Expression Analysis of Circulating miR-21, miR-34a and miR-122 and Redox Status Markers in Metabolic Dysfunction-Associated Steatotic Liver Disease Patients with and Without Type 2 Diabetes
Abstract
1. Introduction
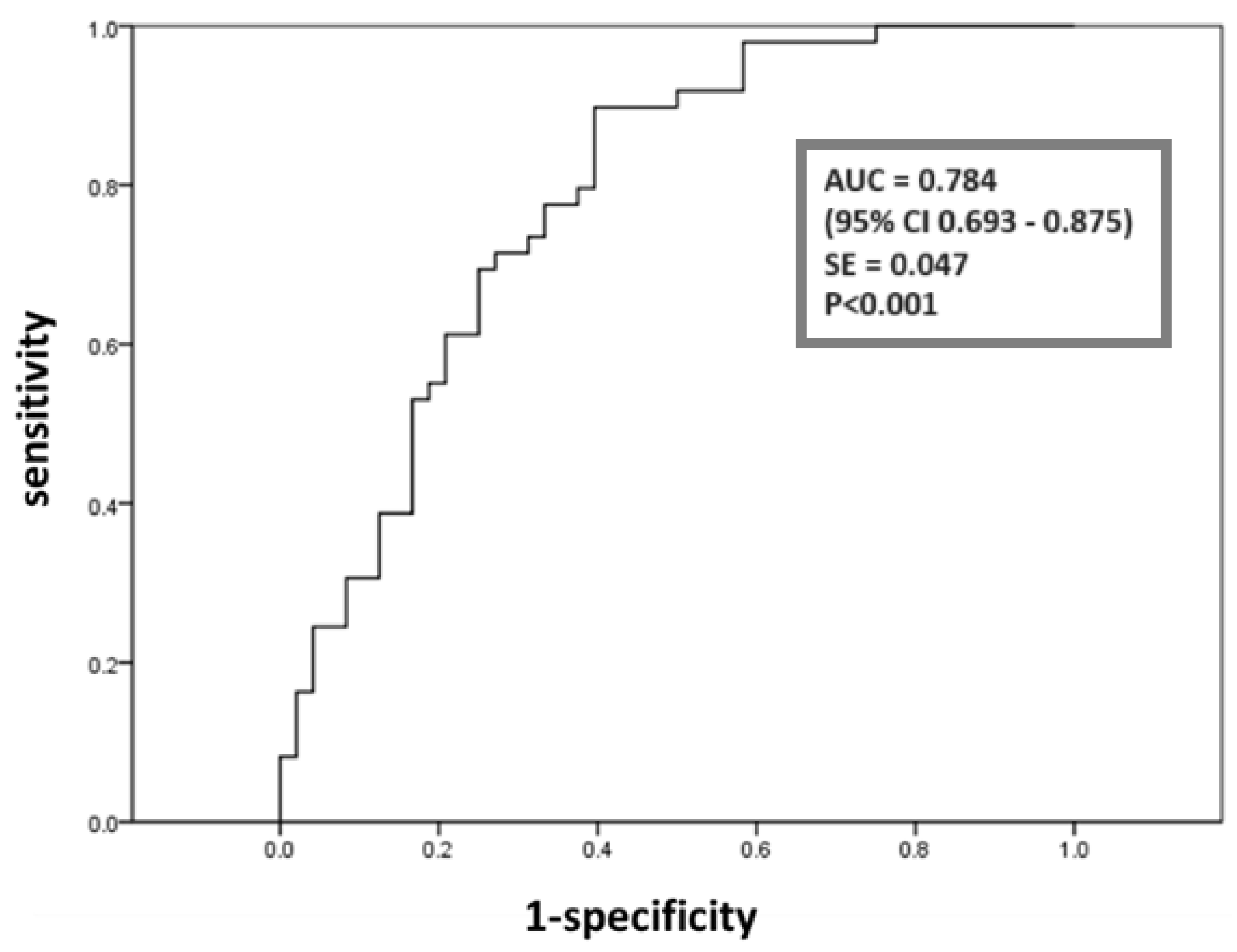
2. Results
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. Blood Biochemistry
4.3. Redox Status Markers
4.4. miRNA Isolation
4.5. miRNA Quantification
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Younossi, Z.M.; Golabi, P.; Paik, J.M.; Henry, A.; Van Dongen, C.; Henry, L. The global epidemiology of nonalcoholic fatty liver disease (NAFLD) and nonalcoholic steatohepatitis (NASH): A systematic review. Hepatology 2023, 77, 1335–1347. [Google Scholar] [CrossRef]
- Rinella, M.E.; Lazarus, J.V.; Ratziu, V.; Francque, S.M.; Sanyal, A.J.; Kanwal, F.; Romero, D.; Abdelmalek, M.F.; Anstee, Q.M.; Arab, J.P.; et al. A multisociety delphi consensus statement on New Fatty Liver Disease Nomenclature. Hepatology 2023, 78, 1966–1986. [Google Scholar] [CrossRef] [PubMed]
- Arora, U.; Biswas, S.; Aggarwal, S.; Duseja, A. MASLD screening and diagnostic algorithms are interchangeable with existing NAFLD literature. J. Hepatol. 2024, 80, e89–e91. [Google Scholar] [CrossRef] [PubMed]
- Clark, J.M.; Brancati, F.L.; Diehl, A.M. Nonalcoholic fatty liver disease. Gastroenterology 2002, 122, 1649–1657. [Google Scholar] [CrossRef]
- Marchesini, G.; Day, C.P.; Dufour, J.-F.; Canbay, A.; Nobili, V.; Ratziu, V.; Tilg, H.; Roden, M.; Gastaldelli, A.; Yki-Järvinen, H.; et al. EASL-EASD-EASO Clinical Practice Guidelines for the management of non-alcoholic fatty liver disease. J. Hepatol. 2016, 64, 1388–1402. [Google Scholar] [CrossRef]
- Méndez-Sánchez, N.; Valencia-Rodríguez, A.; Coronel-Castillo, C.; Vera-Barajas, A.; Contreras-Carmona, J.; Ponciano-Rodríguez, G.; Zamora-Valdés, D. The cellular pathways of liver fibrosis in non-alcoholic steatohepatitis. Ann. Transl. Med. 2020, 8, 400. [Google Scholar] [CrossRef] [PubMed]
- Jou, J.; Choi, S.; Diehl, A. Mechanisms of disease progression in nonalcoholic fatty liver disease. Semin. Liver Dis. 2008, 28, 370–379. [Google Scholar] [CrossRef] [PubMed]
- Targher, G.; Corey, K.E.; Byrne, C.D.; Roden, M. The complex link between NAFLD and type 2 diabetes mellitus—mechanisms and treatments. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 599–612. [Google Scholar] [CrossRef]
- Yamada, H.; Suzuki, K.; Ichino, N.; Ando, Y.; Sawada, A.; Osakabe, K.; Sugimoto, K.; Ohashi, K.; Teradaira, R.; Inoue, T.; et al. Associations between circulating microRNAs (miR-21, miR-34a, miR-122 and miR-451) and non-alcoholic fatty liver. Clin. Chim. Acta 2013, 424, 99–103. [Google Scholar] [CrossRef]
- Tilg, H.; Moschen, A.R. Evolution of inflammation in nonalcoholic fatty liver disease: The multiple parallel hits hypothesis. Hepatology 2010, 52, 1836–1846. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.B. From sugar to liver fat and public health: Systems biology driven studies in understanding non-alcoholic fatty liver disease pathogenesis. Proc. Nutr. Soc. 2019, 78, 290–304. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [PubMed]
