Selection and Validation of Reference Genes in Clinacanthus nutans Under Abiotic Stresses, MeJA Treatment, and in Different Tissues
Abstract
1. Introduction
2. Results
2.1. PCR Specificity and Amplification Efficiency of Candidate Reference Genes
2.2. Expression Profiles of Candidate Reference Genes
2.3. Expression Stability of Candidate Reference Genes
2.4. Comprehensive Ranking of the Candidate Reference Genes
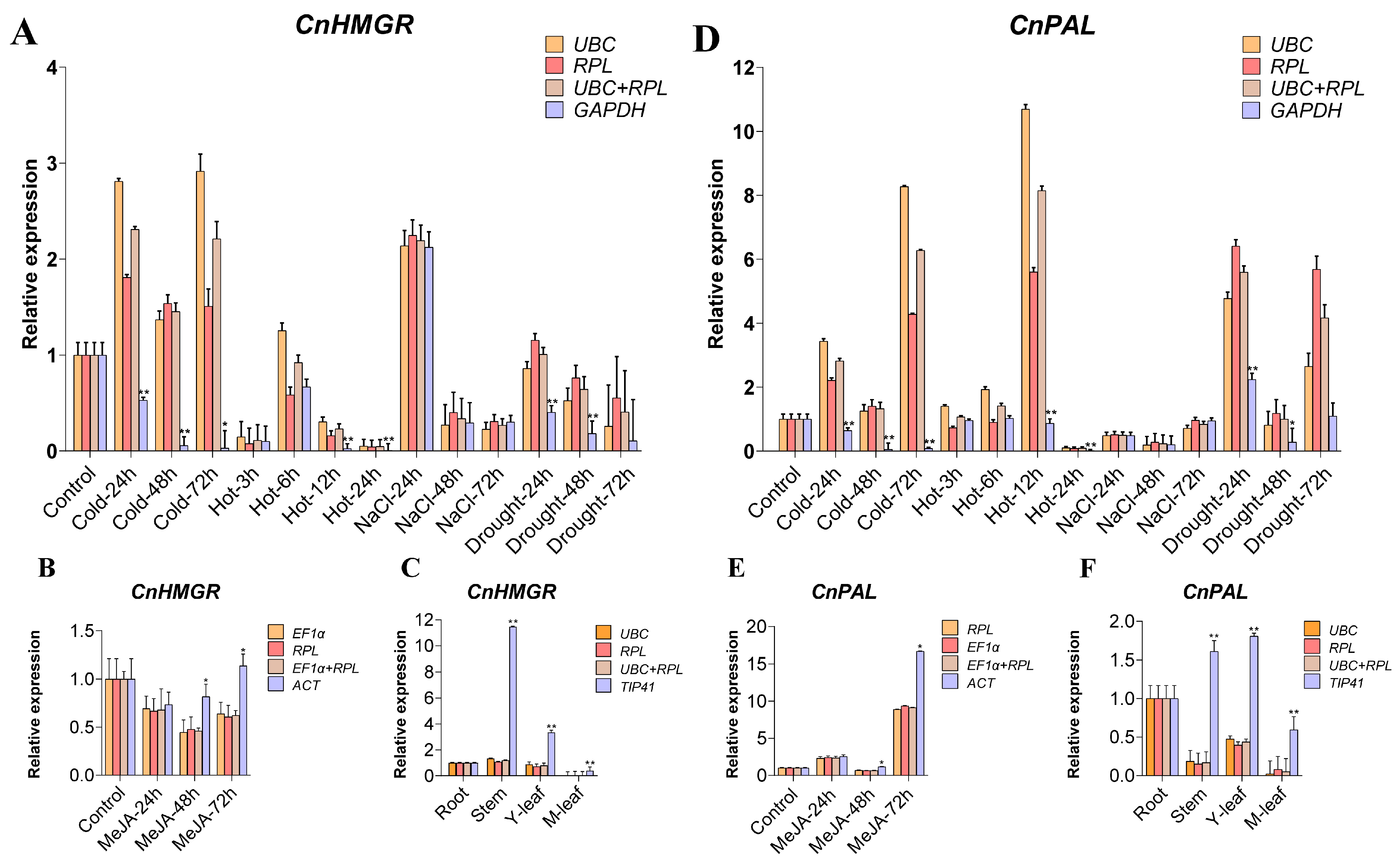
2.5. Validation of the Recommended Candidate Reference Genes
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Stresses/MeJA Treatments
4.2. RNA Isolation and cDNA Synthesis
4.3. Selection of Potential Reference Genes and Primer Design
4.4. Primer’s Specificity, Amplification Efficiency and qRT-PCR Analysis
4.5. Expression Stability Analysis of Candidate Reference Genes
4.6. qRT-PCR Validation of Selected Reference Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alam, A.; Ferdosh, S.; Ghafoor, K.; Hakim, A.; Juraimi, A.S.; Khatib, A.; Sarker, Z.I. Clinacanthus nutans: A review of the medicinal uses, pharmacology and phytochemistry. Asian Pac. J. Trop. Med. 2016, 9, 402–409. [Google Scholar] [CrossRef] [PubMed]
- Zulkipli, I.N.; Rajabalaya, R.; Idris, A.; Sulaiman, N.A.; David, S.R. Clinacanthus nutans: A review on ethnomedicinal uses, chemical constituents and pharmacological properties. Pharm. Biol. 2017, 55, 1093–1113. [Google Scholar] [CrossRef] [PubMed]
- Kamarudin, M.N.A.; Sarker, M.M.R.; Kadir, H.A.; Ming, L.C. Ethnopharmacological uses, phytochemistry, biological activities, and therapeutic applications of Clinacanthus nutans (Burm. f.) Lindau: A comprehensive review. J. Ethnopharmacol. 2017, 206, 245–266. [Google Scholar] [CrossRef]
- Zakaria, Z.A.; Abdul Rahim, M.H.; Roosli, R.A.J.; Mohd Sani, M.H.; Omar, M.H.; Othman, F.; Ching, S.M.; Abdul Kadir, A. Antinociceptive activity of methanolic extract of Clinacanthus nutans leaves: Possible mechanisms of action involved. Pain Res. Manag. 2018, 2018, 9536406. [Google Scholar] [CrossRef]
- Le, C.-F.; Kailaivasan, T.H.; Chow, S.-C.; Abdullah, Z.; Ling, S.-K.; Fang, C.-M. Phytosterols isolated from Clinacanthus nutans induce immunosuppressive activity in murine cells. Int. Immunopharmacol. 2017, 44, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Sakdarat, S.; Shuyprom, A.; Pientong, C.; Ekalaksananan, T.; Thongchai, S. Bioactive constituents from the leaves of Clinacanthus nutans Lindau. Bioorganic Med. Chem. 2009, 17, 1857–1860. [Google Scholar] [CrossRef]
- Mustapa, A.N.; Martin, Á.; Mato, R.B.; Cocero, M.J. Extraction of phytocompounds from the medicinal plant Clinacanthus nutans Lindau by microwave-assisted extraction and supercritical carbon dioxide extraction. Ind. Crops Prod. 2015, 74, 83–94. [Google Scholar] [CrossRef]
- Bong, F.J.; Chear, N.J.Y.; Ramanathan, S.; Mohana-Kumaran, N.; Subramaniam, S.; Chew, B.L. The development of callus and cell suspension cultures of Sabah Snake Grass (Clinacanthus nutans) for the production of flavonoids and phenolics. Biocatal. Agric. Biotechnol. 2021, 33, 101977. [Google Scholar] [CrossRef]
- Ong, W.-Y.; Herr, D.R.; Sun, G.Y.; Lin, T.-N. Anti-inflammatory effects of phytochemical components of Clinacanthus nutans. Molecules 2022, 27, 3607. [Google Scholar] [CrossRef]
