First Complete Mitochondrial Genome Analysis of Tree Frog, Dryophytes flaviventris and Comparison with Dryophytes suweonensis
Abstract
1. Introduction
2. Results and Discussion
2.1. Basic Characteristics of D. flaviventris Mitogenomes
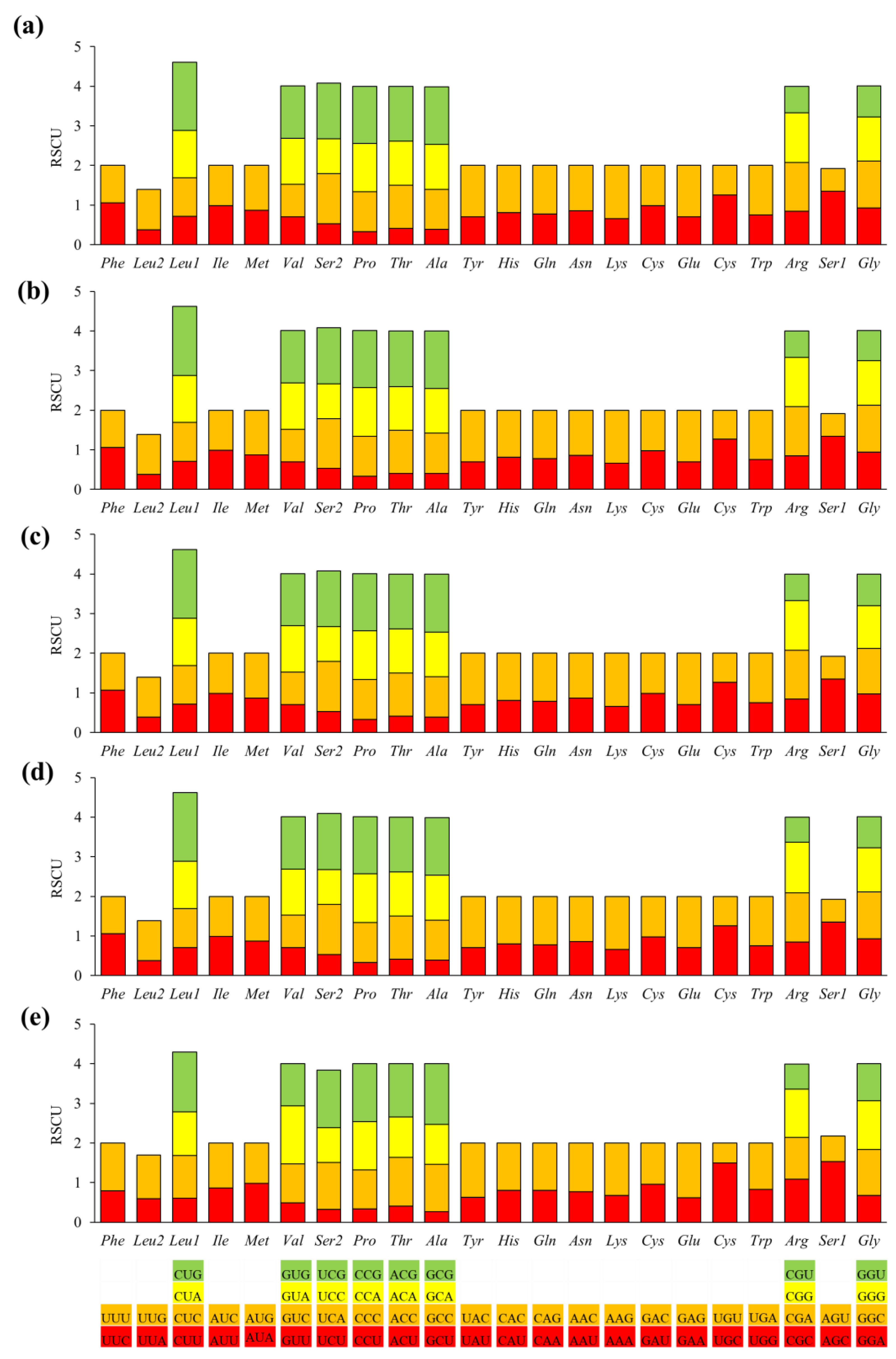
2.2. PCGs and Codon Usage Patterns of D. flaviventris
2.3. rRNA and tRNA Genes of D. flaviventris
2.4. Non-Coding Region of D. flaviventris
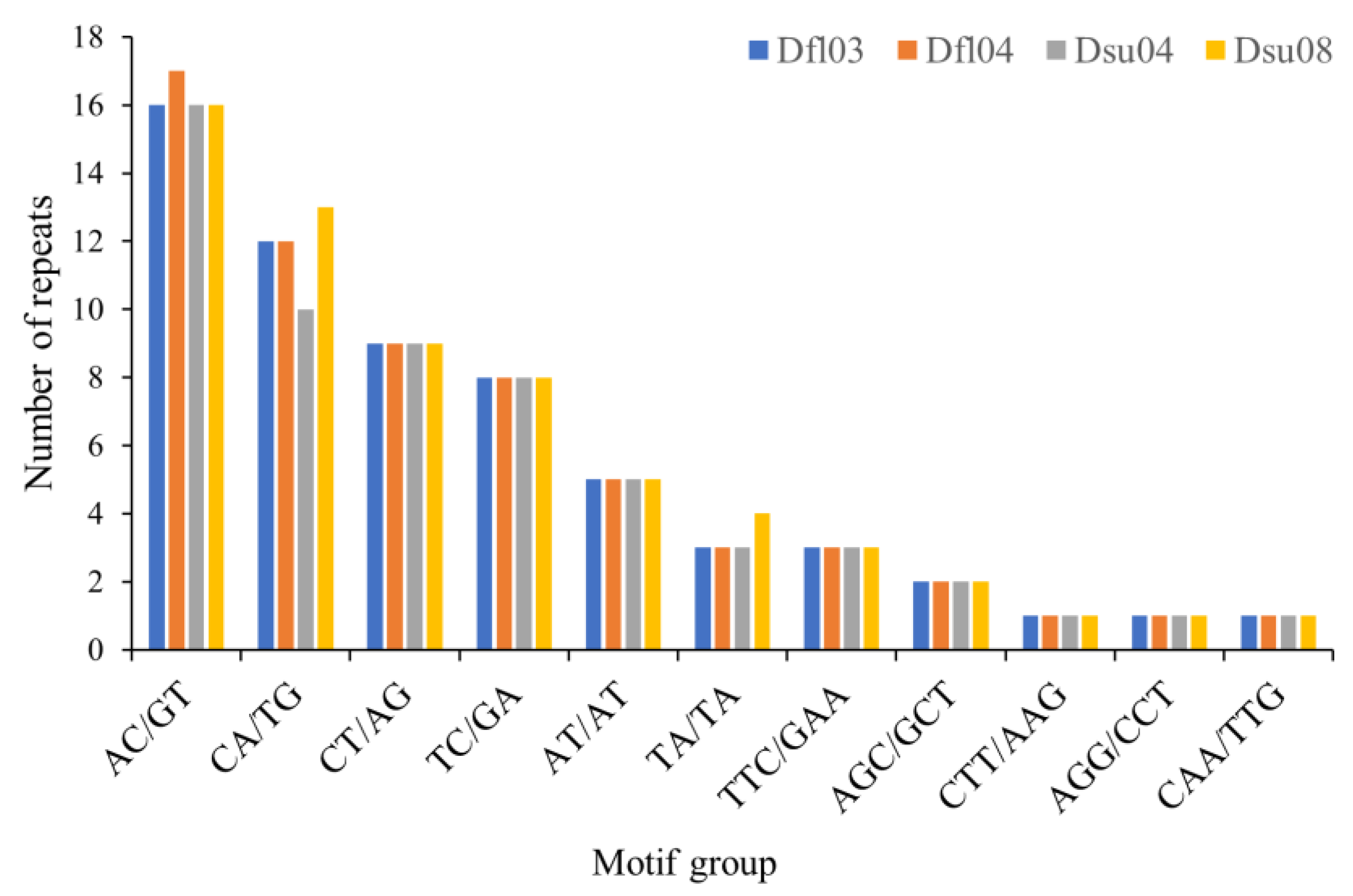
2.5. Repeat Detection in D. flaviventris
2.6. Synonymous and Nonsynonymous Substitution Rate
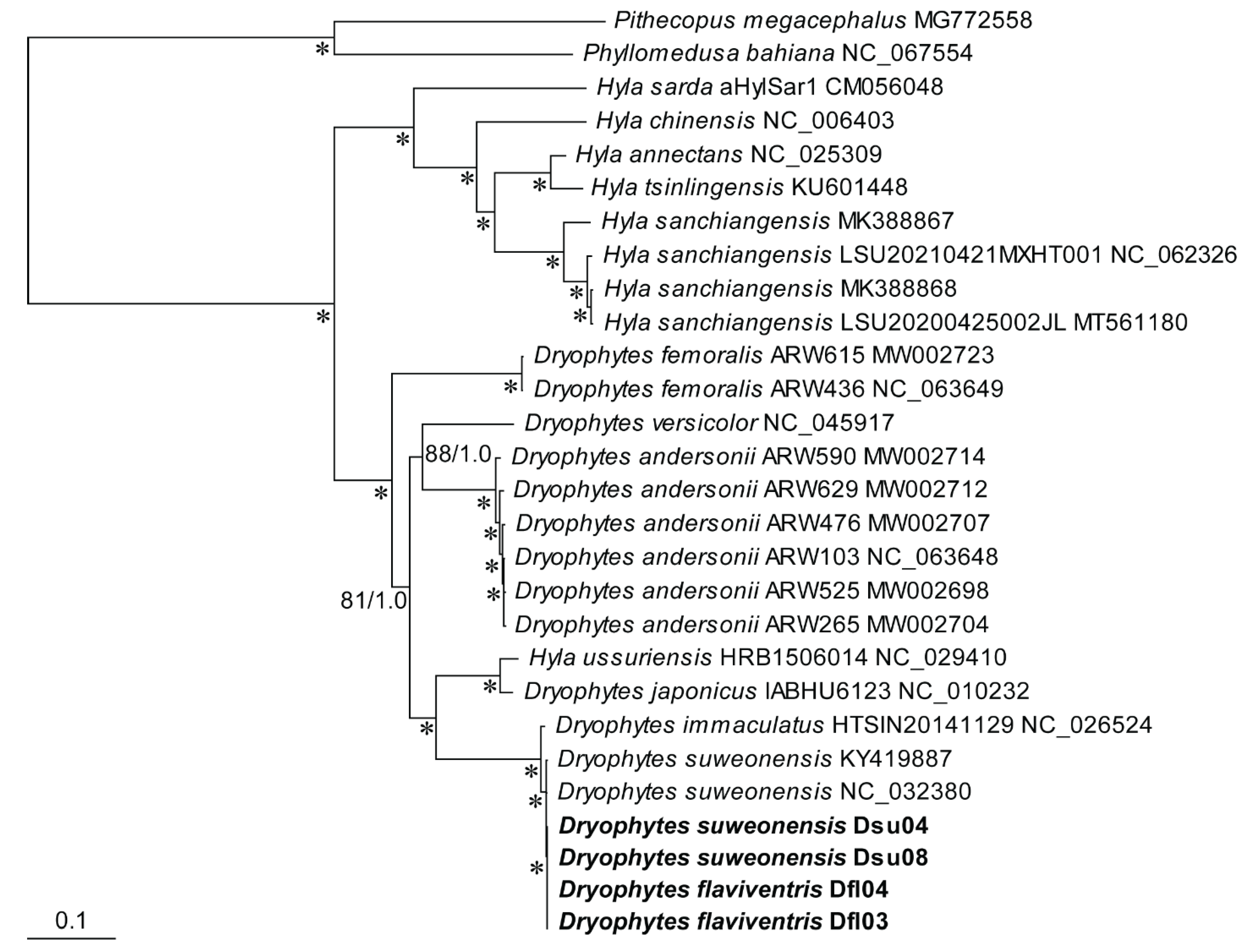
2.7. Phylogenetic Analysis
2.8. Characteristic Analysis of Comparative Genome
3. Materials and Methods
3.1. Sample Collection and Genomic DNA Extraction
3.2. Primer Design and Long-Range PCR
3.3. Library Preparation and Sequencing
3.4. Mitochondrial Genome Assembly
3.5. Mitochondrial Gene Annotation and Codon Usage Analysis
3.6. Repeat Sequence Analysis
3.7. Synonymous and Nonsynonymous Substitution Rate
3.8. Phylogenetic Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Ladoukakis, E.D.; Zouros, E. Evolution and Inheritance of Animal Mitochondrial DNA: Rules and Exceptions. J. Biol. Res.-Thessalon. 2017, 24, 2. [Google Scholar] [CrossRef] [PubMed]