- Carthew, R.W.; Sontheimer, E.J. Origins and mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642–655. [Google Scholar] [CrossRef]
- Pozniak, T.; Shcharbin, D.; Bryszewska, M. Circulating microRNAs in medicine. Int. J. Mol. Sci. 2022, 23, 3996. [Google Scholar] [CrossRef]
- Atic, A.I.; Thiele, M.; Munk, A.; Dalgaard, L.T. Circulating mirnas associated with nonalcoholic fatty liver disease. Am. J. Physiol. Cell Physiol. 2023, 324, C588–C602. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef]
- Chen, X.; Ba, Y.; Ma, L.; Cai, X.; Yin, Y.; Wang, K.; Guo, J.; Zhang, Y.; Chen, J.; Guo, X.; et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008, 18, 997–1006. [Google Scholar] [CrossRef]
- Xu, D.; Di, K.; Fan, B.; Wu, J.; Gu, X.; Sun, Y.; Khan, A.; Li, P.; Li, Z. MicroRNAs in extracellular vesicles: Sorting mechanisms, diagnostic value, isolation, and detection technology. Front. Bioeng. Biotechnol. 2022, 10, 948959. [Google Scholar] [CrossRef]
- Cai, C.; Lin, Y.; Yu, C. Circulating miRNAs as novel diagnostic biomarkers in nonalcoholic fatty liver disease: A systematic review and meta-analysis. Can. J. Gastroenterol. Hepatol. 2019, 1, 2096161. [Google Scholar] [CrossRef]
- Lee, S.S.; Park, S.H. Radiologic evaluation of nonalcoholic fatty liver disease. World J. Gastroenterol. 2014, 20, 7392. [Google Scholar] [CrossRef]
- Baranova, A.; Maltseva, D.; Tonevitsky, A. Adipose may actively delay progression of NAFLD by releasing tumor-suppressing, anti-fibrotic miR-122 into circulation. Obes. Rev. 2019, 20, 108–118. [Google Scholar] [CrossRef]
- Calo, N.; Ramadori, P.; Sobolewski, C.; Romero, Y.; Maeder, C.; Fournier, M.; Rantakari, P.; Zhang, F.-P.; Poutanen, M.; Dufour, J.-F.; et al. Stress-activated miR-21/miR-21*in hepatocytes promotes lipid and glucose metabolic disorders associated with high-fat diet consumption. Gut 2016, 65, 1871–1881. [Google Scholar] [CrossRef]
- Hochreuter, M.Y.; Dall, M.; Treebak, J.T.; Barrès, R. MicroRNAs in non-alcoholic fatty liver disease: Progress and perspectives. Mol. Metab. 2022, 65, 101581. [Google Scholar] [CrossRef] [PubMed]
- Loyer, X.; Paradis, V.; Hénique, C.; Vion, A.-C.; Colnot, N.; Guerin, C.L.; Devue, C.; On, S.; Scetbun, J.; Romain, M.; et al. Liver microRNA-21 is overexpressed in non-alcoholic steatohepatitis and contributes to the disease in experimental models by inhibiting PPARα expression. Gut 2015, 65, 1882–1894. [Google Scholar] [CrossRef]
- Zhao, J.; Tang, N.; Wu, K.; Dai, W.; Ye, C.; Shi, J.; Zhang, J.; Ning, B.; Zeng, X.; Lin, Y. MiR-21 simultaneously regulates ERK1 signaling in HSC activation and hepatocyte EMT in hepatic fibrosis. PLoS ONE 2014, 9, e108005. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Fu, T.; Seok, S.; Kim, D.; Yu, E.; Lee, K.; Kang, Y.; Li, X.; Kemper, B.; Kemper, J.K. Elevated microRNA-34a in obesity reduces NAD+ levels and SIRT1 activity by directly targeting NAMPT. Aging Cell 2013, 12, 1062–1072. [Google Scholar] [CrossRef]
