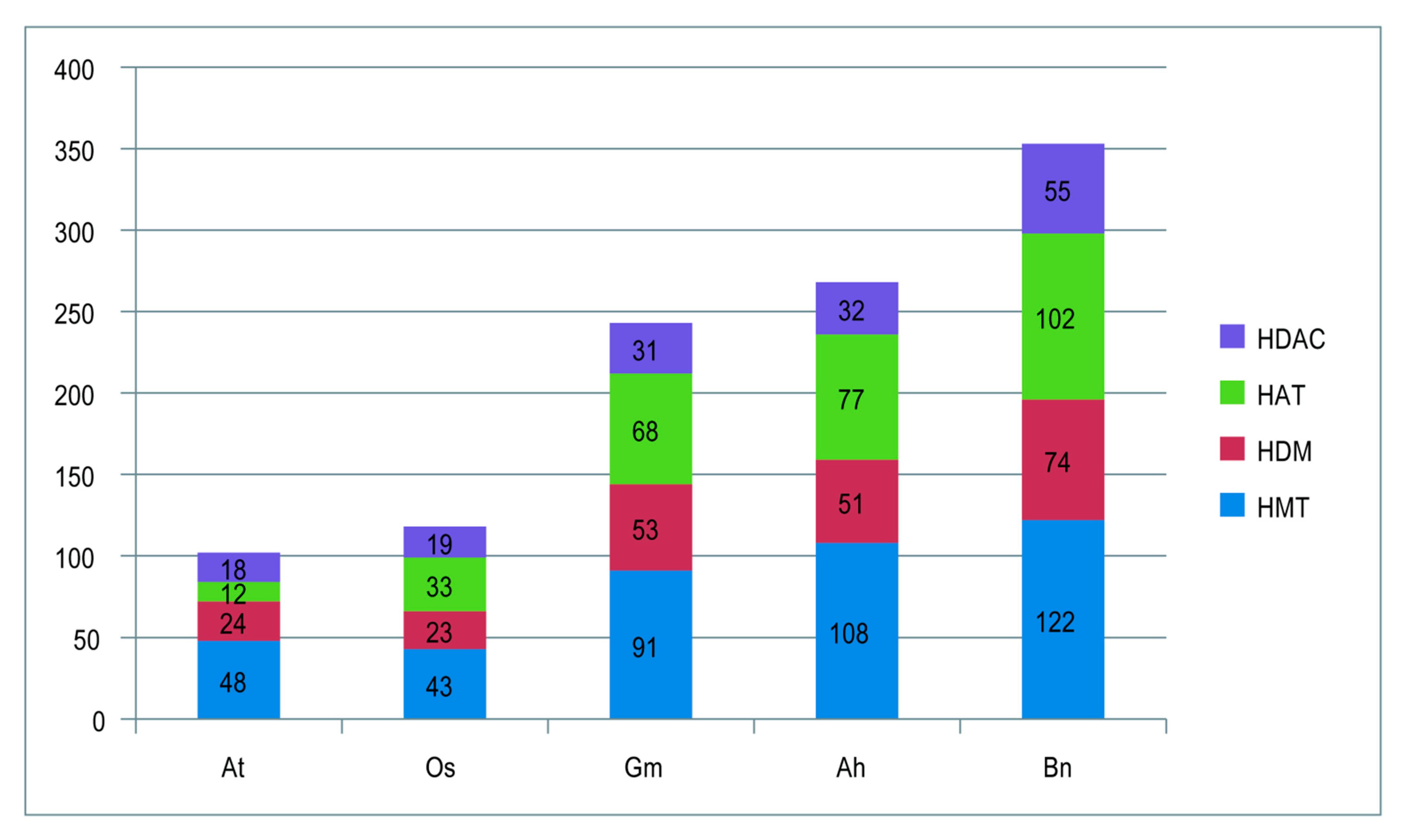

| HMT | HDM | HAT | HDAC | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr | SDG | PRMT | JMJ | HDMA | HAG | HAM | HAC | HAF | HDA | HDT | SRT | Total |
| Chr01 | 6 | 1 | 4 | 1 | 12 | |||||||
| Chr02 | 3 | 1 | 4 | |||||||||
| Chr03 | 3 | 4 | 1 | 2 | 1 | 4 | 1 | 16 | ||||
| Chr04 | 7 | 1 | 8 | |||||||||
| Chr05 | 5 | 2 | 2 | 1 | 10 | |||||||
| Chr06 | 10 | 3 | 1 | 2 | 1 | 1 | 18 | |||||
| Chr07 | 2 | 1 | 3 | |||||||||
| Chr08 | 7 | 1 | 1 | 4 | 1 | 1 | 1 | 16 | ||||
| Chr09 | 6 | 3 | 1 | 10 | ||||||||
| Chr10 | 5 | 1 | 1 | 7 | ||||||||
| Chr11 | 6 | 1 | 3 | 1 | 11 | |||||||
| Chr12 | 5 | 1 | 6 | |||||||||
| Chr13 | 3 | 4 | 1 | 2 | 1 | 3 | 2 | 16 | ||||
| Chr14 | 5 | 1 | 1 | 7 | ||||||||
| Chr15 | 6 | 3 | 2 | 1 | 12 | |||||||
| Chr16 | 9 | 3 | 1 | 1 | 1 | 15 | ||||||
| Chr17 | 3 | 2 | 1 | 6 | ||||||||
| Chr18 | 3 | 1 | 2 | 2 | 2 | 1 | 11 | |||||
| Chr19 | 6 | 4 | 10 | |||||||||
| Chr20 | 6 | 1 | 1 | 1 | 9 | |||||||
| Total | 106 | 2 | 35 | 16 | 4 | 6 | 4 | 2 | 20 | 8 | 4 | 207 |
| Family Name | Subfamily Name | Accession Number |
|---|---|---|
| HMTs | SDGs | PF00856 |
| PRMTs | PF05185 | |
| HDMs | HDMAs | PF04433 |
| JMJs | PF02373 | |
| HATs | HAGs | PF00583 |
| HAMs | PF01853 | |
| HACs | PF08214 | |
| HAFs | PF09247 | |
| HDACs | HDAs | PF00850 |
| SRTs | PF02146 | |
