Genome-Wide Association Study Reveals Growth-Related SNPs and Candidate Genes in Largemouth Bass (Micropterus salmoides) Adapted to Hypertonic Environments
Abstract
1. Introduction
2. Results
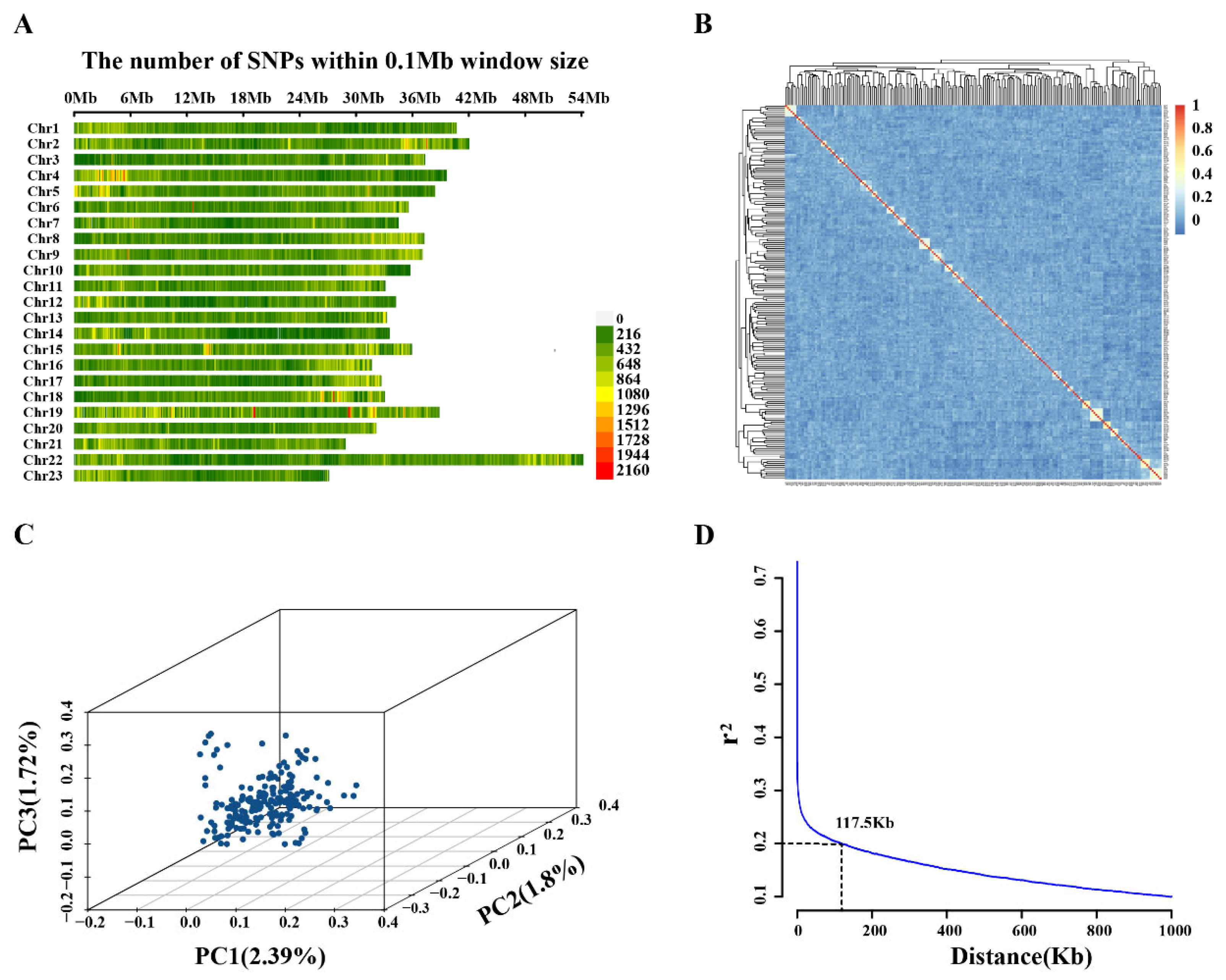
2.1. Sequencing Data Statistics and Population Structure Analysis
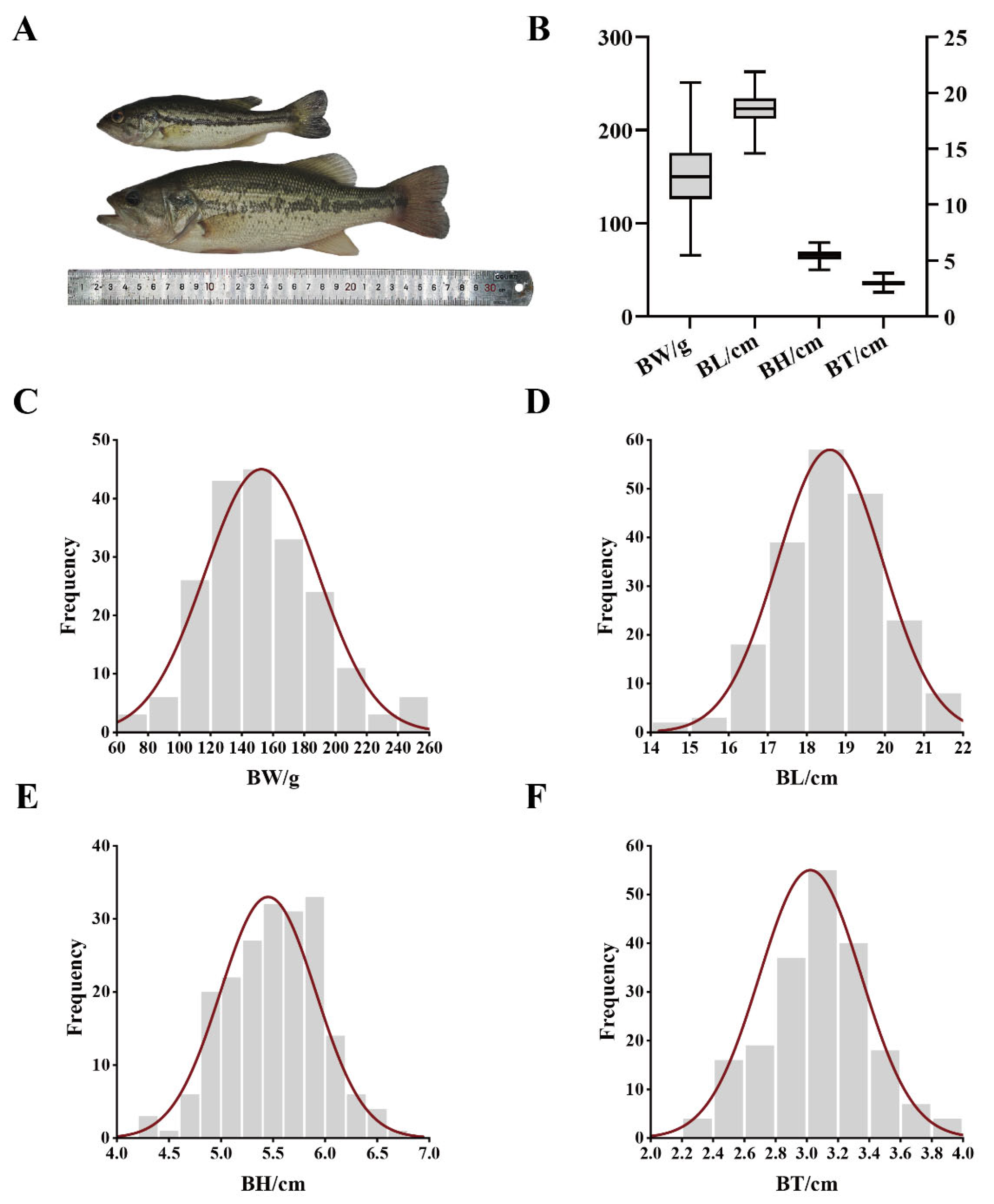
2.2. Statistical Analysis of Growth Data for GWAS and Heritability Assessment of Growth Traits
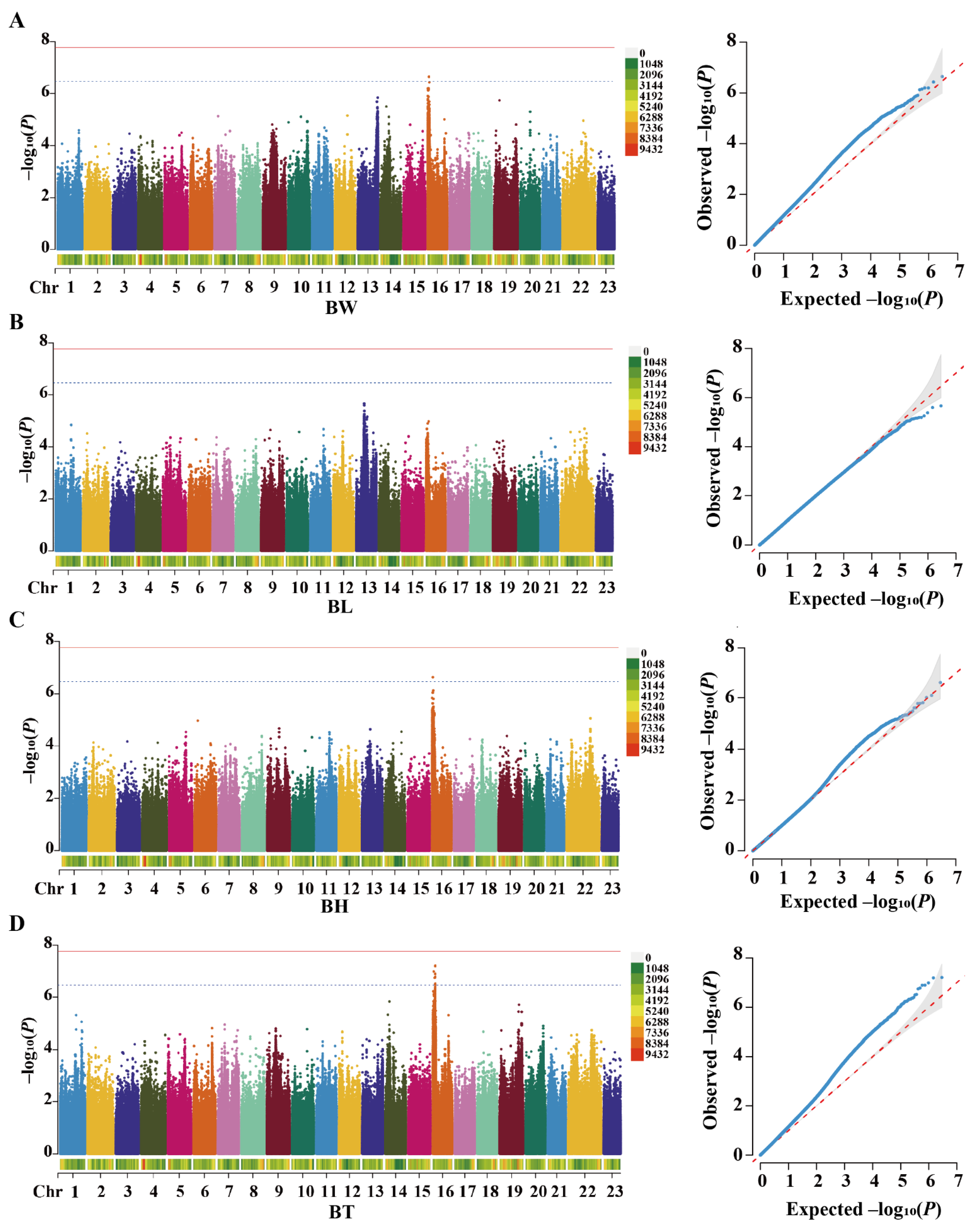
2.3. Screening of Growth-Associated SNPs in Hypertonic Environments
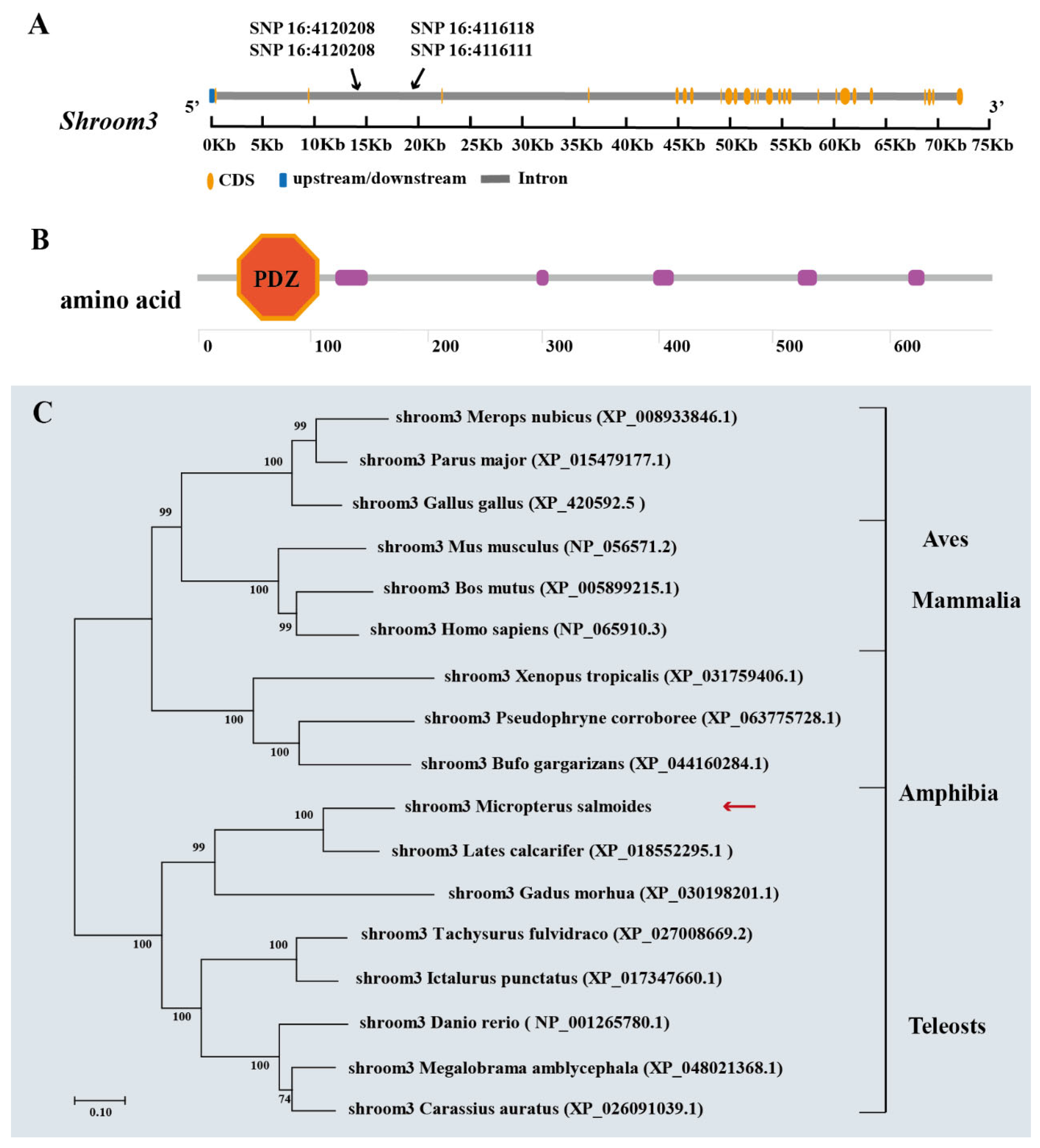
2.4. Candidate Gene Identification and Functional Annotation
2.5. Verification of Growth-Related SNPs
2.6. Joint Analysis of Significant SNP Peak and LD Block
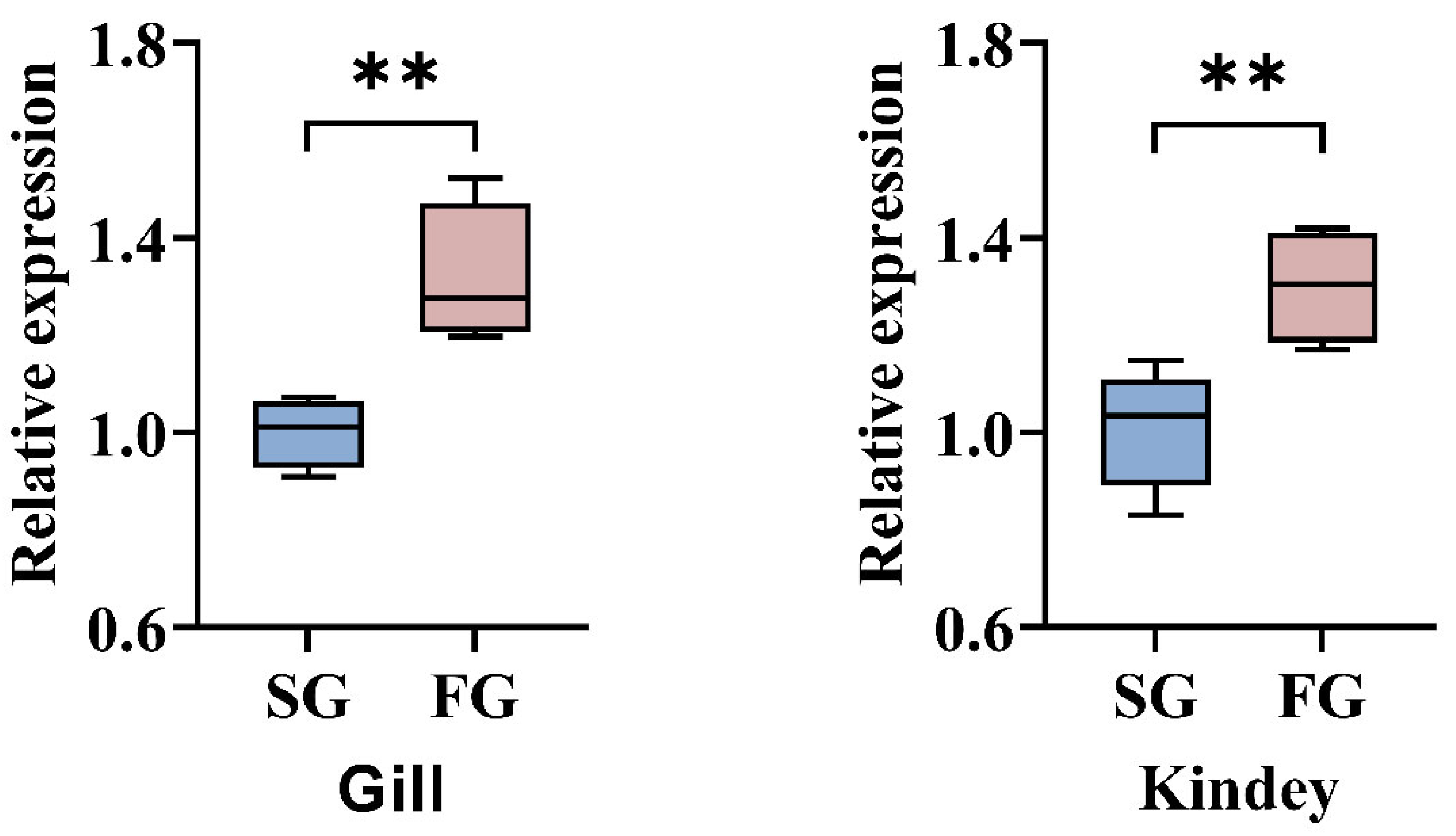
2.7. Molecular Characterization and Expression Analysis of Shroom3 in Different Organizations
3. Discussion
4. Materials and Methods
4.1. Statement of Ethics
4.2. Management and Sample DNA Extraction
4.3. Sequencing Library Preparation, Genotyping, and Data Filtering
4.4. Population Genetic Structure and Linkage Disequilibrium Analysis
4.5. Assessment of Genetic Parameters for Growth Traits
4.6. Growth Trait GWAS
4.7. Annotation of Candidate Genes
4.8. Verification of Growth-Associated SNPs
4.9. LD Block Synthesis and Growth-Advantageous Genotype Statistics
4.10. Analysis of Candidate Gene Sequences and Expression Patterns Under Different Salinities
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hwang, J.; Kim, S.; Seo, Y.; Lee, K.; Park, C.; Choi, Y.; Kim, D.; Gilad, A.A.; Choi, J. Mechanisms of Salinity Control in Sea Bass. Biotechnol. Bioprocess Eng. 2018, 23, 271–277. [Google Scholar] [CrossRef]
- Li, Y.; Gao, P.; Zhou, K.; Yao, Z.; Sun, Z.; Qin, H.; Lai, Q. Effects of saline and alkaline stresses on the survival, growth, and physiological responses in juvenile mandarin fish (Siniperca chuatsi). Aquaculture 2024, 591, 741143. [Google Scholar] [CrossRef]
- Jia, Y.K.; Du, J.; Xi, R.J.; Zhang, Q.; Li, L.; Li, D.P.; Takagi, Y.; Zhang, X. Effects of different culture salinities on the growth and muscle quality of grass carp (Ctenopharyngodon idellus). J. Anim. Sci. 2024, 102, skae281. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Li, E.C.; Suo, Y.T.; Su, Y.J.; Lu, M.H.; Zhao, Q.; Qin, J.G.; Chen, L.Q. Histological and transcriptomic responses of two immune organs, the spleen and head kidney, in Nile tilapia (Oreochromis niloticus) to long-term hypersaline stress. Fish Shellfish. Immunol. 2018, 76, 48–57. [Google Scholar] [CrossRef]
- Dawood, M.A.O.; Noreldin, A.E.; Sewilam, H. Long term salinity disrupts the hepatic function, intestinal health, and gills antioxidative status in Nile tilapia stressed with hypoxia. Ecotoxicol. Environ. Saf. 2021, 220, 112412. [Google Scholar] [CrossRef]
