Genome-Wide Identification of the SWEET Gene Family and Functional Analysis of BraSWEET10 in Winter B. rapa (Brassica rapa L.) Under Low-Temperature Stress
Abstract
1. Introduction
2. Results
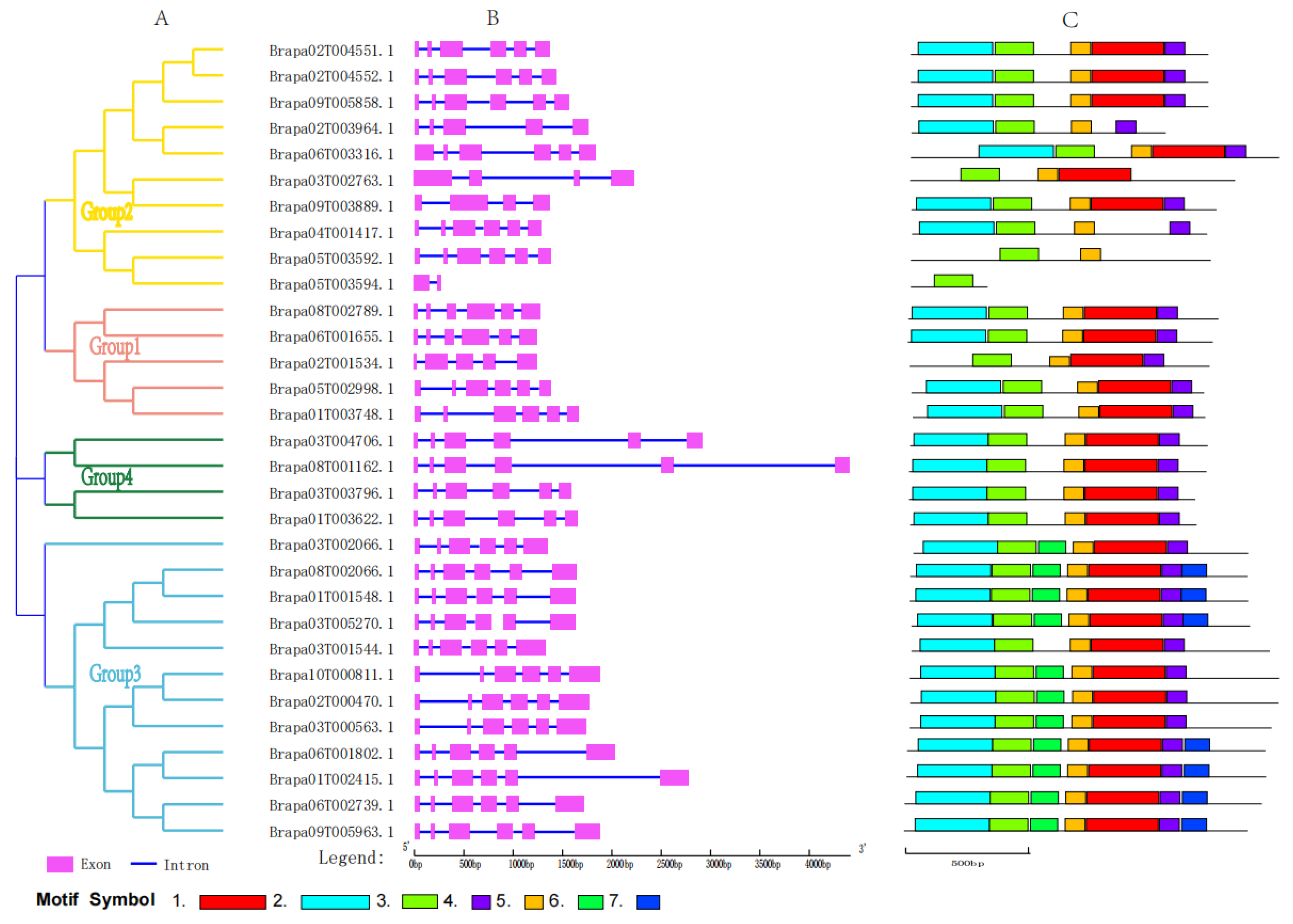
2.1. Identification and Structural Analysis of BraSWEET Genes Family
| Gene Name | Gene ID | Chromosome Number | Transmembrane Domains | CDS (bp) | Protein Length (aa) | MW (kD) | PI | Number of Domains |
|---|---|---|---|---|---|---|---|---|
| BraSWEET1.1 | Brapa06T001655.1 | A06 | 7 | 948 | 246 | 27.155 | 9.3 | 2 |
| BraSWEET1.2 | Brapa08T002789.1 | A08 | 7 | 1006 | 250 | 27.66 | 9.29 | 2 |
| BraSWEET2 | Brapa05T002998.1 | A05 | 7 | 1210 | 236 | 26.644 | 8.95 | 2 |
| BraSWEET2.1 | Brapa01T003748.1 | A01 | 7 | 969 | 236 | 26.3626 | 8.93 | 2 |
| BraSWEET3 | Brapa02T001534.1 | A02 | 6 | 799 | 242 | 27.584 | 8.52 | 2 |
| BraSWEET4.1 | Brapa02T003964.1 | A02 | 6 | 688 | 205 | 22.813 | 7.66 | 2 |
| BraSWEET4.2 | Brapa06T003316.1 | A06 | 8 | 1183 | 297 | 32.983 | 9.22 | 2 |
| BraSWEET5 | Brapa09T005858.1 | A09 | 7 | 723 | 240 | 27.244 | 9.04 | 2 |
| BraSWEET5.1 | Brapa02T004551.1 | A02 | 7 | 723 | 240 | 26.971 | 8.15 | 2 |
| BraSWEET5.2 | Brapa02T004552.1 | A02 | 7 | 723 | 240 | 26.823 | 8.14 | 2 |
| BraSWEET6.1 | Brapa09T003889.1 | A09 | 7 | 741 | 246 | 27.215 | 9.54 | 2 |
| BraSWEET6.2 | Brapa03T002763.1 | A03 | 6 | 789 | 262 | 28.707 | 9.25 | 2 |
| BraSWEET8 | Brapa04T001417.1 | A04 | 6 | 717 | 238 | 26.794 | 8.97 | 2 |
| BraSWEET8.1 | Brapa05T003594.1 | A05 | 1 | 189 | 62 | 7.012 | 4.66 | 1 |
| BraSWEET8.2 | Brapa05T003592.1 | A05 | 6 | 924 | 242 | 27.207 | 8.42 | 2 |
| BraSWEET9 | Brapa03T002066.1 | A03 | 7 | 813 | 270 | 30.125 | 9.18 | 2 |
| BraSWEET10 | Brapa03T001544.1 | A03 | 7 | 952 | 289 | 33.042 | 9.29 | 2 |
| BraSWEET11.1 | Brapa01T002415.1 | A01 | 7 | 1197 | 290 | 32.344 | 9.04 | 2 |
| BraSWEET11.2 | Brapa06T001802.1 | A06 | 7 | 1274 | 289 | 32.067 | 9.25 | 2 |
| BraSWEET12.1 | Brapa06T002739.1 | A06 | 7 | 1318 | 288 | 31.782 | 9.07 | 2 |
| BraSWEET12.2 | Brapa09T005963.1 | A09 | 7 | 1146 | 277 | 30.498 | 9.13 | 2 |
| BraSWEET14 | Brapa03T005270.1 | A03 | 7 | 1048 | 273 | 30.29 | 9.27 | 2 |
| BraSWEET14.1 | Brapa08T002066.1 | A08 | 7 | 930 | 272 | 29.917 | 9.18 | 2 |
| BraSWEET14.2 | Brapa01T001548.1 | A01 | 7 | 1079 | 273 | 30.1 | 9.2 | 2 |
| BraSWEET15.1 | Brapa03T000563.1 | A03 | 7 | 1057 | 292 | 32.848 | 8.16 | 2 |
| BraSWEET15.2 | Brapa10T000811.1 | A10 | 7 | 1155 | 298 | 33.384 | 8.44 | 2 |
| BraSWEET15.3 | Brapa02T000470.1 | A02 | 7 | 1129 | 297 | 33.156 | 8.26 | 2 |
| BraSWEET16.1 | Brapa03T003796.1 | A03 | 7 | 686 | 231 | 25.68 | 9.06 | 2 |
| BraSWEET16.2 | Brapa01T003622.1 | A01 | 7 | 696 | 231 | 25.755 | 8.69 | 2 |
| BraSWEET17.1 | Brapa08T001162.1 | A08 | 7 | 1001 | 240 | 26.561 | 9.24 | 2 |