- Brown, W.M.; George, M.; Wilson, A.C. Rapid Evolution of Animal Mitochondrial DNA. Proc. Natl. Acad. Sci. USA 1979, 76, 1967–1971. [Google Scholar] [CrossRef] [PubMed]
- Mueller, R.L.; Macey, J.R.; Jaekel, M.; Wake, D.B.; Boore, J.L. Morphological Homoplasy, Life History Evolution, and Historical Biogeography of Plethodontid Salamanders Inferred from Complete Mitochondrial Genomes. Proc. Natl. Acad. Sci. USA 2004, 101, 13820–13825. [Google Scholar] [CrossRef] [PubMed]
- Boore, J.L. Animal Mitochondrial Genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef]
- Boore, J.; Brown, W. Big Trees from Little Genomes: Mitochondrial Gene Order as a Phylogenetic Tool. Curr. Opin. Genet. Dev. 1999, 8, 668–674. [Google Scholar] [CrossRef]
- Evans, B.J.; Carter, T.F.; Greenbaum, E.; Gvoždík, V.; Kelley, D.B.; McLaughlin, P.J.; Pauwels, O.S.G.; Portik, D.M.; Stanley, E.L.; Tinsley, R.C.; et al. Genetics, Morphology, Advertisement Calls, and Historical Records Distinguish Six New Polyploid Species of African Clawed frog (Xenopus, Pipidae) from West and Central Africa. PLoS ONE 2015, 10, e0142823. [Google Scholar] [CrossRef]
- Glaw, F.; Köhler, J.; De la Riva, I.; Vieites, D.; Vences, M. Integrative Taxonomy of Malagasy Treefrogs: Combination of Molecular Genetics, Bioacoustics and Comparative Morphology Reveals Twelve Additional Species of Boophis. Zootaxa 2010, 2383, 1–82. [Google Scholar] [CrossRef]