- Takata, R.; Mattioli, C.C.; Bazzoli, N.; Junior, J.D.C.; Luz, R.K. The effects of salinity on growth, gill tissue and muscle cellularity in Lophiosilurus alexandri juvenile, a Neotropical freshwater catfish. Aquac. Res. 2021, 52, 4064–4075. [Google Scholar] [CrossRef]
- Canedo-Arguelles, M.; Kefford, B.J.; Piscart, C.; Prat, N.; Schafer, R.B.; Schulz, C.J. Salinisation of rivers: An urgent ecological issue. Environ. Pollut. 2013, 173, 157–167. [Google Scholar] [CrossRef]
- Patel, R.K.; Verma, A.K.; Krishnani, K.K.; Sreedharan, K.; Chandrakant, M.H. Growth performance, physio-metabolic, and haemato-biochemical status of Labeo rohita (Hamilton, 1822) juveniles reared at varying salinity levels using inland saline groundwater. Aquaculture 2022, 559, 738408. [Google Scholar] [CrossRef]
- Lin, H.L.; Zhou, Z.X.; Zhao, J.; Zhou, T.; Bai, H.Q.; Ke, Q.Z.; Pu, F.; Zheng, W.Q.; Xu, P. Genome-Wide Association Study Identifies Genomic Loci of Sex Determination and Gonadosomatic Index Traits in Large Yellow Croaker (Larimichthys crocea). Mar. Biotechnol. 2021, 23, 127–139. [Google Scholar] [CrossRef]
- San, L.Z.; Liu, B.S.; Liu, B.; Zhu, K.C.; Guo, L.; Guo, H.Y.; Zhang, N.; Jiang, S.G.; Zhang, D.C. Genome-wide association study reveals multiple novel SNPs and putative candidate genes associated with low oxygen tolerance in golden pompano Trachinotus ovatus (Linnaeus 1758). Aquaculture 2021, 544, 737098. [Google Scholar] [CrossRef]
- Wang, L.; Liu, P.; Huang, S.Q.; Ye, B.Q.; Chua, E.; Wan, Z.Y.; Yue, G.H. Genome-Wide Association Study Identifies Loci Associated with Resistance to Viral Nervous Necrosis Disease in Asian Seabass. Mar. Biotechnol. 2017, 19, 255–265. [Google Scholar] [CrossRef]
- Gonzalez-Pena, D.; Gao, G.T.; Baranski, M.; Moen, T.; Cleveland, B.M.; Kenney, P.B.; Vallejo, R.L.; Palti, Y.; Leeds, T.D. Genome-Wide Association Study for Identifying Loci that Affect Fillet Yield, Carcass, and Body Weight Traits in Rainbow Trout (Oncorhynchus mykiss). Front. Genet. 2016, 7, 203. [Google Scholar] [CrossRef] [PubMed]
- Robledo, D.; Fernández, C.; Hermida, M.; Sciara, A.; Alvarez-Dios, J.A.; Cabaleiro, S.; Caamaño, R.; Martínez, P.; Bouza, C. Integrative Transcriptome, Genome and Quantitative Trait Loci Resources Identify Single Nucleotide Polymorphisms in Candidate Genes for Growth Traits in Turbot. Int. J. Mol. Sci. 2016, 17, 243. [Google Scholar] [CrossRef] [PubMed]