| BraSWEET17.2 | Brapa03T004706.1 | A03 | 7 | 1182 | 240 | 26.455 | 8.76 | 2 |
2.2. Phylogenetic Analysis of BraSWEET Proteins
2.3. Chromosomal Localization, Collinearity Analysis, and Tandem Replication of BraSWEETs
2.4. Analysis of Secondary and Tertiary Structures of BraSWEET Proteins
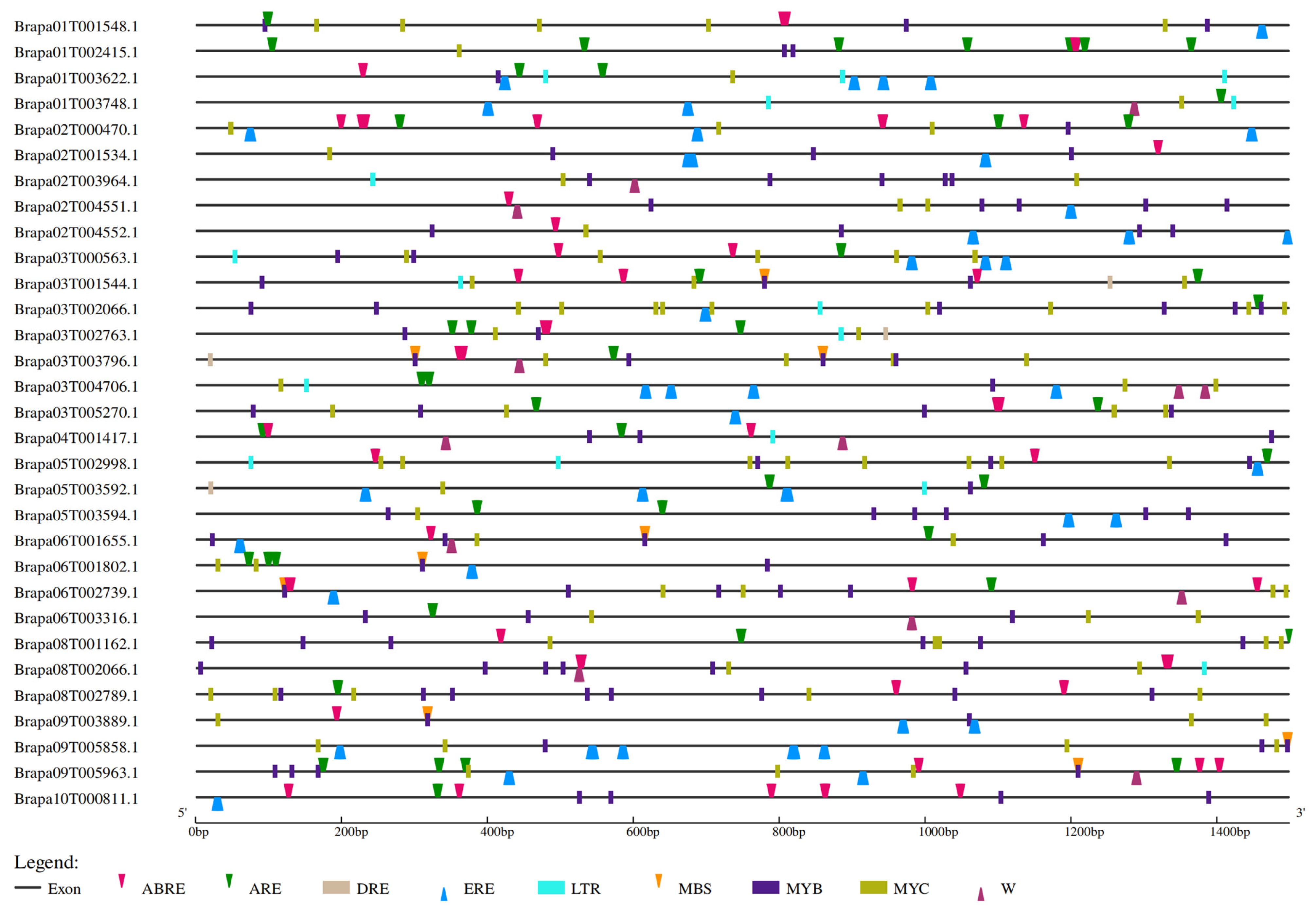
2.5. Cis-Element Analysis for the BraSWEET Gene Family
2.6. Protein–Protein Interaction Network for BraSWEETs
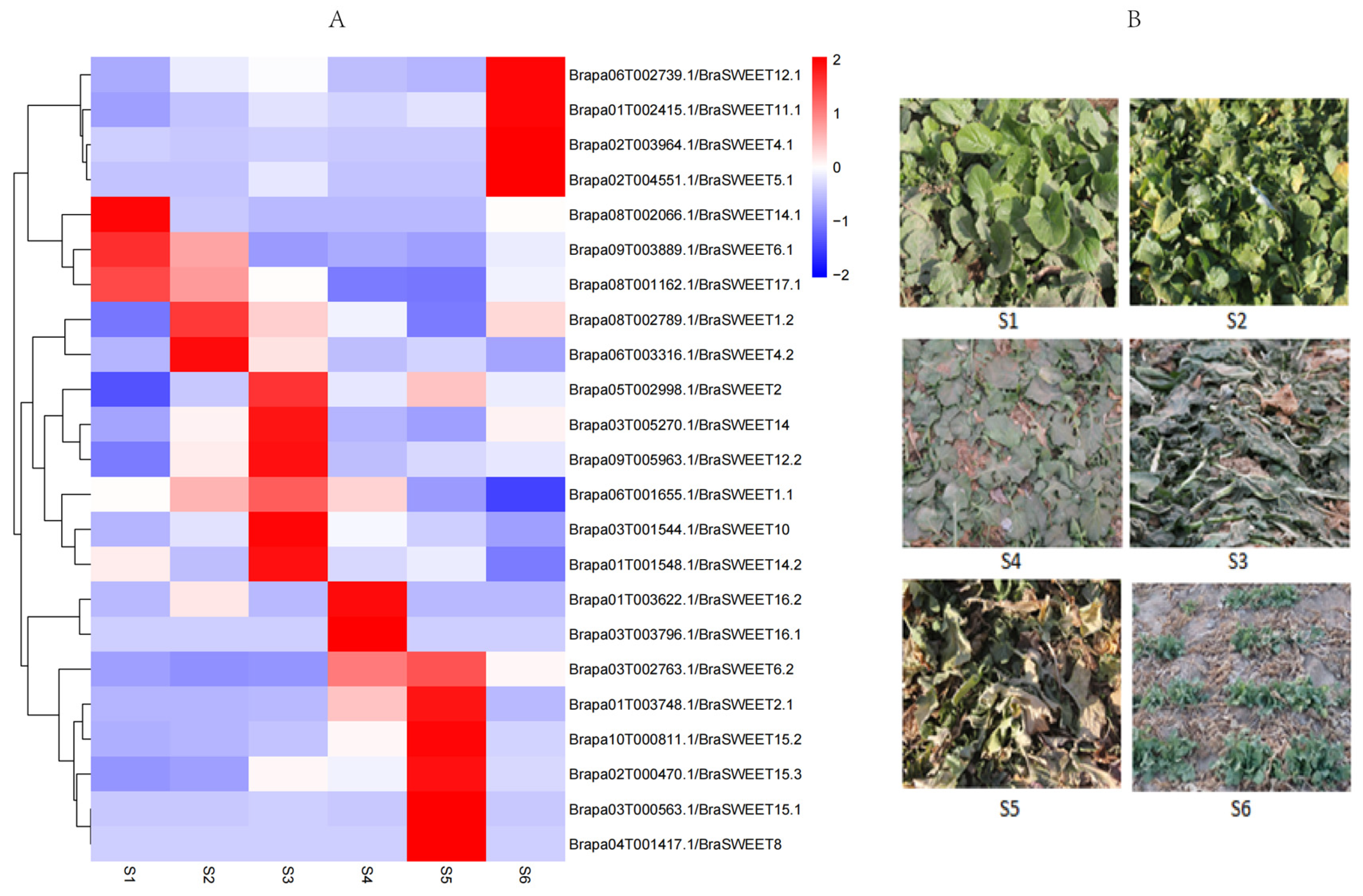
2.7. Analysis of Transcriptional Expression of BraSWEET Genes During Overwintering
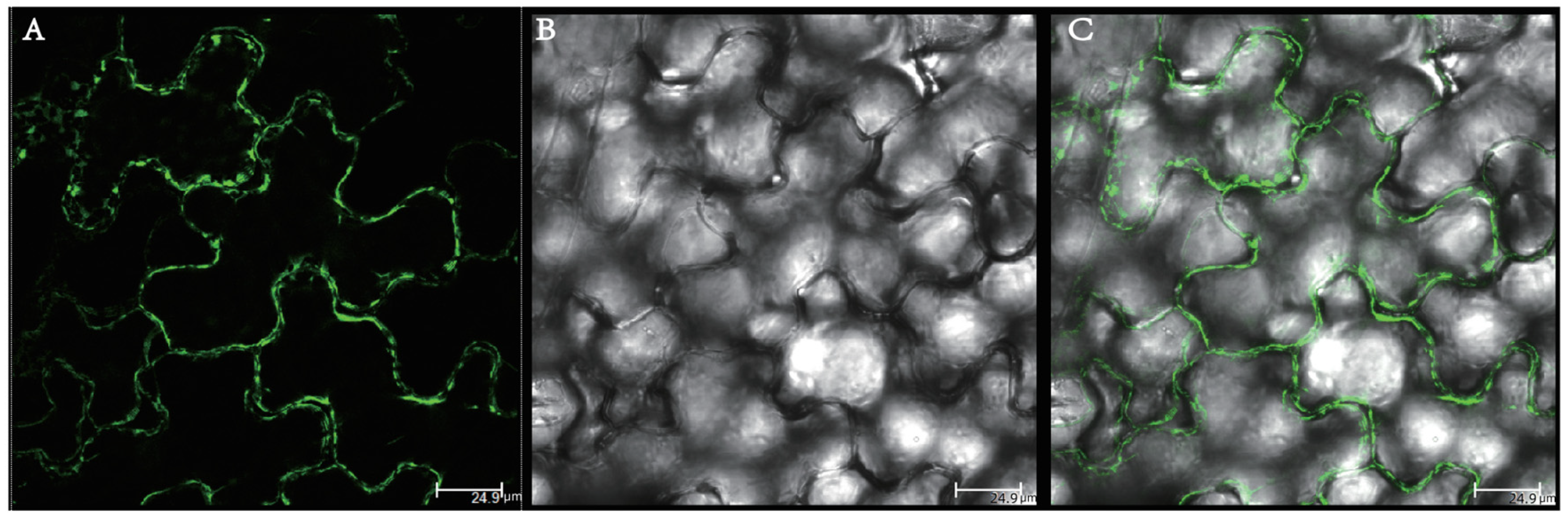
2.8. Subcellular Localization of BraSWEET10 Protein in Tobacco
2.9. Screening and Characterization of BraSWEET10 Overexpressing Transgenic A. thaliana
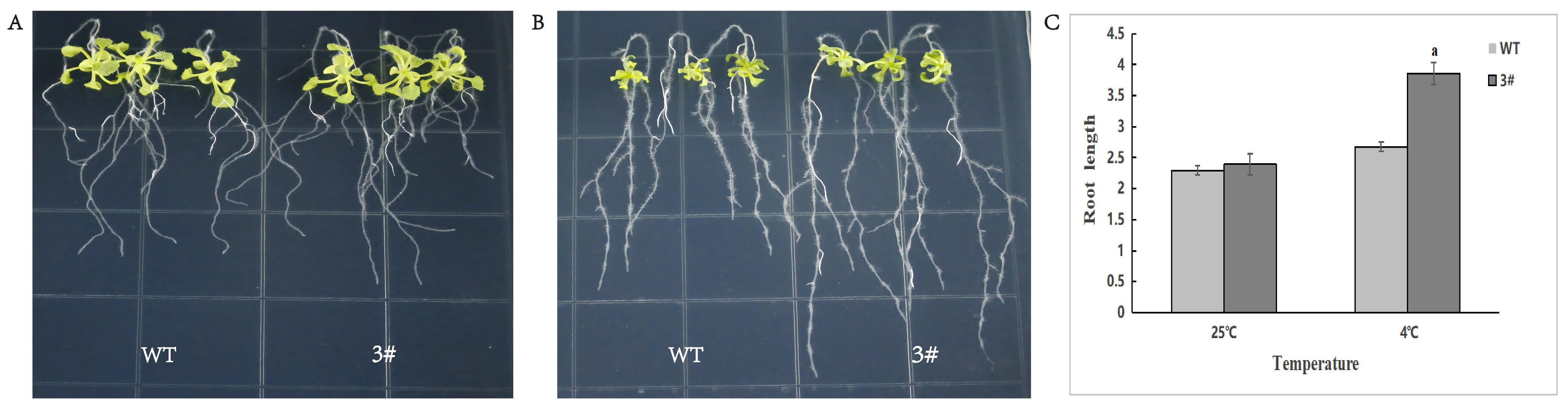
2.10. Resistance Analysis of BraSWEET10 Overexpressing A. thaliana Plants
3. Discussion
3.1. Characteristics of the BraSWEET Gene Family
3.2. Evolutionary Analysis of the BraSWEET Gene Family
3.3. Prediction of Gene Function Based on Protein Interaction and Promoter Analysis
3.4. Analysis of BraSWEET Expression During Overwintering
3.5. Functional Analysis of BraSWEET10
4. Materials and Methods
4.1. Identification of the SWEET Gene Family in B. rapa
4.2. Multiple Sequence Alignment and Phylogenetic Analysis of BraSWEET Genes
4.3. Analysis of Transmembrane Structure, Conserved Motif, and Gene Structure
4.4. Chromosomal Location and Gene Duplication of BraSWEET Genes
4.5. Analysis of the Secondary and Tertiary Structures of BraSWEET Proteins
4.6. Promoter Analysis for BraSWEETs
4.7. Plant Materials, Field Trails, Sample Collection, and Transcriptional Expression Analysis of BraSWEET Genes During Overwintering by RNA-Seq Data Analysis
4.8. Construction of the Overexpression Vector
4.9. Subcellular Localization of the BraSWEET10 Protein in Tobacco (Nicotiana benthamiana)
4.10. Transformation of A. thaliana Using Overexpression Vectors
4.11. Phenotypic Observations of Transgenic A. thaliana
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yamada, K.; Osakabe, Y.; Mizoi, J.; Nakashima, K.; Fujita, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of an Arabidopsis thaliana abiotic stress-inducible facilitated diffusion transporter for monosaccharides. J. Biol. Chem. 2009, 285, 1138–1146. [Google Scholar] [CrossRef] [PubMed]
- Williams, L.E.; Lemoine, R.; Sauer, N. Sugar transporters in higher plants: A diversity of roles and complex regulation. Trends Plant Sci. 2000, 5, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Lalonde, S.; Wipf, D.; Frommer, W.B. Transport mechanisms for organic forms of carbon and nitrogen between source and sink. Annu. Rev. Plant Biol. 2004, 55, 341–372. [Google Scholar] [CrossRef] [PubMed]