- Zhang, P.; Liang, D.; Mao, R.-L.; Hillis, D.M.; Wake, D.B.; Cannatella, D.C. Efficient Sequencing of Anuran mtDNAs and a Mitogenomic Exploration of the Phylogeny and Evolution of Frogs. Mol. Biol. Evol. 2013, 30, 1899–1915. [Google Scholar] [CrossRef]
- Duellman, W.E.; Marion, A.B.; Hedges, S.B. Phylogenetics, Classification, and Biogeography of the Treefrogs (Amphibia: Anura: Arboranae). Zootaxa 2016, 4104, 1–109. [Google Scholar] [CrossRef]
- Borzée, A.; Messenger, K.R.; Chae, S.; Andersen, D.; Groffen, J.; Kim, Y.I.; An, J.; Othman, S.N.; Ri, K.; Nam, T.Y.; et al. Yellow Sea Mediated Segregation between North East Asian Dryophytes Species. PLoS ONE 2020, 15, e0234299. [Google Scholar] [CrossRef]
- IUCN SSC Amphibian Specialist Group. IUCN Red List of Threatened Species: Dryophytes suweonensis. IUCN Red List Threat. Species. 2022. Available online: https://www.iucnredlist.org (accessed on 14 October 2023).
- Frost, D.R. Amphibian Species of the World: An Online Reference. Version 6.2. American Museum of Natural History, New York. References—Scientific Research Publishing. 2024. Available online: https://www.scirp.org/(S(czeh2tfqw2orz553k1w0r45))/reference/referencespapers?referenceid=3692321 (accessed on 17 October 2024).