- Palaiokostas, C.; Kocour, M.; Prchal, M.; Houston, R.D. Accuracy of Genomic Evaluations of Juvenile Growth Rate in Common Carp (Cyprinus carpio) Using Genotyping by Sequencing. Front. Genet. 2018, 9, 82. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.D.; Qin, H.; Zhu, Z.X.; Liang, X.Y.; Xia, J.H. Genome-wide association studies in a F1 tilapia population reveal novel genetic loci associated with salinity tolerance. Aquaculture 2024, 588, 740913. [Google Scholar] [CrossRef]
- Yang, Z.T.; Wang, L.; Sun, F.; Wong, J.; Lee, M.; Yeo, S.; Wen, Y.F.; Yue, G.H. Salt tolerance candidate genes identified by QTL mapping, RNA-seq, and functional analysis in tilapia. Aquaculture 2025, 596, 741762. [Google Scholar] [CrossRef]
- Sun, S.K.; Gong, C.P.; Deng, C.Y.; Yu, H.R.; Zheng, D.; Wang, L.; Sun, J.L.; Song, F.B.; Luo, J. Effects of salinity stress on the growth performance, health status, and intestinal microbiota of juvenile Micropterus salmoides. Aquaculture 2023, 576, 739888. [Google Scholar] [CrossRef]
- Du, X.; Zhang, W.; He, J.; Zhao, M.; Wang, J.; Dong, X.; Fu, Y.; Xie, X.; Miao, S. The Impact of Rearing Salinity on Flesh Texture, Taste, and Fatty Acid Composition in Largemouth Bass Micropterus salmoides. Foods 2022, 11, 3261. [Google Scholar] [CrossRef]
- Yi, H.D.; Chen, X.L.; Liu, S.; Han, L.Q.; Liang, J.H.; Su, Y.Q.; Lai, H.; Bi, S.; Liu, X.G.; Zhang, Y.; et al. Growth, osmoregulatory and hypothalamic-pituitary-somatotropic (HPS) axis response of the juvenile largemouth bass (Micropterus salmoides), reared under different salinities. Aquac. Rep. 2021, 20, 100727. [Google Scholar] [CrossRef]
- He, K.; Zhao, L.L.; Yuan, Z.H.; Canario, A.; Liu, Q.; Chen, S.Y.; Guo, J.Z.; Luo, W.; Yan, H.X.; Zhang, D.M.; et al. Chromosome-level genome assembly of largemouth bass (Micropterus salmoides) using PacBio and Hi-C technologies. Sci. Data 2022, 9, 482. [Google Scholar] [CrossRef]
- Leite, T.; Branco, P.; Ferreira, M.T.; Santos, J.M. Activity, boldness and schooling in freshwater fish are affected by river salinization. Sci. Total. Environ. 2022, 819, 153046. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, N.A.; Saad, M.F.; Shukry, M.; El-Keredy, A.M.S.; Nasif, O.; Van Doan, H.; Dawood, M.A.O. Physiological and ion changes of Nile tilapia (Oreochromis niloticus) under the effect of salinity stress. Aquac. Rep. 2021, 19, 100567. [Google Scholar] [CrossRef]
- Tian, L.; Tan, P.; Yang, L.; Zhu, W.; Xu, D. Effects of salinity on the growth, plasma ion concentrations, osmoregulation, non-specific immunity, and intestinal microbiota of the yellow drum (Nibea albiflora). Aquaculture 2020, 528, 735470. [Google Scholar] [CrossRef]
- Wu, H.F.; Liu, J.L.; Lu, Z.; Xu, L.L.; Ji, C.L.; Wang, Q.; Zhao, J.M. Metabolite and gene expression responses in juvenile flounder Paralichthys olivaceus exposed to reduced salinities. Fish Shellfish. Immunol. 2017, 63, 417–423. [Google Scholar] [CrossRef]