- Haida, Z.; Nakasha, J.J.; Hakiman, M. In vitro responses of plant growth factors on growth, yield, phenolics content and antioxidant activities of Clinacanthus nutans (Sabah Snake Grass). Plants 2020, 9, 1030. [Google Scholar] [CrossRef]
- Md, K.U.; Shamsuzzaman, S.M.; Zi, L.; Mohdselamat, M.; Mahmudul, H. Effects of salinity on growth, antioxidant contents and proximate compositions of Sabah snake grass (Clinacanthus nutans (Burm. F.) Lindau). Bangladesh J. Bot. 2017, 46, 263–269. [Google Scholar]
- Ghasemzadeh, A.; Nasiri, A.; Jaafar, H.Z.; Baghdadi, A.; Ahmad, I. Changes in phytochemical synthesis, chalcone synthase activity and pharmaceutical qualities of Sabah snake grass (Clinacanthus nutans L.) in relation to plant age. Molecules 2014, 19, 17632–17648. [Google Scholar] [CrossRef]
- Fong, S.Y.; Piva, T.; Dekiwadia, C.; Urban, S.; Huynh, T. Comparison of cytotoxicity between extracts of Clinacanthus nutans (Burm. f.) Lindau leaves from different locations and the induction of apoptosis by the crude methanol leaf extract in D24 human melanoma cells. BMC Complement. Altern. Med. 2016, 16, 368. [Google Scholar] [CrossRef]
- Liu, R.; Wang, Z.; Zheng, J.; Xu, Z.; Tang, X.; Huang, Z.; Zhang, N.; Dong, Y.; Li, T. The effects of methyl jasmonate on growth, gene expression and metabolite accumulation in Isatis indigotica Fort. Ind. Crops Prod. 2022, 177, 114482. [Google Scholar] [CrossRef]
- Prinsloo, G.; Nogemane, N. The effects of season and water availability on chemical composition, secondary metabolites and biological activity in plants. Phytochem. Rev. 2018, 17, 889–902. [Google Scholar] [CrossRef]
- Orek, C. A review of the functions of transcription factors and related genes involved in cassava (Manihot Esculenta Crantz) response to drought stress. Trop. Plants 2023, 2, 14. [Google Scholar] [CrossRef]
- Li, Y.; Kong, D.; Fu, Y.; Sussman, M.R.; Wu, H. The effect of developmental and environmental factors on secondary metabolites in medicinal plants. Plant Physiol. Biochem. 2020, 148, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, K.; Urano, K.; Kusano, M.; Sakurai, T.; Takasaki, H.; Kishimoto, M.; Yoshiwara, K.; Kobayashi, M.; Kojima, M.; Sakakibara, H. Metabolite/phytohormone–gene regulatory networks in soybean organs under dehydration conditions revealed by integration analysis. Plant J. 2020, 103, 197–211. [Google Scholar] [CrossRef]
- Jeon, J.; Kim, J.K.; Wu, Q.; Park, S.U. Effects of cold stress on transcripts and metabolites in tartary buckwheat (Fagopyrum tataricum). Environ. Exp. Bot. 2018, 155, 488–496. [Google Scholar] [CrossRef]
- Yang, J.; Guo, C.; Chen, F.; Lv, B.; Song, J.; Ning, G.; He, Y.; Lin, J.; He, H.; Yang, Y. Heat-induced modulation of flavonoid biosynthesis via a lhmybc2-mediated regulatory network in oriental hybrid lily. Plant Physiol. Biochem. 2024, 214, 108966. [Google Scholar] [CrossRef]
- He, J.; Yao, L.; Pecoraro, L.; Liu, C.; Wang, J.; Huang, L.; Gao, W. Cold stress regulates accumulation of flavonoids and terpenoids in plants by phytohormone, transcription process, functional enzyme, and epigenetics. Crit. Rev. Biotechnol. 2023, 43, 680–697. [Google Scholar] [CrossRef] [PubMed]