- Zhang, J.-Y.; Luu, B.E.; Yu, D.-N.; Zhang, L.-P.; Al-Attar, R.; Storey, K.B. The Complete Mitochondrial Genome of Dryophytes versicolor: Phylogenetic Relationship among Hylidae and Mitochondrial Protein-Coding Gene Expression in Response to Freezing and Anoxia. Int. J. Biol. Macromol. 2019, 132, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Ojala, D.; Montoya, J.; Attardi, G. tRNA Punctuation Model of RNA Processing in Human Mitochondria. Nature 1981, 290, 470–474. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.-H.; Yuan, Y.-N.; Li, K.; Storey, K.B.; Zhang, J.-Y.; Zhang, S.-S.; Yu, D.-N. Differential Mitochondrial Genome Expression of Four Hylid frog Species under Low-Temperature Stress and Its Relationship with Amphibian Temperature Adaptation. Int. J. Mol. Sci. 2024, 25, 5967. [Google Scholar] [CrossRef]
- Sano, N.; Kurabayashi, A.; Fujii, T.; Yonekawa, H.; Sumida, M. Complete Nucleotide Sequence and Gene Rearrangement of the Mitochondrial Genome of the Bell-Ring Frog, Buergeria buergeri (Family Rhacophoridae). Genes Genet. Syst. 2004, 79, 151–163. [Google Scholar] [CrossRef]
- Tzeng, C.-S.; Hui, C.-F.; Shen, S.-C.; Huang, P.C. The Complete Nucleotide Sequence of the Crossostoma Lacustre Mitochondrial Genome: Conservation and Variations among Vertebrates. Nucleic Acids Res. 1992, 20, 4853–4858. [Google Scholar] [CrossRef][Green Version]
- Ohtsuki, T.; Kawai, G.; Watanabe, K. The Minimal tRNA: Unique Structure of Ascaris Suum Mitochondrial tRNASerUCU Having a Short T Arm and Lacking the Entire D Arm. FEBS Lett. 2002, 514, 37–43. [Google Scholar] [CrossRef]
- Suzuki, M.; Toyooka, S.; Miyajima, K.; Iizasa, T.; Fujisawa, T.; Bekele, N.B.; Gazdar, A.F. Alterations in the Mitochondrial Displacement Loop in Lung Cancers. Clin. Cancer Res. 2003, 9, 5636–5641. [Google Scholar]
- Ellegren, H. Microsatellites: Simple Sequences with Complex Evolution. Nat. Rev. Genet. 2004, 5, 435–445. [Google Scholar] [CrossRef]
- Wang, J.; Tai, J.; Zhang, W.; He, K.; Lan, H.; Liu, H. Comparison of Seven Complete Mitochondrial Genomes from Lamprologus and Neolamprologus (Chordata, Teleostei, Perciformes) and the Phylogenetic Implications for Cichlidae. ZooKeys 2023, 1184, 115. [Google Scholar] [CrossRef]
- Sun, J.-T.; Jin, P.-Y.; Hoffmann, A.A.; Duan, X.-Z.; Dai, J.; Hu, G.; Xue, X.-F.; Hong, X.-Y. Evolutionary Divergence of Mitochondrial Genomes in Two Tetranychus Species Distributed across Different Climates. Insect Mol. Biol. 2018, 27, 698–709. [Google Scholar] [CrossRef]
- Banguera-Hinestroza, E.; Sawall, Y.; Al-Sofyani, A.; Mardulyn, P.; Fuertes-Aguilar, J.; Cardenas-Henao, H.; Cárdenas-Henao, H.; Jimenez-Infante, F.; Voolstra, C.R.; Flot, J.-F. mtDNA Recombination Indicative of Hybridization Suggests a Role of the Mitogenome in the Adaptation of Reef-Building Corals to Extreme Environments. BioRxiv 2018, 462069. [Google Scholar] [CrossRef]
- Gregory, T.R. Coincidence, Coevolution, or Causation? DNA Content, Cell Size, and the C-Value Enigma. Biol. Rev. 2001, 76, 65–101. [Google Scholar] [CrossRef] [PubMed]
- Peterson, B.K.; Weber, J.N.; Kay, E.H.; Fisher, H.S.; Hoekstra, H.E. Double Digest RADseq: An Inexpensive Method for de Novo SNP Discovery and Genotyping in Model and Non-Model Species. PLoS ONE 2012, 7, e37135. [Google Scholar] [CrossRef] [PubMed]
- Kai, W.; Nomura, K.; Fujiwara, A.; Nakamura, Y.; Yasuike, M.; Ojima, N.; Masaoka, T.; Ozaki, A.; Kazeto, Y.; Gen, K.; et al. A ddRAD-Based Genetic Map and Its Integration with the Genome Assembly of Japanese Eel (Anguilla japonica) Provides Insights into Genome Evolution after the Teleost-Specific Genome Duplication. BMC Genom. 2014, 15, 233. [Google Scholar] [CrossRef]
- Davey, J.W.; Cezard, T.; Fuentes-Utrilla, P.; Eland, C.; Gharbi, K.; Blaxter, M.L. Special Features of RAD Sequencing Data: Implications for Genotyping. Mol. Ecol. 2013, 22, 3151–3164. [Google Scholar] [CrossRef]
- Grover, C.E.; Arick, M.A., II; Thrash, A.; Sharbrough, J.; Hu, G.; Yuan, D.; Snodgrass, S.; Miller, E.R.; Ramaraj, T.; Peterson, D.G.; et al. Dual Domestication, Diversity, and Differential Introgression in Old World Cotton Diploids. Genome Biol. Evol. 2022, 14, evac170. [Google Scholar] [CrossRef]