- Xiong, X.-M.; Huang, W.-J.; Dong, Q.; Zhang, D.-Y.; Wan, S.-M.; Gao, Z.-X. Genetic parameter estimates for intermuscular bone trait in grass carp (Ctenopharyngodon idella). Aquaculture 2023, 563, 739011. [Google Scholar] [CrossRef]
- Yang, J.; Zeng, J.; Goddard, M.E.; Wray, N.R.; Visscher, P.M. Concepts, estimation and interpretation of SNP-based heritability. Nat. Genet. 2017, 49, 1304–1310. [Google Scholar] [CrossRef]
- Hao, Y.H.; Jia, X.W.; Yuan, L.; Liu, Y.T.; Gui, L.; Shen, Y.B.; Li, J.L.; Xu, X.Y. Genome-wide association study reveals growth-related SNPs and candidate genes in grass carp (Ctenopharyngodon). Aquaculture 2023, 577, 739979. [Google Scholar] [CrossRef]
- Chen, G.; Liu, H.Y.; Yu, X.M.; Luo, W.W.; Tong, J.G. Estimation of heritabilities and quantitative trait loci for growth traits of bighead carp (Hypophthalmichthys nobilis). Aquaculture 2023, 566, 739213. [Google Scholar] [CrossRef]
- Li, Y.; Lu, L.J.; Yang, B.; Li, P.F.; Xu, C.X.; Li, Q.; Liu, S.K. Evaluation of genetic parametrs of growth and survival traits and analysis of genotype × environment interaction effects in vibrio-resistant familes of Pacfic Oyster. Acta Hydrobiol. Sin. 2024, 48, 673–682. [Google Scholar]
- Tian, Y.S.; Xu, T.J.; Liang, Y.; Chen, S.L. Estimates of genetic and phenotypic parameters for weight and length in Paralichthys olivaceus (Temminck et Schlegel). Acta Oceanol. Sin. 2011, 30, 58–64. [Google Scholar] [CrossRef]
- Yang, W.; Wang, Y.R.; Jiang, D.N.; Tian, C.X.; Zhu, C.H.; Li, G.L.; Chen, H.P. ddRADseq-assisted construction of a high-density SNP genetic map and QTL fine mapping for growth-related traits in the spotted scat (Scatophagus argus). BMC Genom. 2020, 21, 278. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Chen, B.H.; Ke, Q.Z.; Zhao, J.; Pu, F.; Wu, Y.D.; Chen, L.; Zhou, Z.X.; Bai, Y.L.; Pan, Y.; et al. Development and Evaluation of a High-Throughput Single-Nucleotide Polymorphism Array for Large Yellow Croaker (Larimichthys crocea). Front. Genet. 2020, 11, 571751. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; You, X.; Li, J.; Zhang, X.; Zhang, S.; Jiang, S.; Lin, X.; Lin, H.-R.; Meng, Z.; Shi, Q. A genome-wide association study on growth traits in orange-spotted grouper (Epinephelus coioides) with RAD-seq genotyping. Sci. China Life Sci. 2018, 61, 934–946. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Fu, H.C.; Wang, Y.Y.; Huang, H.Z. Genome-wide association study reveals growth-related SNPs and candidate genes in mandarin fish (Siniperca chuatsi). Aquaculture 2022, 550, 737879. [Google Scholar] [CrossRef]