- Chu, H.Y.; Wegel, E.; Osbourn, A. From hormones to secondary metabolism: The emergence of metabolic gene clusters in plants. Plant J. 2011, 66, 66–79. [Google Scholar] [CrossRef]
- Xiong, M.; Feng, G.-N.; Gao, Q.; Zhang, C.-Q.; Li, Q.-F.; Liu, Q.-Q. Brassinosteroid regulation in rice seed biology. Seed Biol. 2022, 1, 2. [Google Scholar] [CrossRef]
- Xia, Q.; Wang, Z.; Chen, X.; Dong, X.; Cheng, S.; Zhang, S. Effects on the synthesis and accumulation of triterpenes in leaves of Cyclocarya paliurus under MeJA treatment. Forests 2023, 14, 1735. [Google Scholar] [CrossRef]
- Shabani, L.; Ehsanpour, A.A.; Asghari, G.; Emami, J. Glycyrrhizin production by in vitro cultured Glycyrrhiza glabra elicited by methyl jasmonate and salicylic acid. Russ. J. Plant Physiol. 2009, 56, 621–626. [Google Scholar] [CrossRef]
- Li, Y.; Chen, X.; Wang, J.; Zou, G.; Wang, L.; Li, X. Two responses to MeJA induction of R2R3-MYB transcription factors regulate flavonoid accumulation in Glycyrrhiza uralensis Fisch. PLoS ONE 2020, 15, e0236565. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Lin, Y.; Qiu, Z.; Xiang, X.; Shao, D.; Li, Y.; Wu, S. Reference gene selection for qRT-PCR normalization of gene expression analysis in Melaleuca bracteata F. Muell. under abiotic stresses and hormonal stimuli. Sci. Hortic. 2023, 319, 112184. [Google Scholar] [CrossRef]
- Huang, Y.; Tan, H.; Yu, J.; Chen, Y.; Guo, Z.; Wang, G.; Zhang, Q.; Chen, J.; Zhang, L.; Diao, Y. Stable internal reference genes for normalizing real-time quantitative PCR in Baphicacanthus cusia under hormonal stimuli and UV irradiation, and in different plant organs. Front. Plant Sci. 2017, 8, 668. [Google Scholar] [CrossRef]
- Wei, L.; Miao, H.; Zhao, R.; Han, X.; Zhang, T.; Zhang, H. Identification and testing of reference genes for Sesame gene expression analysis by quantitative real-time PCR. Planta 2013, 237, 873–889. [Google Scholar] [CrossRef]
- Nolan, T.; Hands, R.E.; Bustin, S.A. Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 2006, 1, 1559–1582. [Google Scholar] [CrossRef]
- Hu, Y.; Chen, H.; Luo, C.; Dong, L.; Zhang, S.; He, X.; Huang, G. Selection of reference genes for real-time quantitative PCR studies of kumquat in various tissues and under abiotic stress. Sci. Hortic. 2014, 174, 207–216. [Google Scholar] [CrossRef]
- Zhou, X.; Liu, J.; Zhuang, Y. Selection of appropriate reference genes in eggplant for quantitative gene expression studies under different experimental conditions. Sci. Hortic. 2014, 176, 200–207. [Google Scholar] [CrossRef]
- Chen, C.; Wu, J.; Hua, Q.; Tel-Zur, N.; Xie, F.; Zhang, Z.; Chen, J.; Zhang, R.; Hu, G.; Zhao, J. Identification of reliable reference genes for quantitative real-time PCR normalization in pitaya. Plant Methods 2019, 15, 70. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, C.; Harijati, N.; Diao, Y.; Liu, E.; Hu, Z. Selection and Evaluation of Reference Genes for RT-qPCR Analysis in Amorphophallus Konjac Based on Transcriptome Data. Genes 2023, 14, 1513. [Google Scholar] [CrossRef]
- Zhao, M.; Fan, H.; Tu, Z.; Cai, G.; Zhang, L.; Li, A.; Xu, M. Stable reference gene selection for quantitative real-time PCR normalization in passion fruit (Passiflora edulis Sims.). Mol. Biol. Rep. 2022, 49, 5985–5995. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.-M.; Zhang, W.; Zhang, S.-B. Selection and validation of reference genes for quantitative real-time PCR analysis of development and tissue-dependent flower color formation in Cymbidium lowianum. Int. J. Mol. Sci. 2022, 23, 738. [Google Scholar] [CrossRef]
