CircGRB14 Inhibits Proliferation and Promotes Apoptosis of Granulosa Cells in Chicken Follicle Selection Through Sponging miR-12264-3p and miR-6660-3p
Abstract
1. Introduction
2. Results
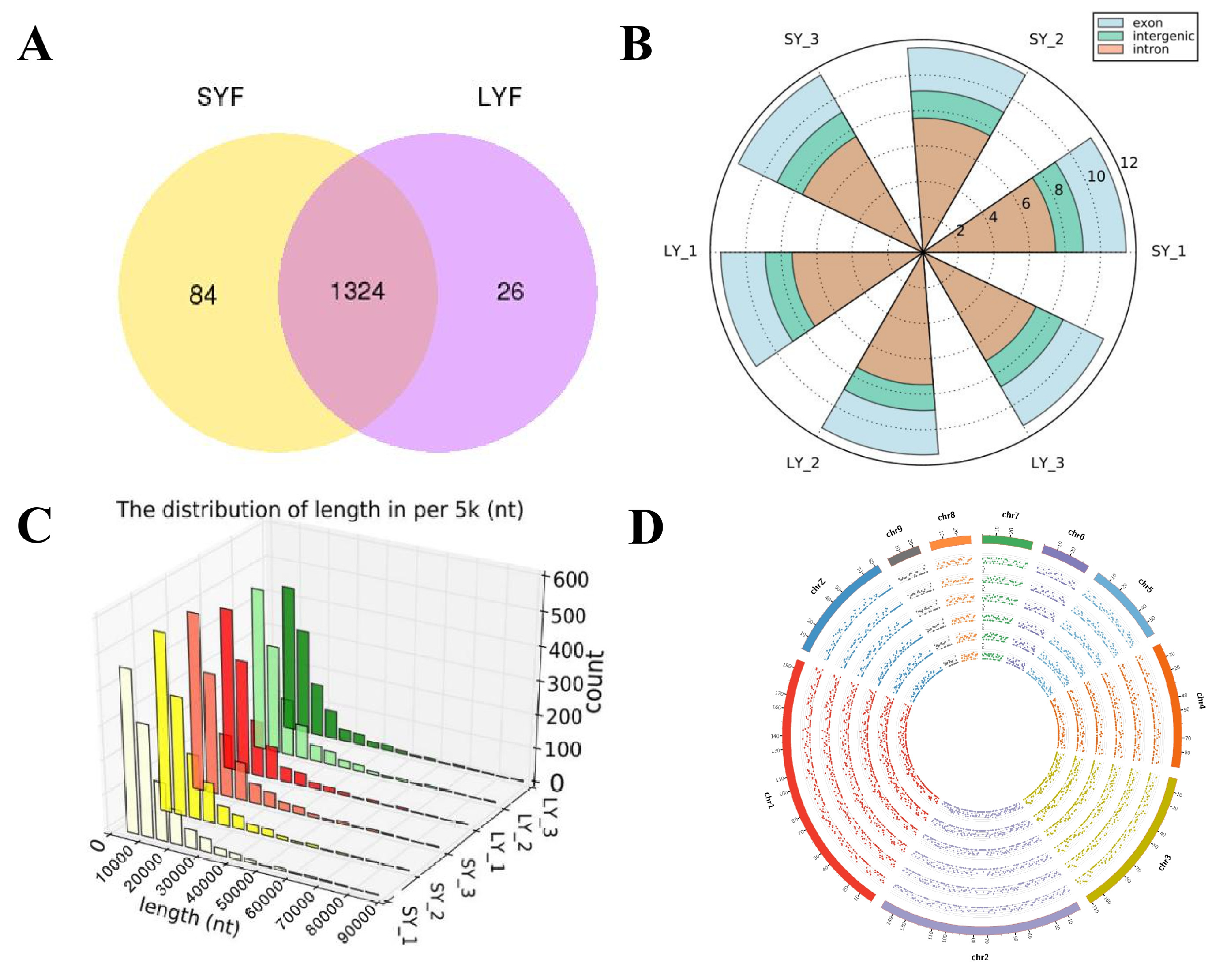
2.1. Overview of circRNAs
2.2. Expression Profiles of circRNAs During Follicular Selection
2.3. Differentially Expressed circRNAs
2.4. Functional Annotation of DE-circRNAs
2.5. CircGRB14 Identification and Expression in Chicken Follicles
2.6. CircGRB14 as an miRNA Sponge for miR-12264-3p and miR-6660-3p
2.7. CircGRB14′s Impact on GC Proliferation and Apoptosis
2.8. miR-12264-3p and miR-6660-3p’s Role in GC Proliferation and Apoptosis
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Chicken Follicle Harvesting
4.3. CircRNA Sequencing and Identification
4.4. Identification of Differentially Expressed circRNAs
4.5. GO and KEGG Enrichment Analysis
4.6. CircGRB14 Validation and qRT-PCR
4.7. Plasmid Construction and Dual-Luciferase Reporter Assay
4.8. GC Isolation and Culture
4.9. Cell Transfection
4.10. Cell Proliferation Assay
4.11. Cell Apoptosis Assay
4.12. Western Blot
4.13. Statistical Analyses
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Burt, D.W. Emergence of the chicken as a model organism: Implications for agriculture and biology. Poultr. Sci. 2007, 86, 1460–1471. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.L. Ovarian follicle selection and granulosa cell differentiation. Poultr. Sci. 2015, 94, 781–785. [Google Scholar] [CrossRef]
- Johnson, P.A. Follicle Selection in the Avian Ovary. Reprod. Domest. Anim. 2012, 47, 283–287. [Google Scholar] [CrossRef] [PubMed]
- McGee, E.A.; Hsueh, A.J.W. Initial and cyclic recruitment of ovarian follicles. Endocr. Rev. 2000, 21, 200–214. [Google Scholar] [PubMed]
- Zhu, G.; Fang, C.; Mo, C.; Wang, Y.; Huang, Y.; Li, J. Transcriptomic analysis of granulosa cell populations proximal and distal to the germinal disc of chicken preovulatory follicles. Sci. Rep. 2021, 11, 4683. [Google Scholar] [CrossRef]
- Jin, Y.; Zhang, C.; Lin, X.; Zeng, W. Prostaglandin involvement in follicle-stimulating hormone-induced proliferation of granulosa cells from chicken prehierarchical follicles. Prostaglandins Other Lipid Mediat. 2006, 81, 45–54. [Google Scholar] [CrossRef]
- Huang, H.; Chen, T.; Li, F.; Jin, D.; Li, C.; Yang, Y.; Liu, X.; Wang, D.; Di, J. The functions, oncogenic roles, and clinical significance of circular RNAs in renal cell carcinoma. Med. Oncol. 2022, 39, 72. [Google Scholar] [CrossRef]
- Hansen, T.B. Signal and noise in circRNA translation. Methods 2021, 196, 68–73. [Google Scholar] [CrossRef]
- Liu, H.; Hu, G.; Wang, Z.; Liu, Q.; Zhang, J.; Chen, Y.; Huang, Y.; Xue, W.; Xu, Y.; Zhai, W. circPTCH1 promotes invasion and metastasis in renal cell carcinoma via regulating miR-485-5p/MMP14 axis. Theranostics 2020, 10, 10791–10807. [Google Scholar] [CrossRef]