- Zhou, Z.X.; Han, K.H.; Wu, Y.D.; Bai, H.Q.; Ke, Q.Z.; Pu, F.; Wang, Y.L.; Xu, P. Genome-Wide Association Study of Growth and Body-Shape-Related Traits in Large Yellow Croaker (Larimichthys crocea) Using ddRAD Sequencing. Mar. Biotechnol. 2019, 21, 655–670. [Google Scholar] [CrossRef]
- Mou, C.Y.; Li, Y.; Zhou, J.; Li, Q.; Zhou, B.; Wei, Z.; Luo, H.; Ke, H.Y.; Duan, Y.L.; Zhai, W.T.; et al. Genome-wide association study reveals growth-related markers and candidate genes for selection in Chinese longsnout catfish (Leiocassis longirostris). Aquaculture 2022, 560, 738513. [Google Scholar] [CrossRef]
- Salem, M.; Al-Tobasei, R.; Ali, A.; Lourenco, D.; Gao, G.T.; Palti, Y.; Kenney, B.; Leeds, T.D. Genome-Wide Association Analysis With a 50K Transcribed Gene SNP-Chip Identifies QTL Affecting Muscle Yield in Rainbow Trout. Front. Genet. 2018, 9, 387. [Google Scholar] [CrossRef]
- Zhu, F.Q.; Sun, H.B.; Jiang, L.M.; Zhang, Q.Q.; Liu, J.X. Genome-wide association study for growth-related traits in golden pompano (Trachinotus. ovatus). Aquaculture 2023, 572, 739549. [Google Scholar] [CrossRef]
- Omeka, W.K.M.; Liyanage, D.S.; Lee, S.; Lim, C.; Yang, H.; Sandamalika, W.M.G.; Udayantha, H.M.V.; Kim, G.; Ganeshalingam, S.; Jeong, T.; et al. Genome-wide association study (GWAS) of growth traits in olive flounder (Paralichthys olivaceus). Aquaculture 2022, 555, 738257. [Google Scholar] [CrossRef]
- Chavali, S.; Barrenas, F.; Kanduri, K.; Benson, M. Network properties of human disease genes with pleiotropic effects. BMC Syst. Biol. 2010, 4, 78. [Google Scholar] [CrossRef]
- Pelis, R.M.; McCormick, S.D. Effects of growth hormone and cortisol on Na+-K+-2Cl− cotransporter localization and abundance in the gills of Atlantic salmon. Gen. Comp. Endocrinol. 2001, 124, 134–143. [Google Scholar] [CrossRef] [PubMed]
- Zikos, A.; Seale, A.P.; Lerner, D.T.; Grau, E.G.; Korsmeyer, K.E. Effects of salinity on metabolic rate and branchial expression of genes involved in ion transport and metabolism in Mozambique tilapia (Oreochromis mossambicus). Comp. Biochem. Phys. A 2014, 178, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Oh, W.J.; Jacinto, E. mTOR complex 2 signaling and functions. Cell Cycle 2011, 10, 2305–2316. [Google Scholar] [CrossRef]
- Guan, Y.J.; Yang, X.; Yang, W.T.; Charbonneau, C.; Chen, Q. Mechanical activation of mammalian target of rapamycin pathway is required for cartilage development. FASEB J. 2014, 28, 4470–4481. [Google Scholar] [CrossRef]
- Yaba, A.; Bianchi, V.; Borini, A.; Johnson, J. A putative mitotic checkpoint dependent on mTOR function controls cell proliferation and survival in ovarian granulosa cells. Reprod. Sci. 2008, 15, 128–138. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Dai, L.Y.; Peng, F.Y.; Tang, L.W.; Wang, X.J.; Chen, J.Y.; Liu, J.H.; Fu, W.; Peng, L.Y.; Liu, W.B.; et al. mTOR signaling pathway regulates embryonic development and rapid growth of triploid crucian carp. Aquac. Rep. 2023, 33, 101860. [Google Scholar] [CrossRef]