- Aron, M.B.; Shennan, L.; Anderson, J.B.; Farideh, C.; Derbyshire, M.K.; Carol, D.W.S.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R. CDD: A Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011, 39, D225–D229. [Google Scholar]
- Büttner, M.; Sauer, N. Monosaccharide transporters in plants: Structure, function and physiology. Biochim. Biophys. Acta Biomembr. 2000, 1465, 263–274. [Google Scholar] [CrossRef]
- Chen, L.Q.; Hou, B.H.; Lalonde, S.; Takanaga, H.; Hartung, M.L.; Qu, X.Q.; Guo, W.J.; Kim, J.G.; Underwood, W.; Chaudhuri, B.; et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 2010, 468, 527–532. [Google Scholar] [CrossRef]
- Yuan, M.; Wang, S. Rice MtN3/Saliva/SWEET family genes and their homologs in cellular organisms. Mol. Plant 2013, 6, 665–674. [Google Scholar] [CrossRef]
- Baker, R.F.; Leach, K.A.; Braun, D.M. SWEET as sugar: New sucrose effluxers in plants. Mol. Plant 2012, 5, 766–768. [Google Scholar] [CrossRef]
- Braun, D.M. SWEET! The pathway is complete. Science 2012, 335, 173–174. [Google Scholar] [CrossRef]
- Seo, P.J.; Park, J.M.; Kang, S.K.; Kim, S.G.; Park, C.M. An Arabidopsis senescence-associated protein SAG29 regulates cell viability under high salinity. Planta 2011, 233, 189–200. [Google Scholar] [CrossRef]
- Chardon, F.; Bedu, M.; Calenge, F.; Klemens, P.W.; Spinner, L.; Clement, G.; Chietera, G.; Léran, S.; Ferrand, M.; Lacombe, B. Leaf fructose content is controlled by the vacuolar transporter SWEET17 in Arabidopsis. Curr. Biol. 2013, 23, 697–702. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.J.; Nagy, R.; Chen, H.Y.; Pfrunder, S.; Yu, Y.C.; Santelia, D.; Frommer, W.B.; Martinoia, E. SWEET17, a facilitative transporter, mediates fructose transport across the tonoplast of Arabidopsis roots and leaves. Plant Physiol. 2014, 164, 777–789. [Google Scholar] [CrossRef] [PubMed]
- Hir, R.L.; Spinner, L.; Klemens, P.A.W.; Chakraborti, D.; Marco, F.d.; Vilaine, F.; Wolff, N.; Lemoine, R.; Porcheron, B.; Géry, C.; et al. Disruption of the Sugar Transporters AtSWEET11 and AtSWEET12 affects vascular development and freezing tolerance in Arabidopsis. Mol. Plant 2015, 8, 1687–1690. [Google Scholar]
- Liu, L.; Pu, Y.; Niu, Z.; Wu, J.; Fang, Y.; Xu, J.; Xu, F.D.; Yue, J.; Ma, L.; Li, X.; et al. Corrigendum: Transcriptomic insights into root development and overwintering transcriptional memory of Brassica rapa L. grown in the field. Front. Plant Sci. 2023, 14, 1195912. [Google Scholar] [CrossRef]
- Gusta, L.V.; Waalen, W.; Øvergaard, S.I.; Åssveen, M.; Eltun, R.; Gusta, L. Winter survival of winter rapeseed and winter turnip rapeseed in field trials, as explained by PPLS regression. Eur. J. Agron. 2013, 51, 81–90. [Google Scholar]
