Genome-Wide Identification and Expression Analysis of the Mediator Complex Subunit Gene Family in Cassava
Abstract
1. Introduction
2. Results
2.1. Identification of the MeMED Gene Family in Cassava
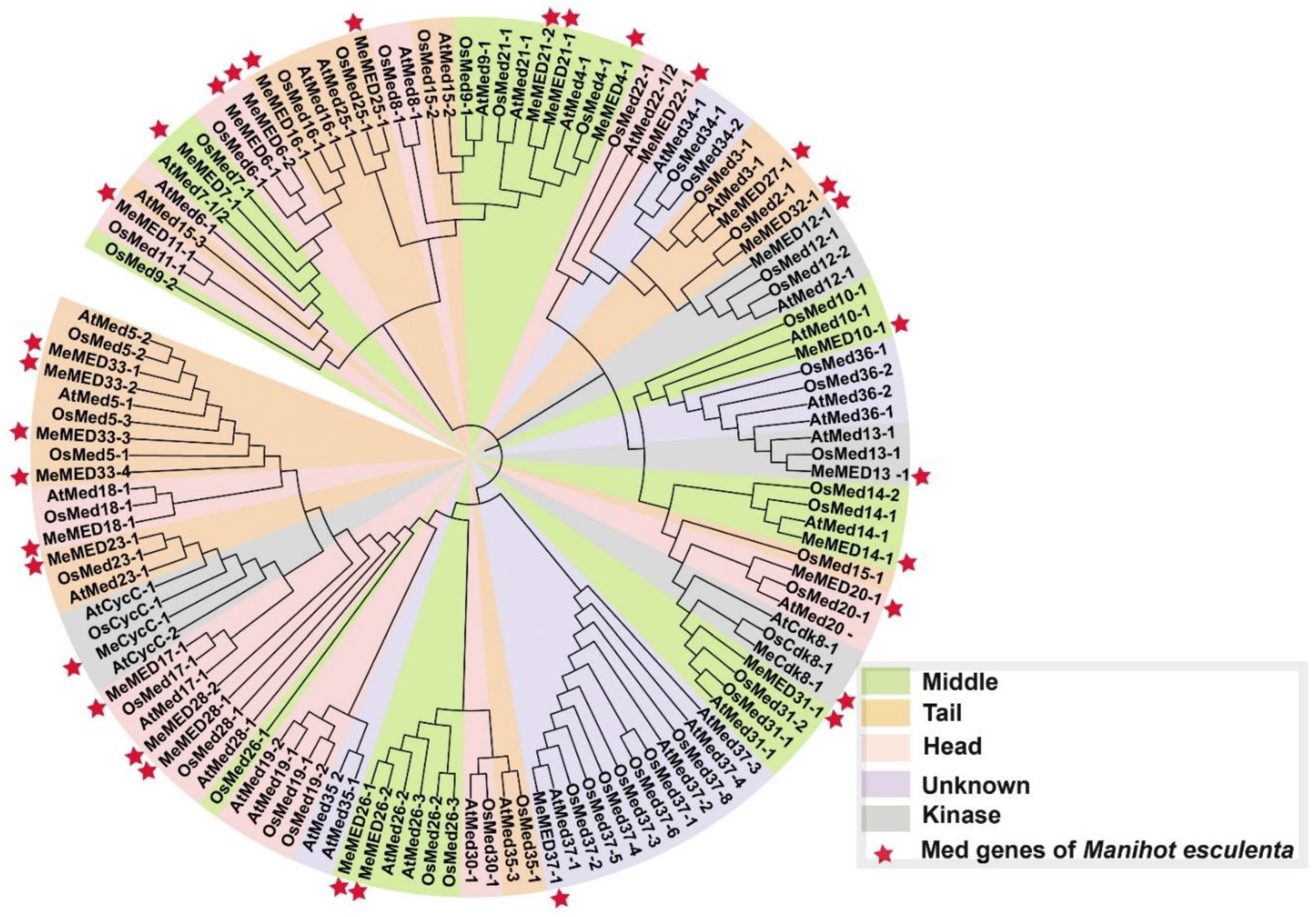
2.2. Phylogenetic Analysis of Cassava MeMED Proteins