- Niu, X.J.; Qian, X.L.; Feng, H.M.; Yi, K.; Li, D.; Chen, W.J.; Ye, J.D. Growth and metabolic responses of grouper juveniles (Epinephelus coioides) fed diets containing varying levels of leucine. Aquaculture 2021, 534, 736281. [Google Scholar] [CrossRef]
- Wu, L.Y.; Li, H.Y.; Xu, W.J.; Dong, B.; Geng, H.C.; Jin, J.Y.; Han, D.; Liu, H.K.; Zhu, X.M.; Yang, Y.X.; et al. Emodin alleviates acute hypoxia-induced apoptosis in gibel carp (Carassius gibelio) by upregulating autophagy through modulation of the AMPK/mTOR pathway. Aquaculture 2022, 548, 737689. [Google Scholar] [CrossRef]
- Wu, L.H.; Liang, H.L.; Hamunjo, C.M.K.; Ge, X.P.; Ji, K.; Yu, H.; Huang, D.Y.; Xu, H.; Ren, M.C. Culture salinity alters dietary protein requirement, whole body composition and nutrients metabolism related genes expression in juvenile Genetically Improved Farmed Tilapia (GIFT) (Oreochromis niloticus). Aquaculture 2020, 531, 735961. [Google Scholar] [CrossRef]
- Lei, C.X.; Li, M.M.; Tian, J.J.; Wen, J.K.; Li, Y.Y. Transcriptome analysis of golden pompano (Trachinotus ovatus) liver indicates a potential regulatory target involved in HUFA uptake and deposition. Comp. Biochem. Physiol. D Genom. Proteom. 2020, 33, 100633. [Google Scholar] [CrossRef]
- Tan, P.; Wabike, E.E.; Qin, G.C.; Lou, B.; Xu, D.D.; Chen, R.Y.; Wang, L.G. Effects of dietary n-3 long-chain polyunsaturated fatty acids (n-3 LC-PUFAs) on growth performance, body composition and subcutaneous adipose tissue transcriptome analysis of juvenile yellow drum (Nibea albiflora). Aquac. Nutr. 2021, 27, 556–567. [Google Scholar] [CrossRef]
- Xu, X.X.; Ji, H.; Belghit, I.; Liland, N.S.; Wu, W.Y.; Li, X.Q. Effects of black soldier fly oil rich in n-3 HUFA on growth performance, metabolism and health response of juvenile mirror carp (Cyprinus carpio var. specularis). Aquaculture 2021, 533, 736144. [Google Scholar] [CrossRef]
- Si, Y.F.; Wen, H.S.; Li, Y.; He, F.; Li, J.F.; Li, S.P.; He, H.W. Liver transcriptome analysis reveals extensive transcriptional plasticity during acclimation to low salinity in Cynoglossus semilaevis. BMC Genom. 2018, 19, 464. [Google Scholar] [CrossRef] [PubMed]
- Kornau, H.C.; Seeburg, P.H.; Kennedy, M.B. Interaction of ion channels and receptors with PDZ domain proteins. Curr. Opin. Neurobiol. 1997, 7, 368–373. [Google Scholar] [CrossRef]
- Khalili, H.; Sull, A.; Sarin, S.; Boivin, F.J.; Halabi, R.; Svajger, B.; Li, A.H.; Cui, V.W.; Drysdale, T.; Bridgewater, D. Developmental Origins for Kidney Disease Due to Deficiency. J. Am. Soc. Nephrol. 2016, 27, 2965–2973. [Google Scholar] [CrossRef]