- Hong, L.; Xiao Nan, L.; Yuan Hu, X.; Jing, J.; Yang Dou, W.; Zhong Yun, P. Genome wide identification and expression profiling of SWEET genes family reveals its role during Plasmodiophora brassicae-induced formation of clubroot in Brassica rapa. Front. Plant Sci. 2018, 9, 207. [Google Scholar]
- Li Ping, H.; Feng, Z.; Shu Hui, S.; Xiao Wei, T.; Hui, X.; Guang Min, L.; Ya Qin, W.; Hong Ju, H. Genome-wide identification, characterization, and expression analysis of the SWEET gene family in cucumber. J. Integr. Agric. 2017, 16, 1486–1501. [Google Scholar]
- Xuan, Y.H.; Hu, Y.B.; Chen, L.Q.; Sosso, D.; Ducat, D.C.; Hou, B.H.; Frommer, W.B. Functional role of oligomerization for bacterial and plant SWEET sugar transporter family. Proc. Natl. Acad. Sci. USA 2013, 110, E3685–E3694. [Google Scholar] [CrossRef]
- Swanson, W.J.; Yang, Z.; Wolfner, M.F. Positive Darwinian selection drives the evolution of several female reproductive proteins in mammals. Proc. Natl. Acad. Sci. USA 2001, 98, 2509–2514. [Google Scholar] [CrossRef]
- Bahn, S.C.; Bae, M.S.; Park, Y.B.; Oh, S.I.; Shin, J.S. Molecular cloning and characterization of a novel low temperature-induced gene, blti2, from barley (Hordeum vulgare L.). Biochim. Biophys. Acta Biomembr. 2001, 1522, 134–137. [Google Scholar] [CrossRef]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 1994, 6, 251–264. [Google Scholar]
- Dang, F.; Wang, Y.; She, J.; Lei, Y.; Liu, Z.; Eulgem, T.; Lai, Y.; Lin, J.; Yu, L.; Lei, D. Overexpression of CaWRKY27, a subgroup IIe WRKY transcription factor of Capsicum annuum, positively regulates tobacco resistance to Ralstonia solanacearum infection. Physiol. Plant. 2014, 150, 397–411. [Google Scholar] [CrossRef]
- Eulgem, T.; Somssich, I.E. Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 2007, 10, 366–371. [Google Scholar] [CrossRef]
- Dou, X.Y.; Yang, K.Z.; Zhang, Y.; Wang, W.; Liu, X.L.; Chen, L.Q.; Zhang, X.Q.; Ye, D. WBC27,an adenosine Tri-phosphate-binding cassette protein, controls pollen wall formation and patterning in Arabidopsis. J. Integr. Plant Biol. 2011, 53, 74–88. [Google Scholar] [CrossRef]
- Huang, X.Y.; Niu, J.; Sun, M.X.; Zhu, J.; Gao, J.F.; Yang, J.; Zhou, Q.; Yang, Z.N. CYCLIN-DEPENDENT KINASE G1 is associated with the spliceosome to regulate CALLOSE SYNTHASE5 splicing and pollen wall formation in Arabidopsis. Plant Cell 2013, 25, 637–648. [Google Scholar] [CrossRef]
- Paxson-Sowders, D.M.; Dodrill, C.H.; Owen, H.A.; Makaroff, C.A. DEX1, a novel plant protein, is required for exine pattern formation during pollen development in Arabidopsis. Plant Physiol. 2001, 127, 1739–1749. [Google Scholar] [CrossRef]
- Huang, K.C.; Lin, W.C.; Cheng, W.H. Salt hypersensitive mutant 9, a nucleolar APUM23 protein, is essential for salt sensitivity in association with the ABA signaling pathway in Arabidopsis. BMC Plant Biol. 2018, 18, 40. [Google Scholar] [CrossRef]