- Li, M.; Hong, W.; Hao, C.; Li, L.; Wu, D.; Shen, A.; Lu, J.; Zheng, Y.; Li, P.; Xu, Y. SIRT1 antagonizes liver fibrosis by blocking hepatic stellate cell activation in mice. FASEB J. 2017, 32, 500–511. [Google Scholar] [CrossRef]
- Wang, Y.; Zhou, F.; Li, M.; Zhang, Y.; Li, N.; Shao, L. Mir-34a-5p promotes hepatic gluconeogenesis by suppressing SIRT1 expression. Exp. Cell Res. 2022, 420, 113336. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Lin, L.; Zhou, W.; Wang, Z.; Ding, G.; Dong, Q.; Qin, L.; Wu, X.; Zheng, Y.; Yang, Y.; et al. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell 2011, 19, 232–243. [Google Scholar] [CrossRef]
- Hosmer, D.W.; Lemeshow, S. Applied Logistic Regression, 2nd ed.; Wiley: Hoboken, NJ, USA, 2000; pp. 156–164. [Google Scholar]
- Cao, L.; An, Y.; Liu, H.; Jiang, J.; Liu, W.; Zhou, Y.; Shi, M.; Dai, W.; Lv, Y.; Zhao, Y.; et al. Global epidemiology of type 2 diabetes in patients with NAFLD or MAFLD: A systematic review and meta-analysis. BMC Med. 2024, 22, 101. [Google Scholar] [CrossRef]
- Wei, J.; Feng, L.; Li, Z.; Xu, G.; Fan, X. MicroRNA-21 activates hepatic stellate cells via PTEN/Akt signaling. Biomed. Pharmacother. 2013, 67, 387–392. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, X.; Huang, Y.; Chen, X.; Lü, M.; Shi, L.; Li, C. Role and mechanisms of action of microRNA-21 as regards the regulation of the wnt/β-catenin signaling pathway in the pathogenesis of non-alcoholic fatty liver disease. Int. J. Mol. Med. 2019, 44, 2201–2212. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zha, Y.; Hu, W.; Huang, Z.; Gao, Z.; Zang, Y.; Chen, J.; Dong, L.; Zhang, J. The autoregulatory feedback loop of microRNA-21/programmed cell death protein 4/activation protein-1 (MiR-21/PDCD4/AP-1) as a driving force for hepatic fibrosis development. J. Biol. Chem. 2013, 288, 37082–37093. [Google Scholar] [CrossRef] [PubMed]
- Seeger, T.; Fischer, A.; Muhly-Reinholz, M.; Zeiher, A.M.; Dimmeler, S. Long-term inhibition of miR-21 leads to reduction of obesity in db/db mice. Obesity 2014, 22, 2352–2360. [Google Scholar] [CrossRef] [PubMed]
- Pillai, S.S.; Lakhani, H.V.; Zehra, M.; Wang, J.; Dilip, A.; Puri, N.; O’Hanlon, K.; Sodhi, K. Predicting nonalcoholic fatty liver disease through a panel of plasma biomarkers and microRNAs in female West Virginia population. Int. J. Mol. Sci. 2020, 21, 6698. [Google Scholar] [CrossRef]
- Zhang, T.; Yang, Z.; Kusumanchi, P.; Han, S.; Liangpunsakul, S. Critical role of microRNA-21 in the pathogenesis of liver diseases. Front. Med. 2020, 7, 7. [Google Scholar] [CrossRef] [PubMed]
- Khokhar, M.; Roy, D.; Bajpai, N.K.; Bohra, G.K.; Yadav, D.; Sharma, P.; Purohit, P. Metformin mediates microRNA-21 regulated circulating matrix metalloproteinase-9 in diabetic nephropathy: An in-silico and clinical study. Arch. Physiol. Biochem. 2021, 129, 1200–1210. [Google Scholar] [CrossRef] [PubMed]
- Understanding Diabetes Diagnosis. Available online: https://diabetes.org/about-diabetes/diagnosis (accessed on 5 February 2025).