- Chen, L.L.; Yang, L. Regulation of circRNA biogenesis. RNA Biol. 2015, 12, 381–388. [Google Scholar] [CrossRef]
- Lu, T.; Cui, L.; Zhou, Y.; Zhu, C.; Fan, D.; Gong, H.; Zhao, Q.; Zhou, C.; Zhao, Y.; Lu, D.; et al. Transcriptome-wide investigation of circular RNAs in rice. RNA 2015, 21, 2076–2087. [Google Scholar] [CrossRef] [PubMed]
- Qu, S.; Yang, X.; Li, X.; Wang, J.; Gao, Y.; Shang, R.; Sun, W.; Dou, K.; Li, H. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015, 365, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, S.; Gunasekaran, K.; Sasidharan, S.; Jeyamanickavel Mathan, V.; Perumal, E. MicroRNAs and xenobiotic toxicity: An overview. Toxicol. Rep. 2020, 7, 583–595. [Google Scholar] [CrossRef] [PubMed]
- Mahesh, G.; Biswas, R. MicroRNA-155: A master regulator of inflammation. J. Interferon. Cytokine. Res. 2019, 39, 321–330. [Google Scholar] [CrossRef]
- Zhang, P.; Li, X.; Zhang, S.; Wu, S.; Xiao, Q.; Gu, Y.; Guo, X.; Lin, X.; Chen, L.; Zhao, Y.; et al. miR-370-3p regulates adipogenesis through targeting Mknk1. Molecules 2021, 26, 6926. [Google Scholar] [CrossRef]
- Ponsuksili, S.; Hadlich, F.; Reyer, H.; Oster, M.; Trakooljul, N.; Iqbal, M.A.; Sommerfeld, V.; Rodehutscord, M.; Wimmers, K. Genetic background and production periods shape the microRNA profiles of the gut in laying hens. Genomics 2021, 113, 1790–1801. [Google Scholar] [CrossRef]
- Zhang, T.; Chen, L.; Ding, H.; Wu, P.; Zhang, G.; Pan, Z.; Xie, K.; Dai, G.; Wang, J. Construction of miRNA-mRNA network in the differentiation of chicken preadipocytes. Br. Poultr. Sci. 2022, 63, 298–306. [Google Scholar] [CrossRef]
- Yang, F.; Chen, Y.; Xue, Z.; Lv, Y.; Shen, L.; Li, K.; Zheng, P.; Pan, P.; Feng, T.; Jin, L.; et al. High-Throughput sequencing and exploration of the lncRNA-circRNA-miRNA-mRNA network in type 2 diabetes mellitus. Biomed. Res. Int. 2020, 20, 8162524. [Google Scholar] [CrossRef]
- Lu, S.; Zhu, N.; Guo, W.; Wang, X.; Li, K.; Yan, J.; Jiang, C.; Han, S.; Xiang, H.; Wu, X.; et al. RNA-Seq revealed a circular RNA-microRNA-mRNA regulatory network in hantaan virus infection. Front. Cell Infect. Microbiol. 2020, 10, 97. [Google Scholar] [CrossRef]
- Wu, P.; Zhou, K.; Zhang, L.; Li, P.; He, M.; Zhang, X.; Ye, H.; Zhang, Q.; Wei, Q.; Zhang, G. High-throughput sequencing reveals crucial miRNAs in skeletal muscle development of Bian chicken. Br. Poult. Sci. 2021, 62, 658–665. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Li, H.; Yang, J.; Hao, D.; Dong, D.; Huang, Y.; Lan, X.; Plath, M.; Lei, C.; Lin, F.; et al. Circular RNA profiling reveals an abundant circLMO7 that regulates myoblasts differentiation and survival by sponging miR-378a-3p. Cell Death Dis. 2017, 8, e3153. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.L.; Ouyang, H.J.; Wang, Z.J.; Chen, B.A.; Nie, Q.H. A novel circular RNA generated by FGFR2 gene promotes myoblast proliferation and differentiation by sponging miR-133a-5p and miR-29b-1-5p. Cells 2018, 7, 199. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Teerds, K.; Tao, J.; Wei, H.; Jaklofsky, M.; Zhao, Z.; Liang, Y.; Li, L.; Wang, C.C.; Zhang, S. Characteristics of circular RNA expression profiles of porcine granulosa cells in healthy and atretic antral follicles. Int. J. Mol. Sci. 2020, 21, 5217. [Google Scholar] [CrossRef]
- Huiming, W.; Zhang, Y.; Zhang, J.B.; Du, X.; Li, Q.F.; Pan, Z.X. circSLC41A1 resists porcine granulosa cell apoptosis and follicular atresia by promoting SRSF1 through miR-9820-5p sponging. Int. J. Mol. Sci. 2022, 23, 1509. [Google Scholar] [CrossRef]
- Wang, Y.; Guo, Z.; Zi, C.; Wu, P.; Lv, X.; Chen, L.; Chen, F.; Zhang, G.; Wang, J. CircRNA expression in chicken granulosa cells illuminated with red light. Poultr. Sci. 2022, 101, 101734. [Google Scholar] [CrossRef]
- Patop, I.L.; Wüst, S.; Kadener, S. Past, present, and future of circRNAs. EMBO J. 2019, 38, 100836. [Google Scholar] [CrossRef]
- Kim, D.H.; Lee, Y.K.; Kim, S.H.; Lee, K.W. The Impact of Temperature and Humidity on the Performance and Physiology of Laying Hens. Animals 2020, 11, 56. [Google Scholar] [CrossRef]
- Fu, Y.; Wang, J.; Zhang, H.J.; Wu, S.G.; Zhou, J.M.; Qi, G.H. The partial replacement of sodium chloride with sodium bicarbonate or sodium sulfate in laying hen diets improved laying performance, and eggshell quality and ultrastructure. Poult. Sci. 2021, 100, 101–102. [Google Scholar] [CrossRef]
- Du, Y.; Liu, L.; He, Y.; Dou, T.; Jia, J.; Ge, C. Endocrine and genetic factors affecting egg laying performance in chickens: A review. Br. Poultr. Sci. 2020, 61, 538–549. [Google Scholar] [CrossRef]
- Salzman, J.; Gawad, C.; Wang, P.L.; Lacayo, N.; Brown, P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 2012, 7, 30733. [Google Scholar] [CrossRef] [PubMed]
- Shen, M.; Li, T.; Zhang, G.; Wu, P.; Chen, F.; Lou, Q.; Chen, L.; Yin, X.; Zhang, T.; Wang, J. Dynamic expression and functional analysis of circRNA in granulosa cells during follicular development in chicken. BMC Genom. 2019, 20, 96. [Google Scholar] [CrossRef] [PubMed]
- Shen, M.; Wu, P.; Li, T.; Wu, P.; Chen, F.; Chen, L.; Xie, K.; Wang, J.; Zhang, G. Transcriptome analysis of circRNA and mRNA in theca cells during follicular development in chickens. Genes 2020, 11, 489. [Google Scholar] [CrossRef] [PubMed]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 2014, 56, 55–66. [Google Scholar] [CrossRef]
- Bohrer, R.C.; Rosa, P.R.; Ferreira, R.; Bordignon, V.; Oliveira, J.F.; Gonçalves, P.B. GRB14 mRNA levels during follicular deviation in cattle are higher in granulosa cells of subordinate compared to dominant follicles. Reprod. Domest. Anim. 2013, 48, 396–401. [Google Scholar] [CrossRef]