- Ye, X.; Zhang, F.; Tao, Y.; Song, S.; Fang, J. Reference gene selection for quantitative real-time PCR normalization in different cherry genotypes, developmental stages and organs. Sci. Hortic. 2015, 181, 182–188. [Google Scholar] [CrossRef]
- Xu, X.; Yang, Z.; Sun, X.; Zhang, L.; Fang, Z. Selection of reference genes for quantitative real-time PCR during flower bud development in CMS7311 of heading Chinese cabbage (Brassica rapa L. ssp. pekinensis). Acta Physiol. Plant. 2014, 36, 809–814. [Google Scholar] [CrossRef]
- An, C.; Liao, J.; Lu, L.; Cai, X.; Liu, R.; Chen, S.; Shen, M.; Wang, X.; Qin, Y.; Zheng, P. From gene expression to flower patterns: Genome-wide characterization of the MADS-box gene family in passion fruit (Passiflora edulis). Trop. Plants 2024, 3, e004. [Google Scholar] [CrossRef]
- Liang, J.; Fang, Y.; An, C.; Yao, Y.; Wang, X.; Zhang, W.; Liu, R.; Wang, L.; Aslam, M.; Cheng, Y. Genome-wide identification and expression analysis of the bHLH gene family in passion fruit (Passiflora edulis) and its response to abiotic stress. Int. J. Biol. Macromol. 2023, 225, 389–403. [Google Scholar] [CrossRef]
- Klumb, E.K.; Rickes, L.N.; Braga, E.J.B.; Bianchi, V.J. Evaluation of stability and validation of reference genes for real time PCR expression studies in leaves and roots of Prunus spp. rootstocks under flooding. Sci. Hortic. 2019, 247, 310–319. [Google Scholar] [CrossRef]
- Prakash, H.; Shukla, P.; Ramesha, A.; Arunakumar, G.S.; Doss, S.G.; Ponnuvel, K.M. Evaluation of reference genes for accurate normalization of qPCR data under biotic stresses in mulberry (Morus indica L.). Sci. Hortic. 2024, 323, 112507. [Google Scholar] [CrossRef]
- Chang, Y.; Hu, S.; Xu, J.; Gong, H.; Guo, X.; Song, Q.; Gong, W.; Yuan, D. Identification of reference genes provides insights into the determinants of self-incompatibility in Camellia oleifera. Sci. Hortic. 2023, 321, 112301. [Google Scholar] [CrossRef]
- LI, J.-y.; Sun, M.-y.; XU, S.-q.; Yu, M.E.I.; Yan, G.U.; Fang, Z.; Wei, S.U.N.; Wang, J.-h. Screening of Reference Genes of Andrographis paniculata Under MeJA and Abiotic Stresses by Real-time Fluorescence-based Quantitative PCR. Chin. J. Exp. Tradit. Med. Formulae 2022, 28, 133–140. [Google Scholar]
- Vogt, T. Phenylpropanoid biosynthesis. Mol. Plant 2010, 3, 2–20. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, C.-J. Multifaceted regulations of gateway enzyme phenylalanine ammonia-lyase in the biosynthesis of phenylpropanoids. Mol. Plant 2015, 8, 17–27. [Google Scholar] [CrossRef]
- Liu, Y.; Tang, Y.; Zhang, W.; Liang, R.; Luo, K.; Jiang, X.; Yang, P.; Xu, L.; Ming, J. Postharvest methyl jasmonate treatment enhanced biological activity by promoting phenylpropanoid metabolic pathways in Lilium brownii var. viridulum. Sci. Hortic. 2023, 308, 111551. [Google Scholar] [CrossRef]
- Haines, B.E.; Wiest, O.; Stauffacher, C.V. The increasingly complex mechanism of HMG-CoA reductase. Acc. Chem. Res. 2013, 46, 2416–2426. [Google Scholar] [CrossRef]