- Cui, L.; Huang, A.; He, Z.; Ao, L.; Ge, F.; Fan, X.; Zeng, B.; Yang, M.; Yang, D.; Ni, Q. Complete Mitogenomes of Polypedates Tree Frogs Unveil Gene Rearrangement and Concerted Evolution within Rhacophoridae. Animals 2022, 12, 2449. [Google Scholar] [CrossRef]
- Tang, Y.; Huo, Z.; Liu, Y.; Wang, Y.; Zuo, L.; Fang, L.; Zhao, W.; Tan, Y.; Yan, X. Full Mitochondrial Genomes Reveal Species Differences between the Venerid Clams Ruditapes philippinarum and R. variegatus. Genes 2022, 13, 2157. [Google Scholar] [CrossRef]
- Csuzdi, C.; Koo, J.; Hong, Y. The Complete Mitochondrial DNA Sequences of Two Sibling Species of Lumbricid earthworms, Eiseniafetida (Savigny, 1826) and Eiseniaandrei (Bouché, 1972) (Annelida, Crassiclitellata): Comparison of Mitogenomes and Phylogenetic Positioning. ZooKeys 2022, 1097, 167. [Google Scholar] [CrossRef]
- Funk, D.J.; Omland, K.E. Species-Level Paraphyly and Polyphyly: Frequency, Causes, and Consequences, with Insights from Animal Mitochondrial DNA. Annu. Rev. Ecol. Evol. Syst. 2003, 34, 397–423. [Google Scholar] [CrossRef]
- Bryson, R.W., Jr.; Smith, B.T.; Nieto-Montes de Oca, A.; García-Vázquez, U.O.; Riddle, B.R. The Role of Mitochondrial Introgression in Illuminating the Evolutionary History of Nearctic Treefrogs. Zool. J. Linn. Soc. 2014, 172, 103–116. [Google Scholar] [CrossRef]
- Li, J.-T.; Wang, J.-S.; Nian, H.-H.; Litvinchuk, S.N.; Wang, J.; Li, Y.; Rao, D.-Q.; Klaus, S. Amphibians Crossing the Bering Land Bridge: Evidence from Holarctic treefrogs (Hyla, Hylidae, Anura). Mol. Phylogenet. Evol. 2015, 87, 80–90. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.M.; Yun, J.; Lee, K.; Lee, S.-H.; Park, S.-M.; Ham, C.-H.; Sung, H.-C. Population-Level Call Properties of Endangered Dryophytes suweonensissensu Lato (Anura: Amphibia) in South Korea. PeerJ 2023, 11, e16492. [Google Scholar] [CrossRef] [PubMed]
- Hillis, D.M.; Moritz, C.; Mable, B.K. Molecular Systematics; Sinauer: Sunderland, MA, USA, 1996; Volume 23. [Google Scholar]
- Holland, B.S.; Hadfield, M.G. Islands within an Island: Phylogeography and Conservation Genetics of the Endangered Hawaiian Tree Snail Achatinella mustelina. Mol. Ecol. 2002, 11, 365–375. [Google Scholar] [CrossRef]
- Rubinoff, D.; Sperling, F.A.H. Mitochondrial DNA Sequence, Morphology and Ecology Yield Contrasting Conservation Implications for Two Threatened Buckmoths (Hemileuca: Saturniidae). Biol. Conserv. 2004, 118, 341–351. [Google Scholar] [CrossRef]
- Haig, S.M. Molecular Contributions to Conservation. Ecology 1998, 79, 413–425. [Google Scholar] [CrossRef]
- Soltis, P.S.; Gitzendanner, M.A. Molecular Systematics and the Conservation of Rare Species. Conserv. Biol. 1999, 13, 471–483. [Google Scholar] [CrossRef]
- Hall, T.; Biosciences, I.; Carlsbad, C. BioEdit: An Important Software for Molecular Biology. GERF Bull. Biosci 2011, 2, 60–61. [Google Scholar]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data; Babraham Bioinformatics: Cambridge, UK, 2010; Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 17 October 2024).
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C. Geneious Basic: An Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Bushnell, B. BBMap: A Fast, Accurate, Splice-Aware Aligner. In Proceedings of the 9th Annual Genomics of Energy & Environment Meeting, Walnut Creek, CA, USA, 17–20 March 2014; Available online: https://sourceforge.net/projects/bbmap/ (accessed on 17 October 2024).
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A Program for Improved Detection of Transfer RNA Genes in Genomic Sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Conant, G.C.; Wolfe, K.H. GenomeVx: Simple Web-Based Creation of Editable Circular Chromosome Maps. Bioinformatics 2008, 24, 861–862. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Chen, Y.; Sun, F.; Waterman, M.S.; Zhang, X. A New Statistic for Efficient Detection of Repetitive Sequences. Bioinformatics 2019, 35, 4596–4606. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, L. GMATA: An Integrated Software Package for Genome-Scale SSR Mining, Marker Development and Viewing. Front. Plant Sci. 2016, 7, 1350. [Google Scholar] [CrossRef]
- Benson, G. Tandem Repeats Finder: A Program to Analyze DNA Sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef]
- Smith, A.F.A.; Hubley, R.; Green, P. RepeatMasker. Available online: https://www.repeatmasker.org (accessed on 4 March 2025).