- Gong, X.; Liu, M.; Zhang, L.; Ruan, Y.; Ding, R.; Ji, Y.; Zhang, N.; Zhang, S.; Farmer, J.; Wang, C. Arabidopsis AtSUC2 and AtSUC4, encoding sucrose transporters, are required for abiotic stress tolerance in an ABA-dependent pathway. Physiol. Plant. 2015, 153, 119–136. [Google Scholar] [CrossRef]
- Vatov, E.; Ludewig, U.; Zentgraf, U. Disparate dynamics of gene body and cis-regulatory element evolution illustrated for the senescence-associated cysteine protease gene SAG12 of plants. Plants 2021, 10, 1380. [Google Scholar] [CrossRef]
- Tran, L.-S.P.; Nakashima, K.; Sakuma, Y.; Simpson, S.D.; Fujita, Y.; Maruyama, K.; Fujita, M.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Isolation and functional analysis of Arabidopsis Stress-inducible NAC transcription factors that bind to a Drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell Online 2004, 16, 2481–2498. [Google Scholar] [CrossRef]
- Wu, H.M.; Cheun, A.Y. Programmed cell death in plant reproduction. Plant Mol. Biol. 2000, 44, 267–281. [Google Scholar] [CrossRef]
- Choi, A.S.; Prokchorchik, M.; Lee, H.; Gupta, R.; Lee, Y.; Chung, E.H.; Cho, B.; Kim, M.S.; Sun, T.K.; Sohn, K.H. Direct acetylation of a conserved threonine of RIN4 by the bacterial effector HopZ5 or AvrBsT activates RPM1-dependent immunity in Arabidopsis. Mol. Plant 2021, 14, 1951–1960. [Google Scholar] [CrossRef]
- Redditt, T.J.; Chung, E.H.; Karimi, H.Z.; Rodibaugh, N.; Innes, R.W. AvrRpm1 functions as an ADP-ribosyl transferase to modify NOI domain-containing proteins, including Arabidopsis and Soybean RPM1-interacting protein4. Plant Cell 2019, 31, 2664–2681. [Google Scholar]
- Gamas, P.; Niebel, F.D.C.; Lescure, N.; Cullimore, J. Use of a subtractive hybridization approach to identify new Medicago truncatula genes induced during root nodule development. Mol. Plant Microbe Interact. 1996, 9, 233–242. [Google Scholar] [CrossRef]
- Zhao, L.; Yao, J.; Chen, W.; Li, Y.; Lü, Y.; Guo, Y.; Wang, J.; Yuan, L.; Liu, Z.; Zhang, Y. A genome-wide analysis of SWEET gene family in cotton and their expressions under different stresses. J. Cotton Res. 2018, 1, 7. [Google Scholar] [CrossRef]
- Hu, B.; Wu, H.; Huang, W.; Song, J.; Lin, Y. SWEET Gene Family in Medicago truncatula: Genome-Wide identification, expression and substrate specificity analysis. Plants 2019, 8, 338. [Google Scholar] [CrossRef]
- Miao, L.; Lv, Y.; Kong, L.; Chen, Q.; Chen, C.; Li, J.; Zeng, F.; Wang, S.; Li, J.; Huang, L. Genome-wide identification, phylogeny, evolution, and expression patterns of MtN3/saliva/SWEET genes and functional analysis of BcNS in Brassica rapa. BMC Genom. 2018, 19, 174. [Google Scholar] [CrossRef]