- Thimmappa, R.; Geisler, K.; Louveau, T.; O’Maille, P.; Osbourn, A. Triterpene biosynthesis in plants. Annu. Rev. Plant Biol. 2014, 65, 225–257. [Google Scholar] [CrossRef]
- Cárdenas, P.D.; Almeida, A.; Bak, S. Evolution of structural diversity of triterpenoids. Front. Plant Sci. 2019, 10, 486054. [Google Scholar] [CrossRef]
- Noushahi, H.A.; Khan, A.H.; Noushahi, U.F.; Hussain, M.; Javed, T.; Zafar, M.; Batool, M.; Ahmed, U.; Liu, K.; Harrison, M.T. Biosynthetic pathways of triterpenoids and strategies to improve their biosynthetic efficiency. Plant Growth Regul. 2022, 97, 439–454. [Google Scholar] [CrossRef] [PubMed]
- De Spiegelaere, W.; Dern-Wieloch, J.; Weigel, R.; Schumacher, V.; Schorle, H.; Nettersheim, D.; Bergmann, M.; Brehm, R.; Kliesch, S.; Vandekerckhove, L. Reference gene validation for RT-qPCR, a note on different available software packages. PLoS ONE 2015, 10, e0122515. [Google Scholar] [CrossRef]
- Manoli, A.; Sturaro, A.; Trevisan, S.; Quaggiotti, S.; Nonis, A. Evaluation of candidate reference genes for qPCR in maize. J. Plant Physiol. 2012, 169, 807–815. [Google Scholar] [CrossRef] [PubMed]
- Uddin, M.J.; Cinar, M.U.; Tesfaye, D.; Looft, C.; Tholen, E.; Schellander, K. Age-related changes in relative expression stability of commonly used housekeeping genes in selected porcine tissues. BMC Res. Notes 2011, 4, 441. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 1–12. [Google Scholar] [CrossRef]
- Kozera, B.; Rapacz, M. Reference genes in real-time PCR. J. Appl. Genet. 2013, 54, 391–406. [Google Scholar] [CrossRef]
- Jin, X.; Hou, Z.; Zhao, L.; Liu, L.; Priyadarshani, S.; Wang, L.; Huang, Y.; Chen, F.; Qin, Y. Genome-wide identification and evaluation of new reference genes in pineapple (Ananas comosus L.) during stamen and ovule development. Trop. Plant Biol. 2020, 13, 371–381. [Google Scholar] [CrossRef]
- Chen, H.; Hu, B.; Zhao, L.; Shi, D.; She, Z.; Huang, X.; Priyadarshani, S.; Niu, X.; Qin, Y. Differential expression analysis of reference genes in Pineapple (Ananas comosus L.) during reproductive development and response to abiotic stress, hormonal stimuli. Trop. Plant Biol. 2019, 12, 67–77. [Google Scholar] [CrossRef]
- Huis, R.; Hawkins, S.; Neutelings, G. Selection of reference genes for quantitative gene expression normalization in flax (Linum usitatissimum L.). BMC Plant Biol. 2010, 10, 71. [Google Scholar] [CrossRef]
- Machado, R.D.; Christoff, A.P.; Loss-Morais, G.; Margis-Pinheiro, M.; Margis, R.; Körbes, A.P. Comprehensive selection of reference genes for quantitative gene expression analysis during seed development in Brassica napus. Plant Cell Rep. 2015, 34, 1139–1149. [Google Scholar] [CrossRef] [PubMed]
- Pihur, V.; Datta, S.; Datta, S. Weighted rank aggregation of cluster validation measures: A Monte Carlo cross-entropy approach. Bioinformatics 2007, 23, 1607–1615. [Google Scholar] [CrossRef] [PubMed]
- Ghasemi, S.; Kumleh, H.H.; Kordrostami, M.; Rezadoost, M.H. Drought stress-mediated alterations in secondary metabolites and biosynthetic gene expression in cumin plants: Insights from gene-specific and metabolite-level analyses. Plant Stress 2023, 10, 100241. [Google Scholar] [CrossRef]