- Zhang, Z.; Li, J.; Zhao, X.-Q.; Wang, J.; Wong, G.K.-S.; Yu, J. KaKs_Calculator: Calculating Ka and Ks through Model Selection and Model Averaging. Genom. Proteom. Bioinform. 2006, 4, 259–263. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A Software for Comprehensive Analysis of DNA Polymorphism Data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Zhang, Z.; Mammola, S.; Liang, Z.; Capinha, C.; Wei, Q.; Wu, Y.; Zhou, J.; Wang, C. Future Climate Change Will Severely Reduce Habitat Suitability of the Critically Endangered Chinese Giant Salamander. Freshw. Biol. 2020, 65, 971–980. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More Models, New Heuristics and High-Performance Computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML-VI-HPC: Maximum Likelihood-Based Phylogenetic Analyses with Thousands of Taxa and Mixed Models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef]
- Stamatakis, A.; Hoover, P.; Rougemont, J. A Rapid Bootstrap Algorithm for the RAxML Web Servers. Syst. Biol. 2008, 57, 758–771. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Page, R.D.M. Visualizing Phylogenetic Trees Using TreeView. Curr. Protoc. Bioinform. 2003, 1, 6.2.1–6.2.6. [Google Scholar] [CrossRef] [PubMed]

| Species ID | Dfl03 | Dfl04 | Dsu04 | Dsu08 | Dj (AB303949) | ||
|---|---|---|---|---|---|---|---|
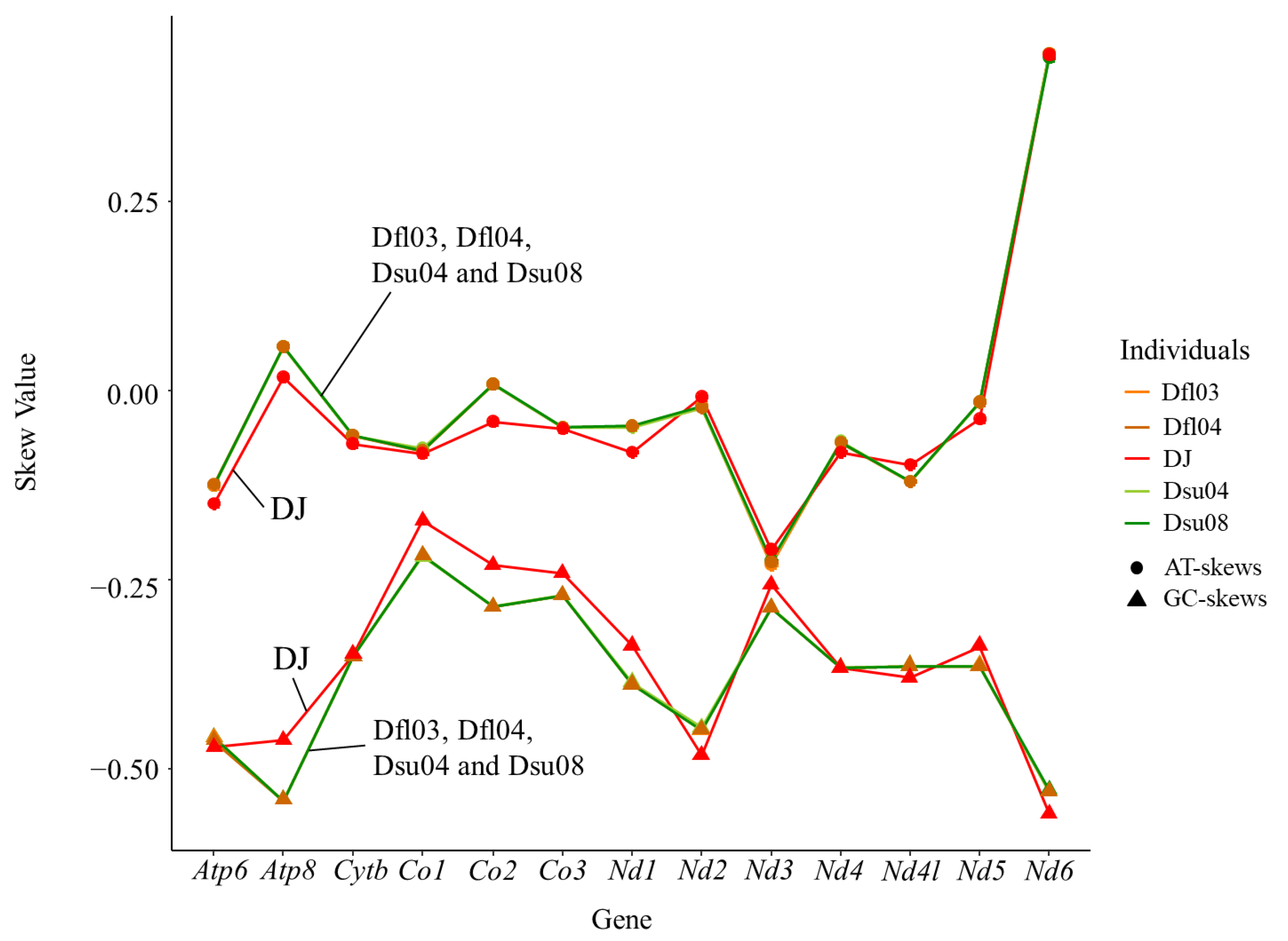
| Nucleotide frequency (%) | Whole | A | 29.12 | 29.13 | 29.14 | 29.14 | 29.59 |
| G | 14.93 | 14.92 | 14.92 | 14.91 | 14.50 | ||
| T | 28.48 | 28.50 | 28.48 | 28.49 | 30.50 | ||
| C | 27.47 | 27.44 | 27.47 | 27.46 | 25.41 | ||
| A + T | 57.60 | 57.64 | 57.61 | 57.63 | 60.09 | ||
| AT-skew | 0.01 | 0.01 | 0.01 | 0.01 | −0.02 | ||
| GC-skew | −0.30 | −0.30 | −0.30 | −0.30 | −0.27 | ||
| PCGs | A | 27.30 | 27.32 | 27.30 | 27.31 | 28.40 | |
| G | 14.07 | 14.05 | 14.07 | 14.06 | 13.45 | ||
| T | 29.18 | 29.20 | 29.19 | 29.20 | 31.12 | ||
| C | 29.45 | 29.43 | 29.43 | 29.43 | 27.03 | ||