- Yeo, N.C.; O’Meara, C.C.; Bonomo, J.A.; Veth, K.N.; Tomar, R.; Flister, M.J.; Drummond, I.A.; Bowden, D.W.; Freedman, B.I.; Lazar, J.; et al. contributes to the maintenance of the glomerular filtration barrier integrity. Genome. Res. 2015, 25, 57–65. [Google Scholar] [CrossRef]
- Cui, W.X.; Ma, A.J.; Huang, Z.H.; Wang, X.N.; Liu, Z.F.; Xia, D.D.; Yang, S.S.; Zhao, T.L. Comparative transcriptomic analysis reveals mechanisms of divergence in osmotic regulation of the turbot Scophthalmus. Fish Physiol. Biochem. 2020, 46, 1519–1536. [Google Scholar] [CrossRef]
- Ding, L.; Liu, Y.J.; Wei, X.F.; Geng, C.Y.; Liu, W.Z.; Han, L.; Yuan, F.Y.; Wang, P.; Sun, Y.C. Effects of Saline-Alkaline Stress on Metabolome, Biochemical Parameters, and Histopathology in the Kidney of Crucian Carp (Carassius auratus). Metabolites 2023, 13, 159. [Google Scholar] [CrossRef]
- Wong, M.K.S.; Woo, N.Y.S. Rapid changes in renal morphometrics in silver sea bream Sparus sarba on exposure to different salinities. J. Fish Biol. 2006, 69, 770–782. [Google Scholar] [CrossRef]
- Matthews, M.; Varga, Z.M. Anesthesia and Euthanasia in Zebrafish. ILAR J. 2012, 53, 192–204. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Zhu, P.Y.; He, L.Y.; Li, Y.Q.; Huang, W.P.; Xi, F.; Lin, L.; Zhi, Q.H.; Zhang, W.W.; Tang, Y.T.; Geng, C.Y.; et al. OTG-snpcaller: An Optimized Pipeline Based on TMAP and GATK for SNP Calling from Ion Torrent Data. PLoS. ONE 2014, 9, e97507. [Google Scholar] [CrossRef] [PubMed]
- Odumpatta, R.; Mohanapriya, A. Next generation sequencing exome data analysis aids in the discovery of SNP and INDEL patterns in Parkinson’s disease. Genomics 2020, 112, 3722–3728. [Google Scholar] [CrossRef]
- Yang, J.A.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A Tool for Genome-wide Complex Trait Analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef]
- Wang, J.B.; Zhang, Z.W. GAPIT Version 3: Boosting Power and Accuracy for Genomic Association and Prediction. Genom. Proteom. Bioinf. 2021, 19, 629–640. [Google Scholar] [CrossRef]
- Yin, L.; Zhang, H.; Tang, Z.; Xu, J.; Yin, D.; Zhang, Z.; Yuan, X.; Zhu, M.; Zhao, S.; Li, X.; et al. rMVP: A Memory-efficient, Visualization-enhanced, and Parallel-accelerated Tool for Genome-wide Association Study. Genom. Proteom. Bioinform. 2021, 19, 619–628. [Google Scholar] [CrossRef]
- Alexa, A.; Rahnenfuhrer, J.; Lengauer, T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 2006, 22, 1600–1607. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Dong, S.S.; He, W.M.; Ji, J.J.; Zhang, C.; Guo, Y.; Yang, T.L. LDBlockShow: A fast and convenient tool for visualizing linkage disequilibrium and haplotype blocks based on variant call format files. Brief. Bioinform. 2021, 22, bbaa227. [Google Scholar] [CrossRef]