- Xiao, Q.L.; Li, Z.; Wang, Y.Y.; Hou, X.B.; Wei, X.M.; Zhao, X.; Huang, L.; Guo, Y.J.; Liu, Z.Z. Genome-wide identification, expression and functional analysis of sugar transporters in sorghum (Sorghum bicolor L.). J. Integr. Agric. 2022, 21, 2848–2864. [Google Scholar] [CrossRef]
- Liu, Q.; Dang, H.; Chen, Z.; Wu, J.; Chen, Y.; Chen, S.; Luo, L. Genome-wide identification, expression, and functional analysis of the sugar transporter gene family in Cassava (Manihot esculenta). Int. J. Mol. Sci. 2018, 19, 987. [Google Scholar] [CrossRef]
- Chen, L.Q.; Qu, X.Q.; Hou, B.H.; Sosso, D.; Osorio, S.; Fernie, A.R.; Frommer, W.B. Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 2012, 335, 207–211. [Google Scholar] [CrossRef]
- Lin, I.W.; Sosso, D.; Chen, L.Q.; Gase, K.; Kim, S.G.; Kessler, D.; Klinkenberg, P.M.; Gorder, M.K.; Hou, B.H.; Qu, X.Q.; et al. Nectar secretion requires sucrose phosphate synthases and the sugar transporter SWEET9. Nature 2014, 508, 546–549. [Google Scholar] [CrossRef]
- Eom, J.S.; Chen, L.Q.; Sosso, D.; Julius, B.T.; Lin, I.W.; Qu, X.Q.; Braun, D.M.; Frommer, W.B. SWEETs, transporters for intracellular and intercellular sugar translocation. Curr. Opin. Plant Biol. 2015, 25, 53–62. [Google Scholar] [CrossRef]
- Patil, G.; Valliyodan, B.; Deshmukh, R.; Prince, S.; Nicander, B.; Zhao, M.; Sonah, H.; Song, L.; Lin, L.; Chaudhary, J.; et al. Soybean (Glycine max) SWEET gene family: Insights through comparative genomics, transcriptome profiling and whole genome re-sequence analysis. BMC Genom. 2015, 16, 520. [Google Scholar] [CrossRef]
- Mizuno, H.; Kasuga, S.; Kawahigashi, H. The sorghum SWEET gene family: Stem sucrose accumulation as revealed through transcriptome profiling. Biotechnol. Biofuels 2016, 9, 127. [Google Scholar] [CrossRef]
- Hu, W.; Hua, X.; Zhang, Q.; Wang, J.; Shen, Q.; Zhang, X.; Wang, K.; Yu, Q.; Lin, Y.R.; Ming, R. New insights into the evolution and functional divergence of the SWEET family in Saccharum based on comparative genomics. BMC Plant Biol. 2018, 18, 270. [Google Scholar] [CrossRef]
- Du, Y.; Li, W.; Geng, J.; Li, S.; Zhang, W.; Liu, X.; Hu, M.; Zhang, Z.; Fan, Y.; Yuan, X.; et al. Genome-wide identification of the SWEET gene family in Phaseolus vulgaris L. and their patterns of expression under abiotic stress. J. Plant Interact. 2022, 17, 390–403. [Google Scholar] [CrossRef]
- Klemens, P.A.W.; Patzke, K.; Deitmer, J.; Spinner, L.; Hir, R.L.; Bellini, C.; Bedu, M.; Chardon, F.; Neuhaus, A.K.E. Overexpression of the vacuolar sugar carrier AtSWEET16 modifies germination, growth, and stress tolerance in Arabidopsis. Plant Physiol. 2013, 163, 1338–1352. [Google Scholar] [CrossRef]