| A + T | 56.48 | 56.52 | 56.49 | 56.51 | 59.52 | ||
| AT-skew | −0.03 | −0.03 | −0.03 | −0.03 | −0.05 | ||
| GC-skew | −0.35 | −0.35 | −0.35 | −0.35 | −0.34 | ||
| rRNAs | A | 33.78 | 33.78 | 33.78 | 33.77 | 34.89 | |
| G | 18.55 | 18.55 | 18.55 | 18.54 | 17.88 | ||
| T | 25.06 | 25.02 | 25.06 | 25.05 | 25.53 | ||
| C | 22.61 | 22.65 | 22.61 | 22.64 | 21.70 | ||
| A + T | 58.84 | 58.80 | 58.84 | 58.82 | 60.42 | ||
| AT-skew | 0.15 | 0.15 | 0.15 | 0.15 | 0.15 | ||
| GC-skew | −0.10 | −0.10 | −0.10 | −0.10 | −0.10 | ||
| tRNAs | A | 31.74 | 31.74 | 31.83 | 31.74 | 31.91 | |
| G | 17.76 | 17.76 | 17.68 | 17.76 | 17.68 | ||
| T | 26.56 | 26.56 | 26.56 | 26.56 | 26.97 | ||
| C | 23.93 | 23.93 | 23.93 | 23.93 | 23.44 | ||
| A + T | 58.31 | 58.31 | 58.39 | 58.31 | 58.88 | ||
| AT-skew | 0.09 | 0.09 | 0.09 | 0.09 | 0.08 | ||
| GC-skew | −0.15 | −0.15 | −0.15 | −0.15 | −0.14 | ||
| D-loop | A | 30.41 | 30.42 | 30.48 | 30.53 | 28.56 | |
| G | 13.95 | 13.96 | 13.89 | 13.87 | 14.28 | ||
| T | 29.57 | 29.7 | 29.54 | 29.59 | 33.12 | ||
| C | 26.06 | 25.92 | 26.09 | 26.02 | 24.03 | ||
| A + T | 59.98 | 60.13 | 60.02 | 60.11 | 61.69 | ||
| AT-skew | 0.01 | 0.01 | 0.02 | 0.02 | −0.07 | ||
| GC-skew | −0.30 | −0.30 | −0.31 | −0.30 | −0.25 | ||
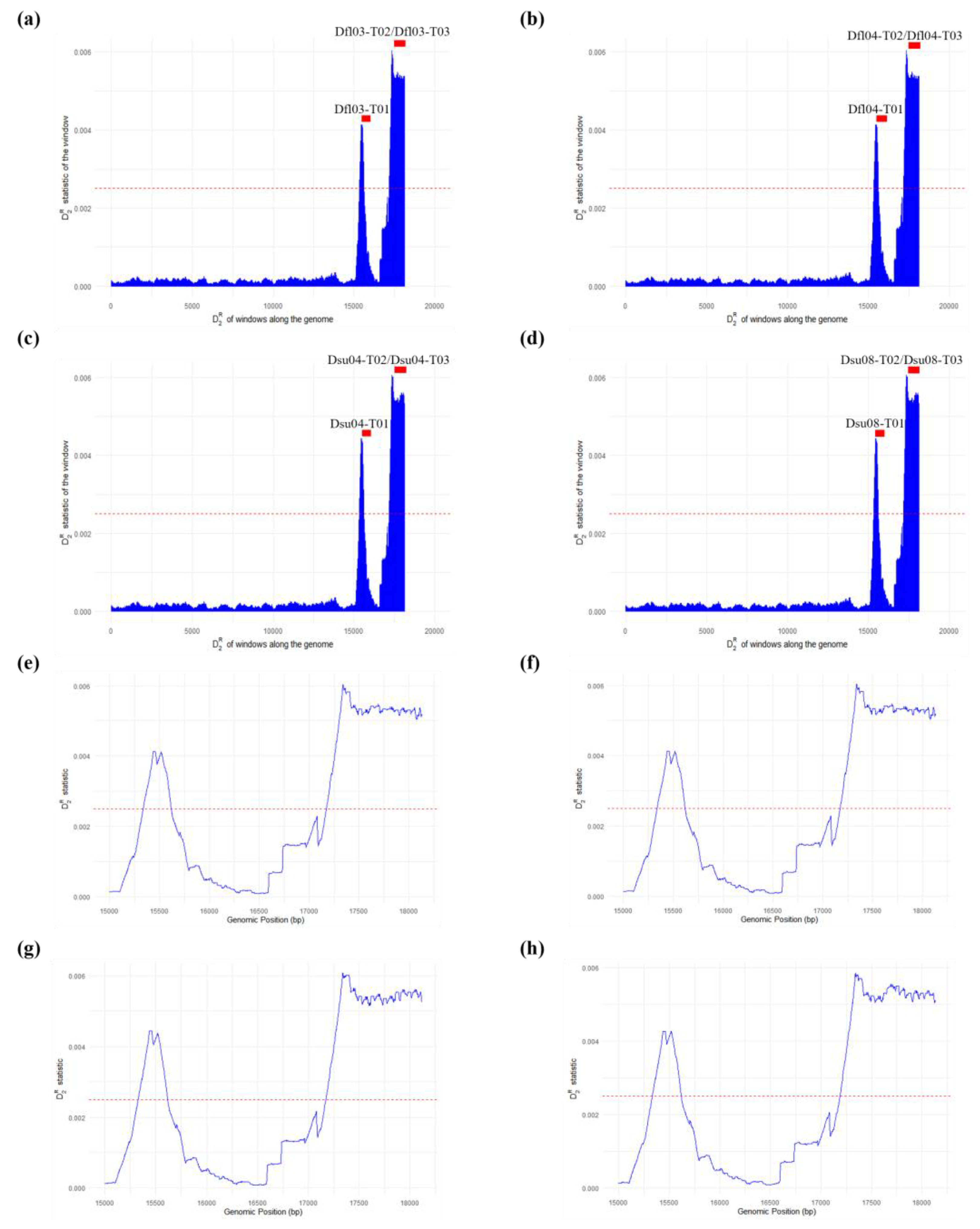
| Sample | Repeat Name | Size | No. Copy | Matches (%) | Start | End | Location |
|---|---|---|---|---|---|---|---|
| Dfl03 | Dfl03-T01 | 160 | 3.3 | 99 | 15,439 | 15,961 | Putative control region |
| Dfl03-T02 | 123 | 10.5 | 95 | 17,336 | 18,615 | Putative control region | |
| Dfl03-T03 | 242 | 5.2 | 94 | 17,336 | 18,615 | Putative control region | |
| Dfl04 | Dfl04-T01 | 160 | 3.3 | 99 | 15,439 | 15,961 | Putative control region |
| Dfl04-T02 | 121 | 10.5 | 95 | 17,335 | 18,614 | Putative control region | |
| Dfl04-T03 | 244 | 5.2 | 96 | 17,335 | 18,614 | Putative control region | |
| Dsu04 | Dsu04-T01 | 160 | 3.3 | 100 | 15,439 | 15,961 | Putative control region |