- Zhang, L.; Qiang, J.; Tao, Y.-F.; Bao, J.-W.; Zhu, H.-J.; He, J.; Xu, P. Cloning of the gene encoding acyl-CoA thioesterase 11 and its functional characterization in hybrid yellow catfish (Pelteobagrus fulvidraco ♀ × Pelteobagrus vachelli ♂) under heat stress. J. Therm. Biol. 2020, 93, 102681. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]

| Trait | SNP | −Log (p Value) | Allele | Position |
|---|---|---|---|---|
| BH | 16:1462653 | 6.63270 | A/T | intergenic |
| BT | 16:1462653 | 6.98889 | A/T | intergenic |
| 16:3192874 | 6.74434 | T/G | intergenic | |
| 16:3967223 | 6.50020 | A/T | Intronic (Dcp2) | |
| 16:3984271 | 6.89572 | C/T | Exonic (Psmb7) | |
| 16:4072454 | 6.77821 | T/A | Intronic (Mfsd14ba) | |
| 16:4116111 | 7.19193 | G/A | intronic (Shroom3) | |
| 16:4116118 | 6.89294 | A/T | intronic (Shroom3) | |
| 16:4120208 | 6.51879 | A/T | intronic (Shroom3) | |
| 16:4120214 | 7.20708 | A/T | intronic (Shroom3) | |
| BW | 16:1964199 | 6.64524 | G/C | Intronic (Ralgps1) |
| SNP | 16:4102436 | 16:4155322 | 16:4173817 | 16:4116285 | 16:4110656 | 16:4121717 | 16:4124047 |
|---|---|---|---|---|---|---|---|
| REF | G | T | A | T | C | A | T |
| ALT | A | C | G | A | A | C | C |
| D’ | 0.855 | 0.807 | 0.856 | 0.813 | 0.816 | 0.826 | 0.809 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, M.; Tao, Y.; Hua, J.; Dong, Y.; Lu, S.; Qiang, J.; He, J. Genome-Wide Association Study Reveals Growth-Related SNPs and Candidate Genes in Largemouth Bass (Micropterus salmoides) Adapted to Hypertonic Environments. Int. J. Mol. Sci. 2025, 26, 1834. https://doi.org/10.3390/ijms26051834
Ding M, Tao Y, Hua J, Dong Y, Lu S, Qiang J, He J. Genome-Wide Association Study Reveals Growth-Related SNPs and Candidate Genes in Largemouth Bass (Micropterus salmoides) Adapted to Hypertonic Environments. International Journal of Molecular Sciences. 2025; 26(5):1834. https://doi.org/10.3390/ijms26051834
Chicago/Turabian StyleDing, Miaomiao, Yifan Tao, Jixiang Hua, Yalun Dong, Siqi Lu, Jun Qiang, and Jixiang He. 2025. "Genome-Wide Association Study Reveals Growth-Related SNPs and Candidate Genes in Largemouth Bass (Micropterus salmoides) Adapted to Hypertonic Environments" International Journal of Molecular Sciences 26, no. 5: 1834. https://doi.org/10.3390/ijms26051834
APA StyleDing, M., Tao, Y., Hua, J., Dong, Y., Lu, S., Qiang, J., & He, J. (2025). Genome-Wide Association Study Reveals Growth-Related SNPs and Candidate Genes in Largemouth Bass (Micropterus salmoides) Adapted to Hypertonic Environments. International Journal of Molecular Sciences, 26(5), 1834. https://doi.org/10.3390/ijms26051834