| HDTs | / |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, Y.; Gelaye, Y.; Yao, R.; Yang, P.; Li, J.; Liu, N.; Huang, L.; Zhou, X.; Chen, W.; Yu, B.; et al. Identification and Characterization of Histone Modification Gene Families and Their Expression Patterns During Pod and Seed Development in Peanut. Int. J. Mol. Sci. 2025, 26, 2591. https://doi.org/10.3390/ijms26062591
Chang Y, Gelaye Y, Yao R, Yang P, Li J, Liu N, Huang L, Zhou X, Chen W, Yu B, et al. Identification and Characterization of Histone Modification Gene Families and Their Expression Patterns During Pod and Seed Development in Peanut. International Journal of Molecular Sciences. 2025; 26(6):2591. https://doi.org/10.3390/ijms26062591
Chicago/Turabian StyleChang, Yingying, Yohannes Gelaye, Ruonan Yao, Ping Yang, Jihua Li, Nian Liu, Li Huang, Xiaojing Zhou, Weigang Chen, Bolun Yu, and et al. 2025. "Identification and Characterization of Histone Modification Gene Families and Their Expression Patterns During Pod and Seed Development in Peanut" International Journal of Molecular Sciences 26, no. 6: 2591. https://doi.org/10.3390/ijms26062591
APA StyleChang, Y., Gelaye, Y., Yao, R., Yang, P., Li, J., Liu, N., Huang, L., Zhou, X., Chen, W., Yu, B., Jiang, H., Liao, B., Lei, Y., & Luo, H. (2025). Identification and Characterization of Histone Modification Gene Families and Their Expression Patterns During Pod and Seed Development in Peanut. International Journal of Molecular Sciences, 26(6), 2591. https://doi.org/10.3390/ijms26062591