- Salvoza, N.C.; Klinzing, D.C.; Gopez-Cervantes, J.; Baclig, M.O. Association of circulating serum miR-34a and miR-122 with dyslipidemia among patients with non-alcoholic fatty liver disease. PLoS ONE 2016, 11, e0153497. [Google Scholar] [CrossRef]
- Cermelli, S.; Ruggieri, A.; Marrero, J.A.; Ioannou, G.N.; Beretta, L. Circulating microRNAs in patients with chronic hepatitis C and non-alcoholic fatty liver disease. PLoS ONE 2011, 6, e23937. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Chen, Y.; Wu, S.; He, J.; Lou, L.; Ye, W.; Wang, J. MicroRNA-34a and microRNA-34c promote the activation of human hepatic stellate cells by targeting peroxisome proliferator-activated receptor γ. Mol. Med. Rep. 2014, 11, 1017–1024. [Google Scholar] [CrossRef] [PubMed]
- Ye, D.; Zhang, T.; Lou, G.; Xu, W.; Dong, F.; Chen, G.; Liu, Y. Plasma miR-17, miR-20a, miR-20b and miR-122 as potential biomarkers for diagnosis of NAFLD in type 2 diabetes mellitus patients. Life Sci. 2018, 208, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Willeit, P.; Skroblin, P.; Moschen, A.R.; Yin, X.; Kaudewitz, D.; Zampetaki, A.; Barwari, T.; Whitehead, M.; Ramírez, C.M.; Goedeke, L.; et al. Circulating microRNA-122 Is Associated With the Risk of New-Onset Metabolic Syndrome and Type 2 Diabetes. Diabetes 2016, 66, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Pirola, C.J.; Fernández Gianotti, T.; Castaño, G.O.; Mallardi, P.; San Martino, J.; Ledesma, M.M.G.L.; Flichman, D.; Mirshahi, F.; Sanyal, A.J.; Sookoian, S. Circulating microRNA signature in non-alcoholic fatty liver disease: From serum non-coding RNAs to liver histology and disease pathogenesis. Gut 2015, 64, 800–812. [Google Scholar] [CrossRef] [PubMed]
- Tobaruela-Resola, A.L.; Milagro, F.I.; Elorz, M.; Benito-Boillos, A.; Herrero, J.I.; Mogna-Peláez, P.; Tur, J.A.; Martínez, J.A.; Abete, I.; Zulet, M.Á. Circulating miR-122-5p, miR-151a-3p, miR-126-5p and miR-21-5p as potential predictive biomarkers for Metabolic Dysfunction-Associated Steatotic Liver Disease assessment. J. Physiol. Biochem. 2024. Online ahead of print. [Google Scholar] [CrossRef]
- Chai, C.; Rivkin, M.; Berkovits, L.; Simerzin, A.; Zorde-Khvalevsky, E.; Rosenberg, N.; Klein, S.; Yaish, D.; Durst, R.; Shpitzen, S.; et al. Metabolic circuit involving free fatty acids, microRNA-122, and triglyceride synthesis in liver and muscle tissues. Gastroenterology 2017, 153, 1404–1415. [Google Scholar] [CrossRef] [PubMed]
- Silvestrini, A.; Meucci, E.; Ricerca, B.M.; Mancini, A. Total antioxidant capacity: Biochemical aspects and clinical significance. Int. J. Mol. Sci. 2023, 24, 10978. [Google Scholar] [CrossRef] [PubMed]
- Koek, G.H.; Liedorp, P.R.; Bast, A. The role of oxidative stress in non-alcoholic steatohepatitis. Clin. Chim. Acta 2011, 412, 1297–1305. [Google Scholar] [CrossRef] [PubMed]
- Ciuti, R.; Liguri, G. A novel assay for measuring total antioxidant capacity in whole blood and other biological samples. J. Biomed. Eng. 2017, 10, 60–76. [Google Scholar] [CrossRef]