| Dsu04-T02 | 121 | 10.5 | 95 | 17,335 | 18,608 | Putative control region | |
| Dsu04-T03 | 242 | 5.2 | 95 | 17,335 | 18,608 | Putative control region | |
| Dsu08 | Dsu08-T01 | 160 | 3.3 | 99 | 15,440 | 15,962 | Putative control region |
| Dsu08-T02 | 123 | 10.5 | 95 | 17,337 | 18,614 | Putative control region | |
| Dsu08-T03 | 244 | 5.2 | 94 | 17,337 | 18,614 | Putative control region |
| Primer | Oligonucleotide (5′ → 3′) | Expected Amplicon Size (bp) |
|---|---|---|
| Set 1 | ||
| HYL-MT-02336f | AGACGAGAAGACCCTDTGGA | ca. 7700 |
| HYL-MT-10028r | TRAGYCGAAATCAGCTGTCTTT | |
| Set 2 | ||
| HYL-MT-07929f | AGCGACAGCCTTTTAAGCT | ca. 13,400 bp |
| HYL-MT-02706r | TGATCTGAGTTCAGACCGGA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yoo, N.; Kim, K.-R.; Tran, B.T.; Kim, K.-Y.; Min, M.-S.; Yoon, J.-D.; Kim, K.-S. First Complete Mitochondrial Genome Analysis of Tree Frog, Dryophytes flaviventris and Comparison with Dryophytes suweonensis. Int. J. Mol. Sci. 2025, 26, 2423. https://doi.org/10.3390/ijms26062423
Yoo N, Kim K-R, Tran BT, Kim K-Y, Min M-S, Yoon J-D, Kim K-S. First Complete Mitochondrial Genome Analysis of Tree Frog, Dryophytes flaviventris and Comparison with Dryophytes suweonensis. International Journal of Molecular Sciences. 2025; 26(6):2423. https://doi.org/10.3390/ijms26062423
Chicago/Turabian StyleYoo, Nakyung, Kang-Rae Kim, Biet Thanh Tran, Keun-Yong Kim, Mi-Sook Min, Ju-Duk Yoon, and Keun-Sik Kim. 2025. "First Complete Mitochondrial Genome Analysis of Tree Frog, Dryophytes flaviventris and Comparison with Dryophytes suweonensis" International Journal of Molecular Sciences 26, no. 6: 2423. https://doi.org/10.3390/ijms26062423
APA StyleYoo, N., Kim, K.-R., Tran, B. T., Kim, K.-Y., Min, M.-S., Yoon, J.-D., & Kim, K.-S. (2025). First Complete Mitochondrial Genome Analysis of Tree Frog, Dryophytes flaviventris and Comparison with Dryophytes suweonensis. International Journal of Molecular Sciences, 26(6), 2423. https://doi.org/10.3390/ijms26062423