- García-Palmero, I.; Pompas-Veganzones, N.; Villalobo, E.; Gioria, S.; Haiech, J.; Villalobo, A. The adaptors GRB10 and GRB14 are calmodulin-binding proteins. FEBS Lett. 2017, 591, 1176–1186. [Google Scholar] [CrossRef]
- Rosa, P.R.A.; Bohrer, R.C.; De Cesaro, M.P.; Gutierrez, K.; Ferreira, R.; Pereira, G.R.; Oliveira, J.F.C.; Gonçalves, P.B.D. Growth factor receptor-bound protein 14: A potential new gene associated with oocyte competence. Zygote 2014, 22, 103–109. [Google Scholar] [CrossRef]
- Chen, S.; Huang, V.; Xu, X.; Livingstone, J.; Soares, F.; Jeon, J.; Zeng, Y.; Hua, J.T.; Petricca, J.; Guo, H.; et al. Widespread and functional RNA circularization in localized prostate cancer. Cell 2019, 176, 831–843. [Google Scholar] [CrossRef]
- Yao, Z.; Luo, J.; Hu, K.; Lin, J.; Huang, H.; Wang, Q.; Zhang, P.; Xiong, Z.; He, C.; Huang, Z.; et al. ZKSCAN1 gene and its related circular RNA (circZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways. Mol. Oncol. 2017, 11, 422–437. [Google Scholar] [CrossRef]
- Liu, Z.; Zhang, L.; Chen, W.; Yuan, F.; Yang, Z.; Liu, S.; Le, F. miR-195-5p regulates cell proliferation, apoptosis, and invasion of thyroid cancer by targeting telomerase reverse transcriptase. Bioengineered 2021, 12, 6201–6209. [Google Scholar] [CrossRef]
- Xie, X.K.; Xiao, Y.X.; Huang, X. Homeobox C10 knockdown suppresses cell proliferation and promotes cell apoptosis in osteosarcoma cells through regulating caspase 3. OncoTargets Ther. 2018, 11, 473–482. [Google Scholar] [CrossRef] [PubMed]
- Malumbres, M.; Barbacid, M. Cell cycle, CDKs and cancer: A changing paradigm. Nat. Rev. Cancer 2009, 9, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Legnini, I.; Di Timoteo, G.; Rossi, F.; Morlando, M.; Briganti, F.; Sthandier, O.; Fatica, A.; Santini, T.; Andronache, A.; Wade, M.; et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell 2017, 66, 22–37. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liang, W.; Wang, S.; Wang, Z.; Bai, H.; Jiang, Y.; Bi, Y.; Chen, G.; Chang, G. Circular RNA expression profiling reveals that circ-PLXNA1 functions in duck adipocyte differentiation. PLoS ONE 2020, 15, 0236069. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Gao, Y.; Zhang, J.Y.; Zhao, F.Q. Circular RNA identification based on multiple seed matching. Brief. Bioinform. 2018, 19, 803–810. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Zhou, L.; Chen, J.; Li, Z.; Li, X.; Hu, X.; Huang, Y.; Zhao, X.; Liang, C.; Wang, Y.; Sun, L.; et al. Integrated profiling of microRNAs and mRNAs: microRNAs located on Xq27.3 associate with clear cell renal cell carcinoma. PLoS ONE 2010, 5, 15224. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef] [PubMed]