- Rubio, C.P.; Cerón, J.J. Spectrophotometric assays for evaluation of reactive oxygen species (ROS) in serum: General Concepts and applications in dogs and humans. BMC Vet. Res. 2021, 17, 226. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.A. The role of ischemia modified albumin as a biomarker in patients with chronic liver disease. J. Clin. Diagn. Res. 2016, 10, BC09–BC12. [Google Scholar] [CrossRef] [PubMed]
- Sushith, S.; Krishnamurthy, H.N.; Reshma, S.; Janice, D.; Madan, G.; Ashok, K.J.; Prathima, M.B.; Kalal, B.S. Serum ischemia-modified albumin, fibrinogen, high sensitivity C- reactive proteins in type-2 diabetes mellitus without hypertension and diabetes mellitus with hypertension: A case-control study. Rep. Biochem. Mol. Biol. 2020, 9, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Sanyal, D.; Mukherjee, P.; Raychaudhuri, M.; Ghosh, S.; Mukherjee, S.; Chowdhury, S. Profile of liver enzymes in non-alcoholic fatty liver disease in patients with impaired glucose tolerance and newly detected untreated type 2 diabetes. Indian. J. Endocrinol. Metab. 2015, 19, 597–601. [Google Scholar] [CrossRef] [PubMed]
- Brouwers, M.C.; Simons, N.; Stehouwer, C.D.; Isaacs, A. Non-alcoholic fatty liver disease and cardiovascular disease: Assessing the evidence for causality. Diabetologia 2019, 63, 253–260. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, D.; Kim, H.J.; Lee, C.H.; Yang, J.I.; Kim, W.; Kim, Y.J.; Yoon, J.H.; Cho, S.H.; Sung, M.W.; et al. Hepatic steatosis index: A simple screening tool reflecting nonalcoholic fatty liver disease. Dig. Liver Dis. 2010, 42, 503–508. [Google Scholar] [CrossRef]
- Bedogni, G.; Bellentani, S.; Miglioli, L.; Masutti, F.; Passalacqua, M.; Castiglione, A.; Tiribelli, C. The Fatty Liver Index: A simple and accurate predictor of hepatic steatosis in the general population. BMC Gastroenterol. 2006, 6, 33. [Google Scholar] [CrossRef] [PubMed]
- World Medical Association. World Medical Association Declaration of Helsinki: Ethical Principles for Medical Research Involving Human Subjects. JAMA 2013, 310, 2191–2194. [Google Scholar] [CrossRef] [PubMed]
- Friedewald, W.T.; Levy, R.I.; Fredrickson, D.S. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin. Chem. 1972, 18, 499–502. [Google Scholar] [CrossRef]
- Erel, O. A novel automated direct measurement method for total antioxidant capacity using a new generation, more stable ABTS radical cation. Clin. Biochem. 2004, 37, 277–285. [Google Scholar] [CrossRef]
- Erel, O. A new automated colorimetric method for measuring total oxidant status. Clin. Biochem. 2005, 38, 1103–1111. [Google Scholar] [CrossRef]
- Bar–Or, D.; Lau, E.; Winkler, J.V. A novel assay for cobalt-albumin binding and its potential as a marker for myocardial ischemia-a preliminary report. J. Emerg. Med. 2000, 19, 311–315. [Google Scholar] [CrossRef] [PubMed]
- Auclair, C.; Voisin, E. Nitroblue tetrazolium reduction. In Handbook of Methods for Oxygen Radical Research; Greenwald, R.A., Ed.; CRC Press: Boca Raton, FL, USA, 1985. [Google Scholar]

| Marker | CG N = 49 | MASLD N = 50 | MASLD + T2D N = 48 | p |
|---|---|---|---|---|
| Age, years | 50 ± 14 | 50 ± 13 | 58 ± 13 a†, b† | 0.006 |
| Sex (males), N (%) ** | 13 (26.5) | 28 (56.0) a# | 22 (45.8) a† | 0.011 |