- He, F.; Kang, J.; Zhou, X.; Su, Z.; Qu, L. Variation at the transcriptional level among Chinese natural populations of Arabidopsis thaliana in response to cold stress. Chin. Sci. Bull. 2008, 53, 2989–2999. [Google Scholar] [CrossRef]
- Wang, L.; Yao, L.; Hao, X.; Li, N.; Qian, W.; Yue, C.; Ding, C.; Zeng, J.; Yang, Y.; Wang, X. Tea plant SWEET transporters: Expression profiling, sugar transport, and the involvement of CsSWEET16 in modifying cold tolerance in Arabidopsis. Plant Mol. Biol. 2018, 96, 577–592. [Google Scholar] [CrossRef]
- Yue, C.; Cao, H.L.; Wang, L.; Zhou, Y.H.; Huang, Y.T.; Hao, X.Y.; Wang, Y.C.; Wang, B.; Yang, Y.J.; Wang, X.C. Effects of cold acclimation on sugar metabolism and sugar-related gene expression in tea plant during the winter season. Plant Mol. Biol. 2015, 88, 591–608. [Google Scholar] [CrossRef]
- Wu, Y.; Peng, W.; Xiong, F. Sucrose transport involves in disease response to Xanthomonas oryzae pathovar oryzae. Plant Signal. Behav. 2019, 14, 1656949. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Elisabeth, G.; Alexandre, G.; Christine, H.; Ivan, I.; Appel, R.D.; Amos, B. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; Mcgettigan, P.A.; Mcwilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Sudhir, K.; Glen, S.; Koichiro, T. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar]
- Sonnhammer, E.L.L.; Heijne, G.V.; Krogh, A.A. Hidden Markov Model for Predicting Transmembrane Helices in Protein Sequences. Int. Conf. Intell. Syst. Mol. Biol. 1998, 6, 175–182. [Google Scholar]
- Paul, B.; Laura, B.; Richard, H.; Jim, B.; Moore, J.D.; Sascha, O. MEME-LaB: Motif analysis in clusters. Bioinformatics 2013, 29, 1696–1697. [Google Scholar]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2014, 31, 1296–1297. [Google Scholar] [CrossRef]
- Voorrips, R.E. MapChart: Software for the Graphical Presentation of Linkage Maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Tae Ho, L.; Jin, H.; Barry, M.; Guo, H. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]



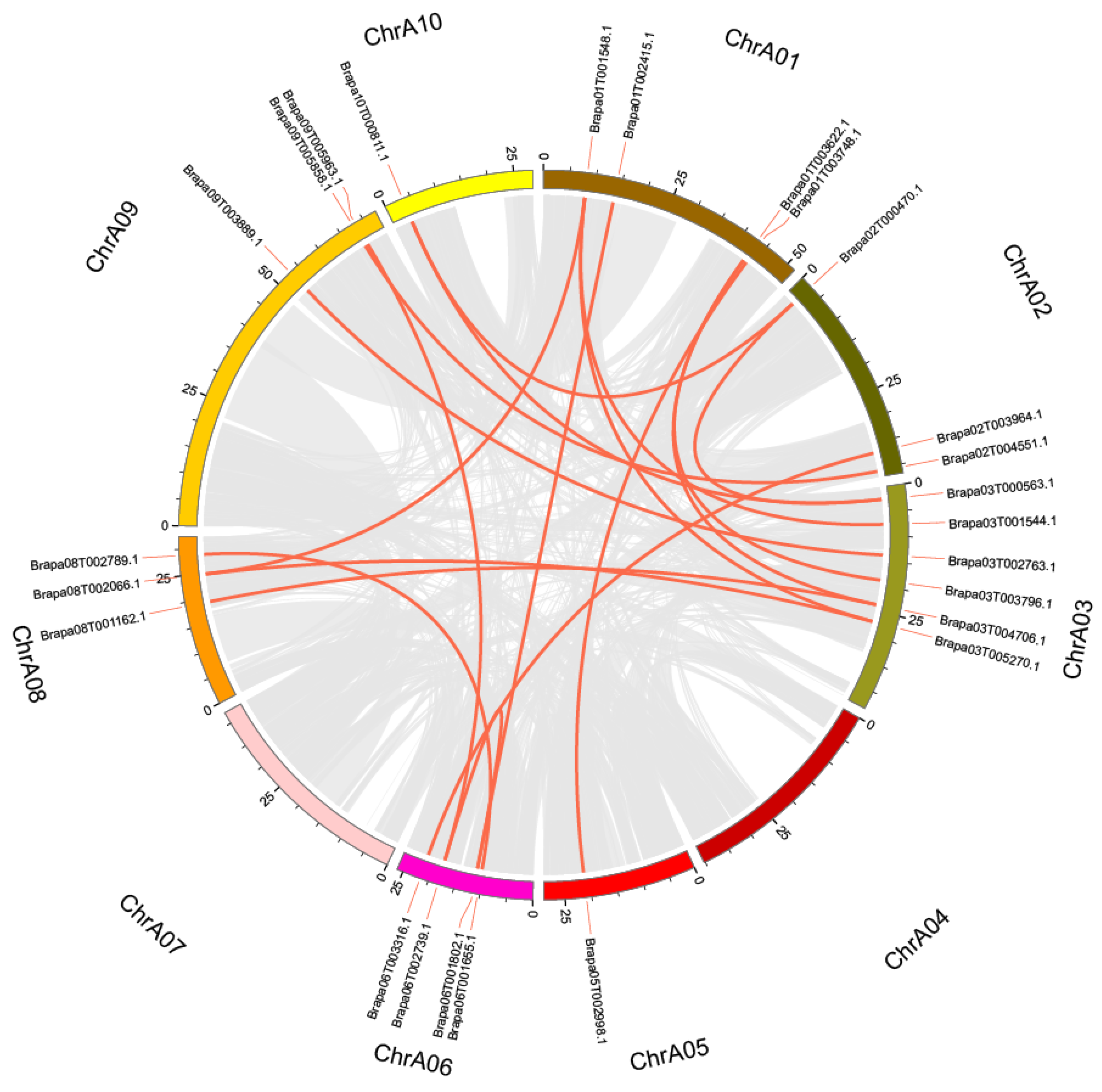




| Orthologous Gene Pairs | Non-Synonymous Substitution Rate (Ka) | Synonymous Substitution Rate (Ks) | Ka/Ks | Duplication Date (MYA) |
|---|---|---|---|---|
| Brapa06T001655/BraSWEET1.1 | 0.0302 | 0.2264 | 0.1332 | 7.55 |
| Brapa08T002789/BraSWEET1.2 | ||||
| Brapa01T003748/BraSWEET2.1 | 0.0714 | 0.3188 | 0.2241 | 10.63 |
| Brapa05T002998/BraSWEET2 | ||||
| Brapa02T003964/BraSWEET4.1 | 0.0375 | 0.2520 | 0.1488 | 8.40 |
| Brapa06T003316/BraSWEET4.2 | ||||
| Brapa02T004551/BraSWEET5.1 | 0.0764 | 0.0928 | 0.8239 | 3.09 |
| Brapa02T004552/BraSWEET5.2 | ||||
| Brapa02T004551/BraSWEET5.1 | 0.1144 | 0.2685 | 0.4261 | 8.95 |
| Brapa09T005858/BraSWEET5 | ||||
| Brapa03T002763/BraSWEET6.2 | 0.2182 | 0.5115 | 0.4267 | 17.05 |
| Brapa09T003889/BraSWEET6.1 | ||||