- Ghasemi, S.; Kumleh, H.H.; Kordrostami, M. Changes in the expression of some genes involved in the biosynthesis of secondary metabolites in Cuminum cyminum L. under UV stress. Protoplasma 2019, 256, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Jha, Y.; Mohamed, H.I. Plant secondary metabolites as a tool to investigate biotic stress tolerance in plants: A review. Gesunde Pflanz. 2022, 74, 771–790. [Google Scholar] [CrossRef]
- Hampton, R.; Dimster-Denk, D.; Rine, J. The biology of HMG-CoA reductase: The pros of contra-regulation. Trends Biochem. Sci. 1996, 21, 140–145. [Google Scholar] [CrossRef]
- Stermer, B.A.; Bianchini, G.M.; Korth, K.L. Regulation of HMG-CoA reductase activity in plants. J. Lipid Res. 1994, 35, 1133–1140. [Google Scholar] [CrossRef]
- Darabi, M.; Masoudi-Nejad, A.; Nemat-Zadeh, G. Bioinformatics study of the 3-hydroxy-3-methylglotaryl-coenzyme A reductase (HMGR) gene in Gramineae. Mol. Biol. Rep. 2012, 39, 8925–8935. [Google Scholar] [CrossRef]
- Wang, Q.J.; Zheng, L.P.; Zhao, P.F.; Zhao, Y.L.; Wang, J.W. Cloning and characterization of an elicitor-responsive gene encoding 3-hydroxy-3-methylglutaryl coenzyme A reductase involved in 20-hydroxyecdysone production in cell cultures of Cyanotis arachnoidea. Plant Physiol. Biochem. 2014, 84, 1–9. [Google Scholar] [CrossRef]
- Bansal, S.; Narnoliya, L.K.; Mishra, B.; Chandra, M.; Yadav, R.K.; Sangwan, N.S. HMG-CoA reductase from Camphor Tulsi (Ocimum kilimandscharicum) regulated MVA dependent biosynthesis of diverse terpenoids in homologous and heterologous plant systems. Sci. Rep. 2018, 8, 3547. [Google Scholar] [CrossRef]
- Wang, J.-J.; Han, S.; Yin, W.; Xia, X.; Liu, C. Comparison of reliable reference genes following different hormone treatments by various algorithms for qRT-PCR analysis of Metasequoia. Int. J. Mol. Sci. 2018, 20, 34. [Google Scholar] [CrossRef]
- Robledo, D.; Hernández-Urcera, J.; Cal, R.M.; Pardo, B.G.; Sánchez, L.; Martínez, P.; Viñas, A. Analysis of qPCR reference gene stability determination methods and a practical approach for efficiency calculation on a turbot (Scophthalmus maximus) gonad dataset. BMC Genom. 2014, 15, 648. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, L.; Cheng, K.; Wang, X.; Ren, G.; Xie, P. Identification of suitable plasma-based reference genes for miRNAome analysis of major depressive disorder. J. Affect. Disord. 2014, 163, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Komili, S.; Farny, N.G.; Roth, F.P.; Silver, P.A. Functional specificity among ribosomal proteins regulates gene expression. Cell 2007, 131, 557–571. [Google Scholar] [CrossRef]
- Deng, L.-T.; Wu, Y.-L.; Li, J.-C.; OuYang, K.-X.; Ding, M.-M.; Zhang, J.-J.; Li, S.-Q.; Lin, M.-F.; Chen, H.-B.; Hu, X.-S. Screening reliable reference genes for RT-qPCR analysis of gene expression in Moringa oleifera. PLoS ONE 2016, 11, e0159458. [Google Scholar] [CrossRef] [PubMed]
- Kumar, G.; Singh, A.K. Reference gene validation for qRT-PCR based gene expression studies in different developmental stages and under biotic stress in apple. Sci. Hortic. 2015, 197, 597–606. [Google Scholar] [CrossRef]
- Xu, M.; Zhang, B.; Su, X.; Zhang, S.; Huang, M. Reference gene selection for quantitative real-time polymerase chain reaction in Populus. Anal. Biochem. 2011, 408, 337–339. [Google Scholar] [CrossRef]