| BMI, kg/m2 * | 25.1 (22.2–27.0) | 29.1 (26.4–31.3) a# | 30.4 (26.8–31.4) a# | <0.001 |
| Waist circumference, cm | 84.7 ± 10.1 | 96.9 ± 12.7 a# | 99.6 ± 14.4 a# | <0.001 |
| Systolic blood pressure, mmHg * | 120 (115–130) | 130 (120–140) a‡ | 130 (120–140) a† | 0.016 |
| Diastolic blood pressure, mmHg * | 80 (70–83) | 80 (70–90) | 80 (70–86) | 0.153 |
| Smoking status (yes), N (%) ** | 13 (26.5%) | 20 (40.0%) | 22 (45.8%) | 0.130 |
| Alcohol consumption (yes), N (%) ** | 18 (36.7%) | 9 (18.0%) | 11 (22.9%) | 0.088 |
| Physical activity (yes), N (%) ** | 27 (55.1%) | 14 (28.0%) a# | 17 (35.4%) | 0.017 |
| Hypertension and/or CVD (yes), N (%) ** | 11 (22.4%) | 31 (62.0%) a# | 25 (52.1%) a‡ | <0.001 |
| Insulin therapy (yes), N (%) ** | 0 (0.0%) | 0 (0.0%) | 19 (39.6%) a#, b# | <0.001 |
| Oral antidiabetic therapy (yes), N (%) ** | 0 (0.0%) | 0 (0.0%) | 36 (75.0%) a#, b# | <0.001 |
| Antihyperlipidemic therapy (yes), N (%) ** | 2 (4.1%) | 5 (10.0%) | 7 (14.6%) | 0.210 |
| Antihypertensives and/or CVD therapy (yes), N (%) ** | 10 (20.4%) | 30 (60.0%) a# | 21 (43.8%) a† | <0.001 |
| Marker | CG N = 49 | MASLD N = 50 | MASLD + T2D N = 48 | p |
|---|---|---|---|---|
| Glucose, mmol/L * | 5.1 (4.8–5.4) | 5.4 (4.9–5.9) a† | 7.5 (6.0–10.2) a#, b# | <0.001 |
| HbA1C, % | 5.1 ± 0.34 | 5.4 ± 0.41 | 8.5 ± 2.32 a#, b# | <0.001 |
| Total cholesterol, mmol/L | 5.46 ± 1.33 | 5.31 ± 1.01 | 4.91 ± 1.22 | 0.081 |
| TG, mmol/L * | 0.90 (0.72–1.46) | 1.30 (0.80–1.63) | 1.88 (1.40–2.63) a#, b# | <0.001 |
| HDL-cholesterol, mmol/L | 1.68 ± 0.43 | 1.40 ± 0.27 a# | 1.17 ± 0.29 a#, b† | <0.001 |
| LDL-cholesterol, mmol/L | 3.38 ± 1.14 | 3.28 ± 0.975 | 2.88 ± 1.05 | 0.081 |
| Total bilirubin, µmol/L * | 12.5 (9.3–17.7) | 11.8 (7.4–17.1) | 10.7 (8.5–14.7) | 0.644 |
| Direct bilirubin, µmol/L * | 2.1 (1.6–2.7) | 2.4 (1.8–4.0) | 2.0 (1.5–2.5) | 0.075 |
| Total protein, g/L * | 71 (68–75) | 75 (72–77) a‡ | 68 (64–74) a‡, b# | <0.001 |
| Albumin, g/L * | 44 (43–46) | 45 (44–49) | 42 (39–44) a#, b# | <0.001 |
| Uric acid, µmol/L * | 268 (203–308) | 373 (286–416) a# | 308 (287–347) a#, b† | <0.001 |
| Creatinine, µmol/L * | 68 (58–85) | 77 (63–87) | 72 (59–80) | 0.393 |
| Urea, mmol/L * | 5.0 (4.3–5.7) | 5.2 (4.2–6.1) | 5.1 (4.2–6.2) | 0.789 |
| ALT, U/L * | 19 (16–25) | 32 (19–51) a# | 25 (17–47) a‡ | <0.001 |
| GGT, U/L * | 15 (12–22) | 36 (25–56) a# | 30 (22–51) a# | <0.001 |
| CRP, mg/L * | 0.90 (0.40–2.25) | 2.70 (1.35–5.30) a# | 4.00 (2.00–6.90) a# | <0.001 |
| TAS, µmol/L * | 721 (695–745) | 981 (950–1022) a# | 870 (835–901) a#, b# | <0.001 |
| TOS, µmol/L * | 6.7 (3.2–8.3) | 19.3 (14.6–25.5) a# | 9.7 (5.8–13.3) a#, b# | <0.001 |
| O2•−,μmol/L NBT/min/L * | 41 (33–47) | 50 (46–56) a# | 42 (36–49) b# | <0.001 |
| IMA, ABSU * | 0.347 (0.335–0.359) | 0.473 (0.460–0.485) a# | 0.467 (0.452–0.478) a# | <0.001 |
| Marker | OR | 95% CI | Nagelkerke R2 | p |
|---|---|---|---|---|
| Age, years | 1.000 | 0.970–1.031 | <0.001 | 0.995 |
| BMI, kg/m2 | 1.392 | 1.196–1.620 | 0.345 | <0.001 |
| Waist circumference, cm | 1.095 | 1.044–1.148 | 0.293 | <0.001 |
| Systolic blood pressure, mmHg | 1.033 | 1.002–1.065 | 0.070 | 0.038 |
| Diastolic blood pressure, mmHg | 1.037 | 0.997–1.078 | 0.051 | 0.069 |