- Obi, P.; Chen, Y.G. The design and synthesis of circular RNAs. Methods 2021, 196, 85–103. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, A.B.; Evans, A.J.; Perry, M.M.; Davidson, M.H. A method for separating the granulosa cells, the basal lamina and the theca of the preovulatory ovarian follicle of the domestic fowl (Gallus domesticus). J. Reprod. Fertil. 1977, 50, 179–181. [Google Scholar] [CrossRef]








| Sample | raw_reads | clean_reads | clean_bases | error_rate (%) | Q20 (%) | Q30 (%) | GC_content (%) |
|---|---|---|---|---|---|---|---|
| SYF_1 | 96589128 | 81847658 | 12.28G | 0.02 | 96.86 | 92.22 | 50.21 |
| SYF_2 | 103783026 | 98453096 | 14.77G | 0.02 | 96.35 | 91.01 | 48.52 |
| SYF_3 | 95081398 | 90236062 | 13.54G | 0.02 | 96.41 | 91.12 | 49.65 |
| LYF_1 | 95699154 | 89900182 | 13.49G | 0.02 | 95.35 | 89.12 | 48.18 |
| LYF_2 | 99396460 | 93902260 | 14.09G | 0.02 | 96.12 | 90.62 | 46.96 |
| LYF_3 | 103617614 | 96549922 | 14.48G | 0.03 | 95.05 | 88.13 | 47.23 |
| Primer | Sequence | Annealing Temp (°C) | Product Size (bp) |
|---|---|---|---|
| PCR-circGRB14 | F: 5′-AACTGTAAAACACTCTGGGA-3′ R: 5′-CTAAGCCTGTATGAGTAAGT-3′ | 60 | 210 |
| circGRB14 | F: 5′-CAAACTGGGGGATGGAAGAG-3′ R(RT): 5′-CAGTTGCATTGGTCTCACTTGAA-3′ | 59 | 132 |
| GRB14 | F: 5′-GGAGGAACCAGAGACCTGAAACAAC-3′ R: 5′-GCAGCCACTTCTACCTTGATACGG-3′ | 60 | 141 |
| 18s rRNA | F: 5′-TAGTTGGTGGAGCGATTTGTCT-3′ R: 5′-CGGACATCTAAGGGCATCACA-3′ | 60 | 169 |
| GAPDH | F: 5′-CTGTGCCCATCTATGAAGGCTA-3′ R: 5′-ATTTCTCTCTCGGCT-GTGGTG-3′ | 60 | 139 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, H.; Li, M.; Zhang, B.; Zhang, J.; Shi, Y.; Ma, T.; Sun, Y. CircGRB14 Inhibits Proliferation and Promotes Apoptosis of Granulosa Cells in Chicken Follicle Selection Through Sponging miR-12264-3p and miR-6660-3p. Int. J. Mol. Sci. 2025, 26, 2214. https://doi.org/10.3390/ijms26052214
Yang H, Li M, Zhang B, Zhang J, Shi Y, Ma T, Sun Y. CircGRB14 Inhibits Proliferation and Promotes Apoptosis of Granulosa Cells in Chicken Follicle Selection Through Sponging miR-12264-3p and miR-6660-3p. International Journal of Molecular Sciences. 2025; 26(5):2214. https://doi.org/10.3390/ijms26052214
Chicago/Turabian StyleYang, Huanqi, Mengxiao Li, Beibei Zhang, Jinming Zhang, Yuxiang Shi, Tenghe Ma, and Yanyan Sun. 2025. "CircGRB14 Inhibits Proliferation and Promotes Apoptosis of Granulosa Cells in Chicken Follicle Selection Through Sponging miR-12264-3p and miR-6660-3p" International Journal of Molecular Sciences 26, no. 5: 2214. https://doi.org/10.3390/ijms26052214
APA StyleYang, H., Li, M., Zhang, B., Zhang, J., Shi, Y., Ma, T., & Sun, Y. (2025). CircGRB14 Inhibits Proliferation and Promotes Apoptosis of Granulosa Cells in Chicken Follicle Selection Through Sponging miR-12264-3p and miR-6660-3p. International Journal of Molecular Sciences, 26(5), 2214. https://doi.org/10.3390/ijms26052214