- Tong, Z.; Gao, Z.; Wang, F.; Zhou, J.; Zhang, Z. Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Mol. Biol. 2009, 10, 71. [Google Scholar] [CrossRef]
- Zhao, Y.; Luo, J.; Xu, S.; Wang, W.; Liu, T.; Han, C.; Chen, Y.; Kong, L. Selection of reference genes for gene expression normalization in Peucedanum praeruptorum Dunn under abiotic stresses, hormone treatments and different tissues. PLoS ONE 2016, 11, e0152356. [Google Scholar] [CrossRef]
- Xiao, X.; Ma, J.; Wang, J.; Wu, X.; Li, P.; Yao, Y. Validation of suitable reference genes for gene expression analysis in the halophyte Salicornia europaea by real-time quantitative PCR. Front. Plant Sci. 2015, 5, 788. [Google Scholar] [CrossRef]
- Fan, C.; Ma, J.; Guo, Q.; Li, X.; Wang, H.; Lu, M. Selection of reference genes for quantitative real-time PCR in bamboo (Phyllostachys edulis). PLoS ONE 2013, 8, e56573. [Google Scholar] [CrossRef] [PubMed]
- Mafra, V.; Kubo, K.S.; Alves-Ferreira, M.; Ribeiro-Alves, M.; Stuart, R.M.; Boava, L.P.; Rodrigues, C.M.; Machado, M.A. Reference genes for accurate transcript normalization in citrus genotypes under different experimental conditions. PLoS ONE 2012, 7, e31263. [Google Scholar] [CrossRef] [PubMed]



| Gene Name | Annealing Temperature (°C) | Amplicon Size (bp) | Primer Efficiency (%) | R2 Value |
|---|---|---|---|---|
| CnUBQ | 60.1 | 70 | 92.02 | 0.9985 |
| CnRPL | 60 | 90 | 93.44 | 0.9983 |
| CnRPS | 60 | 125 | 96.1 | 0.9986 |
| CnPTB1 | 59.8 | 145 | 98.35 | 0.9976 |
| CnTIP41 | 60 | 101 | 95.83 | 0.9953 |
| CnACT | 59.9 | 178 | 102.4 | 0.9979 |
| CnUBC | 59.9 | 90 | 92.54 | 0.9981 |
| CnGAPDH | 60 | 91 | 93.86 | 0.9953 |
| Cn18S | 60 | 180 | 94.5 | 0.9925 |
| CnCYP | 60.2 | 141 | 97.97 | 0.9989 |
| CnEF1α | 60 | 132 | 92.12 | 0.998 |
| CnTUB | 60 | 160 | 89.78 | 0.9964 |
| Group | Rank | geNorm | NormFinder | BestKeeper | Rank Aggreg | |||
|---|---|---|---|---|---|---|---|---|
| Gene | MV Value | Gene | Stability | Gene | SD Value | |||
| Abiotic stresses | 1 | CnUBQ | 0.338135 | CnEF1α | 0.19 | CnRPL | 0.44 | CnUBC |
| 2 | CnUBC | 0.338135 | CnUBC | 0.24 | CnUBC | 0.579 | CnRPL | |
| 3 | CnEF1α | 0.387752 | CnUBQ | 0.33 | CnEF1α | 0.678 | CnEF1α | |
| 4 | CnRPL | 0.474062 | CnRPL | 0.35 | CnUBQ | 0.701 | CnUBQ | |
| 5 | CnACT | 0.637643 | CnACT | 0.6 | CnTUB | 0.851 | CnACT | |
| 6 | CnRPS | 0.742216 | CnCYP | 0.62 | Cn18S | 0.945 | CnRPS | |
| 7 | Cn18S | 0.821862 | CnRPS | 0.73 | CnACT | 0.968 | Cn18S | |
| 8 | CnPTB1 | 0.895786 | Cn18S | 0.8 | CnCYP | 0.994 | CnCYP | |
| 9 | CnCYP | 1.017078 | CnTIP41 | 0.89 | CnRPS | 1.117 | CnPTB1 | |
| 10 | CnTUB | 1.123396 | CnPTB1 | 0.96 | CnPTB1 | 1.148 | CnTUB | |
| 11 | CnTIP41 | 1.215425 | CnTUB | 1.08 | CnTIP41 | 1.149 | CnTIP41 | |
| 12 | CnGAPDH | 1.365533 | CnGAPDH | 1.18 | CnGAPDH | 1.182 | CnGAPDH | |
| Hormonal stimulus | 1 | CnRPL | 0.083542 | CnRPL | 0.09 | CnUBC | 0.32 | CnRPL |
| 2 | CnEF1α | 0.083542 | CnGAPDH | 0.11 | Cn18S | 0.328 | CnEF1α | |
| 3 | CnGAPDH | 0.130138 | CnEF1α | 0.11 | CnRPL | 0.37 | CnGAPDH | |
| 4 | CnRPS | 0.161941 | CnRPS | 0.17 | CnUBQ | 0.399 | CnRPS | |
| 5 | CnTIP41 | 0.197269 | CnTIP41 | 0.21 | CnEF1α | 0.412 | CnUBC | |
| 6 | CnUBQ | 0.218152 | CnUBC | 0.22 | CnRPS | 0.425 | CnUBQ | |
| 7 | CnUBC | 0.234476 | CnCYP | 0.26 | CnPTB1 | 0.434 | Cn18S | |