| Glucose, mmol/L | 2.611 | 1.182–5.766 | 0.091 | 0.018 |
| HbA1C, % | 7.290 | 1.779–29.871 | 0.151 | 0.006 |
| Total cholesterol, mmol/L | 0.899 | 0.626–1.293 | 0.005 | 0.567 |
| TG, mmol/L | 2.142 | 1.057–4.341 | 0.078 | 0.035 |
| HDL-cholesterol, mmol/L | 0.068 | 0.012–0.372 | 0.202 | 0.002 |
| LDL-cholesterol, mmol/L | 0.915 | 0.591–1.417 | 0.003 | 0.690 |
| Total bilirubin, µmol/L | 0.986 | 0.924–1.052 | 0.003 | 0.670 |
| Direct bilirubin, µmol/L | 1.369 | 0.947–1.979 | 0.049 | 0.094 |
| Total protein, g/L | 1.028 | 0.963–1.097 | 0.013 | 0.410 |
| Albumin, g/L | 0.994 | 0.975–1.013 | 0.011 | 0.558 |
| Uric acid, µmol/L | 1.014 | 1.007–1.021 | 0.302 | <0.001 |
| Creatinine, µmol/L | 1.016 | 0.988–1.045 | 0.019 | 0.263 |
| Urea, mmol/L | 1.145 | 0.846–1.548 | 0.012 | 0.381 |
| ALT, U/L | 1.085 | 1.035–1.137 | 0.313 | 0.001 |
| GGT, U/L | 1.065 | 1.031–1.101 | 0.317 | <0.001 |
| CRP, mg/L | 1.201 | 1.030–1.401 | 0.122 | 0.019 |
| TAS, µmol/L | 1.004 | 1.002–1.006 | 0.288 | <0.001 |
| TOS, µmol/L | 1.127 | 1.020–1.245 | 0.158 | 0.019 |
| O2•−, μmol/L NBT/min/L | 1.014 | 0.997–1.031 | 0.098 | 0.040 |
| IMA, ABSU | 1.006 | 1.003–1.009 | 0.194 | <0.001 |
| miR-21 expression | 8.337 | 1.707–40.722 | 0.126 | 0.009 |
| miR-34a expression | 7.512 | 1.495–37.742 | 0.149 | 0.014 |
| miR-122 expression | 1.849 | 0.970–3.526 | 0.070 | 0.062 |
| Marker | OR | 95% CI | Nagelkerke R2 | p |
|---|---|---|---|---|
| Age, years | 1.047 | 1.013–1.083 | 0.105 | 0.007 |
| BMI, kg/m2 | 1.040 | 0.948–1.140 | 0.010 | 0.405 |
| Waist circumference, cm | 1.016 | 0.976–1.057 | 0.013 | 0.450 |
| Systolic blood pressure, mmHg | 0.997 | 0.971–1.024 | 0.001 | 0.835 |
| Diastolic blood pressure, mmHg | 1.000 | 0.970–1.031 | 0.000 | 0.991 |
| Glucose, mmol/L | 3.535 | 1.924–6.495 | 0.519 | <0.001 |
| HbA1C, % | 13.187 | 3.242–53.635 | 0.728 | <0.001 |
| Total cholesterol, mmol/L | 0.731 | 0.496–1.079 | 0.040 | 0.115 |
| TG, mmol/L | 2.409 | 1.325–4.381 | 0.164 | 0.004 |
| HDL-cholesterol, mmol/L | 0.063 | 0.010–0.386 | 0.179 | 0.003 |
| LDL-cholesterol, mmol/L | 0.673 | 0.419–1.083 | 0.051 | 0.103 |
| Total bilirubin, µmol/L | 1.001 | 0.953–1.052 | 0.000 | 0.960 |
| Direct bilirubin, µmol/L | 0.573 | 0.370–0.886 | 0.118 | 0.012 |
| 5Total protein, g/L | 0.920 | 0.854–0.991 | 0.098 | 0.028 |
| Albumin, g/L | 0.624 | 0.500–0.780 | 0.470 | <0.001 |
| Uric acid, µmol/L | 0.993 | 0.986–1.000 | 0.073 | 0.047 |
| Creatinine, µmol/L | 0.992 | 0.969–1.016 | 0.007 | 0.501 |
| Urea, mmol/L | 1.034 | 0.792–1.349 | 0.001 | 0.807 |
| ALT, U/L | 1.001 | 0.990–1.012 | 0.000 | 0.878 |
| GGT, U/L | 1.002 | 0.993–1.010 | 0.002 | 0.707 |
| CRP, mg/L | 1.054 | 0.968–1.149 | 0.032 | 0.226 |
| TAS, µmol/L | 0.998 | 0.996–1.000 | 0.066 | 0.033 |
| TOS, µmol/L | 0.982 | 0.949–1.016 | 0.038 | 0.300 |
| O2•−, μmol/L NBT/min/L | 0.994 | 0.982–1.007 | 0.012 | 0.360 |
| IMA, ABSU | 1.000 | 0.997–1.003 | <0.001 | 0.939 |
| miR-21 expression | 0.036 | 0.005–0.256 | 0.218 | 0.001 |
| miR-34a expression | 0.932 | 0.463–1.877 | 0.001 | 0.844 |
| miR-122 expression | 0.995 | 0.702–1.411 | <0.001 | 0.978 |
| Factors | Variables (Loadings) | Factor Variability (%) |