| Brapa01T002415/BraSWEET11.1 | 0.0738 | 0.2497 | 0.2955 | 8.32 |
| Brapa06T001802/BraSWEET11.2 | ||||
| Brapa06T001802/BraSWEET11.2 | 0.0777 | 0.7063 | 0.1100 | 23.54 |
| Brapa06T002739/BraSWEET12.1 | ||||
| Brapa06T002739/BraSWEET12.1 | 0.0460 | 0.1753 | 0.2625 | 5.84 |
| Brapa09T005963/BraSWEET12.2 | ||||
| Brapa03T005270/BraSWEET14 | 0.0431 | 0.2317 | 0.1861 | 7.72 |
| Brapa08T002066/BraSWEET14.1 | ||||
| Brapa01T001548/BraSWEET14.2 | 0.0340 | 0.2175 | 0.1565 | 7.25 |
| Brapa03T005270/BraSWEET14 | ||||
| Brapa01T001548/BraSWEET14.2 | 0.0323 | 0.2122 | 0.1521 | 7.07 |
| Brapa08T002066/BraSWEET14.1 | ||||
| Brapa01T001548/BraSWEET14.2 | 0.4723 | 2.1231 | 0.2224 | 70.77 |
| Brapa03T001544/BraSWEET10 | ||||
| Brapa03T000563/BraSWEET15.1 | 0.0782 | 0.2395 | 0.3263 | 7.98 |
| Brapa10T000811/BraSWEET15.2 | ||||
| Brapa01T003622/BraSWEET16.2 | 0.0452 | 0.3249 | 0.1392 | 10.83 |
| Brapa03T003796/BraSWEET16.1 | ||||
| Brapa01T003622/BraSWEET16.2 | 0.1716 | 0.6453 | 0.2659 | 21.51 |
| Brapa03T004706/BraSWEET17.2 | ||||
| Brapa02T000470/BraSWEET17.2 | 0.0967 | 0.2848 | 0.3394 | 9.49 |
| Brapa03T000563/BraSWEET15.1 | ||||
| Brapa02T000470/BraSWEET17.2 | 0.0937 | 0.2711 | 0.3458 | 9.04 |
| Brapa10T000811/BraSWEET15.2 | ||||
| Brapa03T004706/BraSWEET17.2 | 0.0520 | 0.1836 | 0.2831 | 6.12 |
| Brapa08T001162/BraSWEET17.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yue, J.; Yuan, S.; Liu, L.; Niu, Z.; Ma, L.; Pu, Y.; Wu, J.; Fang, Y.; Sun, W. Genome-Wide Identification of the SWEET Gene Family and Functional Analysis of BraSWEET10 in Winter B. rapa (Brassica rapa L.) Under Low-Temperature Stress. Int. J. Mol. Sci. 2025, 26, 2398. https://doi.org/10.3390/ijms26062398
Yue J, Yuan S, Liu L, Niu Z, Ma L, Pu Y, Wu J, Fang Y, Sun W. Genome-Wide Identification of the SWEET Gene Family and Functional Analysis of BraSWEET10 in Winter B. rapa (Brassica rapa L.) Under Low-Temperature Stress. International Journal of Molecular Sciences. 2025; 26(6):2398. https://doi.org/10.3390/ijms26062398
Chicago/Turabian StyleYue, Jinli, Shunjie Yuan, Lijun Liu, Zaoxia Niu, Li Ma, Yuanyuan Pu, Junyan Wu, Yan Fang, and Wancang Sun. 2025. "Genome-Wide Identification of the SWEET Gene Family and Functional Analysis of BraSWEET10 in Winter B. rapa (Brassica rapa L.) Under Low-Temperature Stress" International Journal of Molecular Sciences 26, no. 6: 2398. https://doi.org/10.3390/ijms26062398
APA StyleYue, J., Yuan, S., Liu, L., Niu, Z., Ma, L., Pu, Y., Wu, J., Fang, Y., & Sun, W. (2025). Genome-Wide Identification of the SWEET Gene Family and Functional Analysis of BraSWEET10 in Winter B. rapa (Brassica rapa L.) Under Low-Temperature Stress. International Journal of Molecular Sciences, 26(6), 2398. https://doi.org/10.3390/ijms26062398