| 8 | Cn18S | 0.249184 | CnUBQ | 0.28 | CnGAPDH | 0.479 | CnTIP41 | |
| 9 | CnCYP | 0.269206 | Cn18S | 0.33 | CnTIP41 | 0.483 | CnCYP | |
| 10 | CnTUB | 0.344994 | CnTUB | 0.59 | CnCYP | 0.53 | CnPTB1 | |
| 11 | CnPTB1 | 0.434288 | CnPTB1 | 0.67 | CnACT | 0.919 | CnTUB | |
| 12 | CnACT | 0.512254 | CnACT | 0.72 | CnTUB | 0.951 | CnACT | |
| Different tissues | 1 | CnRPL | 0.220736 | CnUBC | 0.2 | CnTUB | 0.252 | CnUBC |
| 2 | CnCYP | 0.220736 | CnEF1α | 0.34 | CnGAPDH | 0.31 | CnRPL | |
| 3 | CnUBC | 0.282248 | CnRPL | 0.46 | CnCYP | 0.381 | CnCYP | |
| 4 | CnEF1α | 0.346926 | CnUBQ | 0.57 | CnRPL | 0.395 | CnTUB | |
| 5 | CnTUB | 0.464707 | CnCYP | 0.57 | CnUBC | 0.564 | CnEF1α | |
| 6 | CnGAPDH | 0.519896 | Cn18S | 0.84 | CnPTB1 | 0.596 | CnGAPDH | |
| 7 | CnPTB1 | 0.617063 | CnPTB1 | 0.91 | CnEF1α | 0.652 | CnPTB1 | |
| 8 | Cn18S | 0.74218 | CnRPS | 0.92 | CnUBQ | 1.203 | CnUBQ | |
| 9 | CnUBQ | 0.818679 | CnTUB | 0.98 | Cn18S | 1.287 | Cn18S | |
| 10 | CnRPS | 0.918168 | CnGAPDH | 0.98 | CnRPS | 1.571 | CnRPS | |
| 11 | CnACT | 1.024491 | CnACT | 1.34 | CnACT | 1.863 | CnACT | |
| 12 | CnTIP41 | 1.209021 | CnTIP41 | 1.45 | CnTIP41 | 1.97 | CnTIP41 | |
| Total | 1 | CnUBC | 0.404232 | CnEF1α | 0.2 | CnRPL | 0.444 | CnRPL |
| 2 | CnEF1α | 0.404232 | CnUBC | 0.21 | CnUBC | 0.559 | CnUBC | |
| 3 | CnRPL | 0.464657 | CnRPL | 0.31 | CnEF1α | 0.648 | CnEF1α | |
| 4 | CnUBQ | 0.506615 | CnUBQ | 0.37 | CnTUB | 0.73 | CnUBQ | |
| 5 | Cn18S | 0.669644 | CnCYP | 0.57 | CnUBQ | 0.781 | Cn18S | |
| 6 | CnRPS | 0.768979 | CnRPS | 0.7 | CnCYP | 0.841 | CnCYP | |
| 7 | CnPTB1 | 0.865782 | Cn18S | 0.72 | CnGAPDH | 0.923 | CnRPS | |
| 8 | CnACT | 0.949271 | CnACT | 0.74 | Cn18S | 0.938 | CnPTB1 | |
| 9 | CnCYP | 1.034368 | CnTIP41 | 0.81 | CnPTB1 | 0.968 | CnTUB | |
| 10 | CnTUB | 1.121927 | CnPTB1 | 0.9 | CnRPS | 1.145 | CnACT | |
| 11 | CnTIP41 | 1.21541 | CnTUB | 0.98 | CnACT | 1.201 | CnTIP41 | |
| 12 | CnGAPDH | 1.328414 | CnGAPDH | 1.02 | CnTIP41 | 1.211 | CnGAPDH | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
An, C.; Lu, L.; Yao, Y.; Liu, R.; Cheng, Y.; Lin, Y.; Qin, Y.; Zheng, P. Selection and Validation of Reference Genes in Clinacanthus nutans Under Abiotic Stresses, MeJA Treatment, and in Different Tissues. Int. J. Mol. Sci. 2025, 26, 2483. https://doi.org/10.3390/ijms26062483
An C, Lu L, Yao Y, Liu R, Cheng Y, Lin Y, Qin Y, Zheng P. Selection and Validation of Reference Genes in Clinacanthus nutans Under Abiotic Stresses, MeJA Treatment, and in Different Tissues. International Journal of Molecular Sciences. 2025; 26(6):2483. https://doi.org/10.3390/ijms26062483
Chicago/Turabian StyleAn, Chang, Lin Lu, Yixin Yao, Ruoyu Liu, Yan Cheng, Yanxiang Lin, Yuan Qin, and Ping Zheng. 2025. "Selection and Validation of Reference Genes in Clinacanthus nutans Under Abiotic Stresses, MeJA Treatment, and in Different Tissues" International Journal of Molecular Sciences 26, no. 6: 2483. https://doi.org/10.3390/ijms26062483
APA StyleAn, C., Lu, L., Yao, Y., Liu, R., Cheng, Y., Lin, Y., Qin, Y., & Zheng, P. (2025). Selection and Validation of Reference Genes in Clinacanthus nutans Under Abiotic Stresses, MeJA Treatment, and in Different Tissues. International Journal of Molecular Sciences, 26(6), 2483. https://doi.org/10.3390/ijms26062483