|---|---|---|
| Epigenetic liver-specific-related factor | miR-122 (0.877) | 23.3 |
| miR-34a (0.846) | ||
| ALT (0.841) | ||
| GGT (0.625) | ||
| TG (0.531) | ||
| Cardiometabolic antioxidant-related factor | HDL-cholesterol (−0.710) | 14.4 |
| TAS (0.709) | ||
| BMI (0.615) | ||
| CRP (0.570) | ||
| Redox-related factor | O2•− (−0.711) | 10.2 |
| TOS (0.561) | ||
| IMA (0.551) | ||
| Age–epigenetic-related factor | Age (0.758) | 10.1 |
| miR-21 (−0.743) |
| Predictors Towards MASLD | B | SE | Unadjusted OR (95%CI) | p | Nagelkerke R2 |
| Epigenetic liver-specific-related factor | 1.071 | 0.481 | 2.918 (1.138–7.486) | 0.026 | 0.165 |
| Cardiometabolic antioxidant-related factor | 1.524 | 0.432 | 4.592 (1.968–10.711) | <0.001 | 0.348 |
| Redox-related factor | 0.073 | 0.277 | 1.075 (0.625–1.850) | 0.793 | 0.001 |
| Age–epigenetic-related factor | −0.279 | 0.260 | 0.756 (0.454–1.259) | 0.283 | 0.025 |
| Predictors towards T2D in MASLD | B | SE | Unadjusted OR (95%CI) | p | Nagelkerke R2 |
| Epigenetic liver-specific-related factor | 0.060 | 0.227 | 1.062 (0.680–1.658) | 0.793 | 0.002 |
| Cardiometabolic antioxidant-related factor | 0.374 | 0.315 | 1.454 (0.784–2.698) | 0.235 | 0.034 |
| Redox-related factor | 0.163 | 0.239 | 1.178 (0.738–1.879) | 0.493 | 0.011 |
| Age–epigenetic-related factor | 1.187 | 0.390 | 3.279 (1.527–7.040) | 0.002 | 0.277 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Erceg, S.; Munjas, J.; Sopić, M.; Tomašević, R.; Mitrović, M.; Kotur-Stevuljević, J.; Mamić, M.; Vujčić, S.; Klisic, A.; Ninić, A. Expression Analysis of Circulating miR-21, miR-34a and miR-122 and Redox Status Markers in Metabolic Dysfunction-Associated Steatotic Liver Disease Patients with and Without Type 2 Diabetes. Int. J. Mol. Sci. 2025, 26, 2392. https://doi.org/10.3390/ijms26062392
Erceg S, Munjas J, Sopić M, Tomašević R, Mitrović M, Kotur-Stevuljević J, Mamić M, Vujčić S, Klisic A, Ninić A. Expression Analysis of Circulating miR-21, miR-34a and miR-122 and Redox Status Markers in Metabolic Dysfunction-Associated Steatotic Liver Disease Patients with and Without Type 2 Diabetes. International Journal of Molecular Sciences. 2025; 26(6):2392. https://doi.org/10.3390/ijms26062392
Chicago/Turabian StyleErceg, Sanja, Jelena Munjas, Miron Sopić, Ratko Tomašević, Miloš Mitrović, Jelena Kotur-Stevuljević, Milica Mamić, Sanja Vujčić, Aleksandra Klisic, and Ana Ninić. 2025. "Expression Analysis of Circulating miR-21, miR-34a and miR-122 and Redox Status Markers in Metabolic Dysfunction-Associated Steatotic Liver Disease Patients with and Without Type 2 Diabetes" International Journal of Molecular Sciences 26, no. 6: 2392. https://doi.org/10.3390/ijms26062392
APA StyleErceg, S., Munjas, J., Sopić, M., Tomašević, R., Mitrović, M., Kotur-Stevuljević, J., Mamić, M., Vujčić, S., Klisic, A., & Ninić, A. (2025). Expression Analysis of Circulating miR-21, miR-34a and miR-122 and Redox Status Markers in Metabolic Dysfunction-Associated Steatotic Liver Disease Patients with and Without Type 2 Diabetes. International Journal of Molecular Sciences, 26(6), 2392. https://doi.org/10.3390/ijms26062392