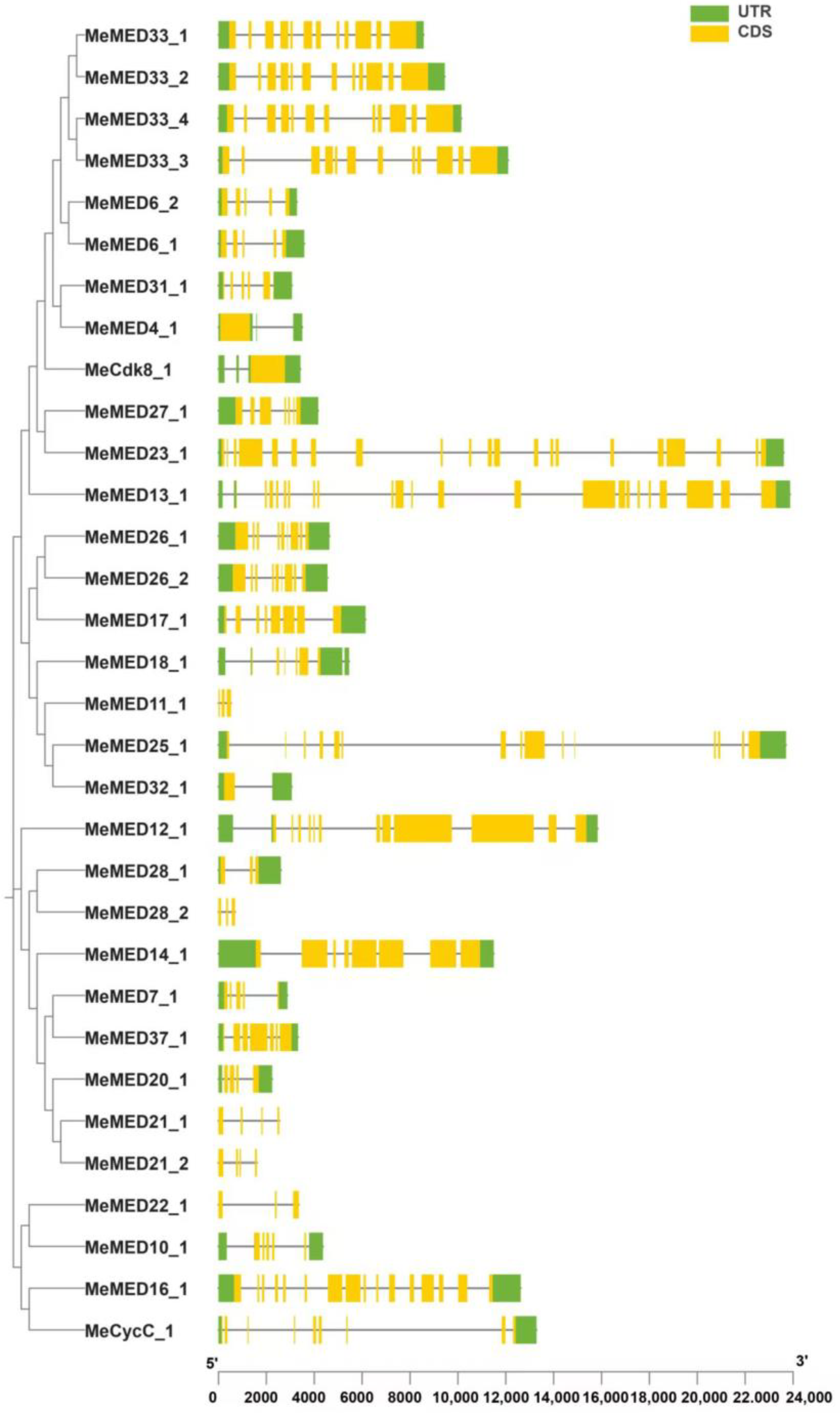
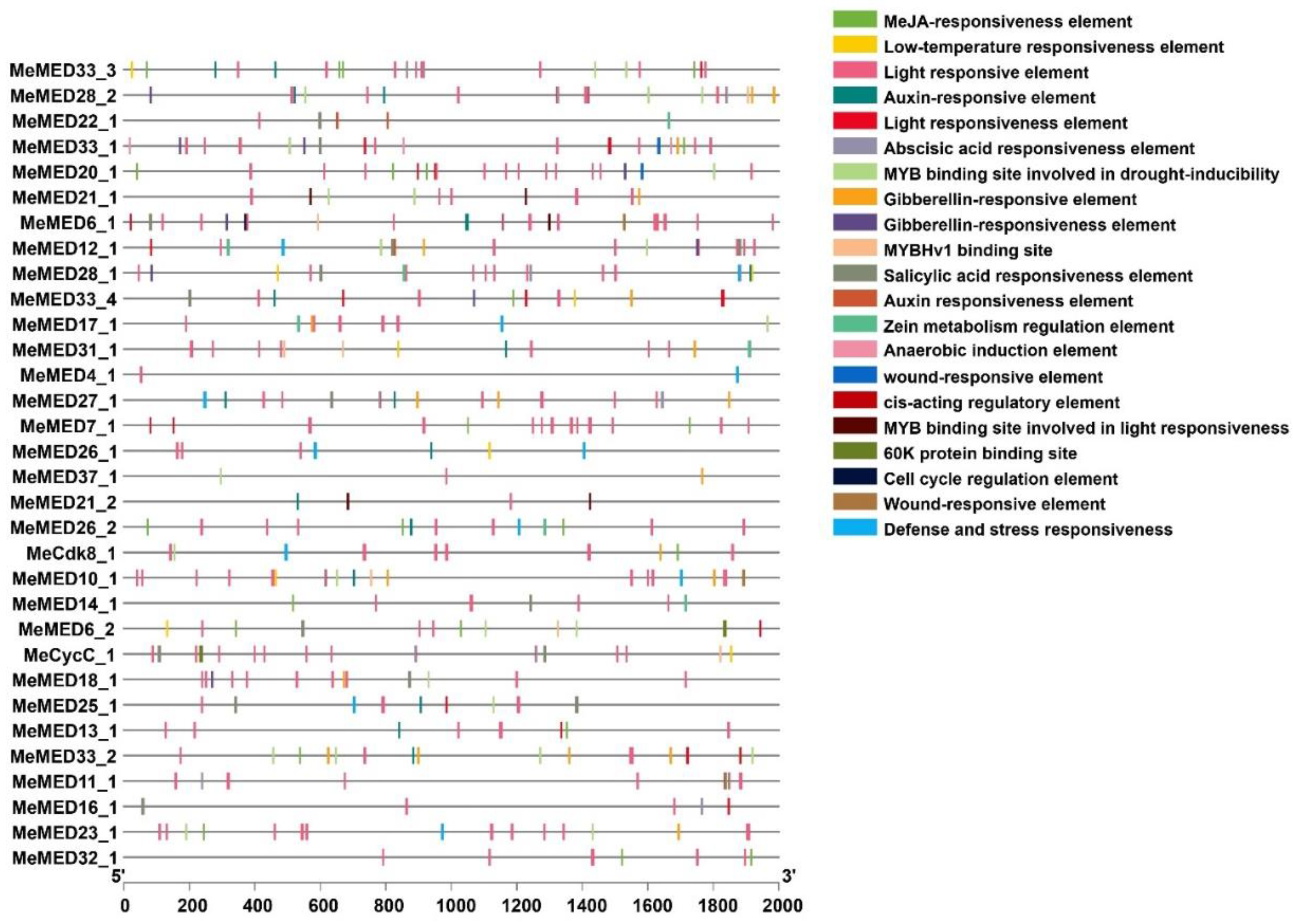
2.3. Analysis of Gene Structure and Cis-Acting Elements of the Cassava MeMED Genes
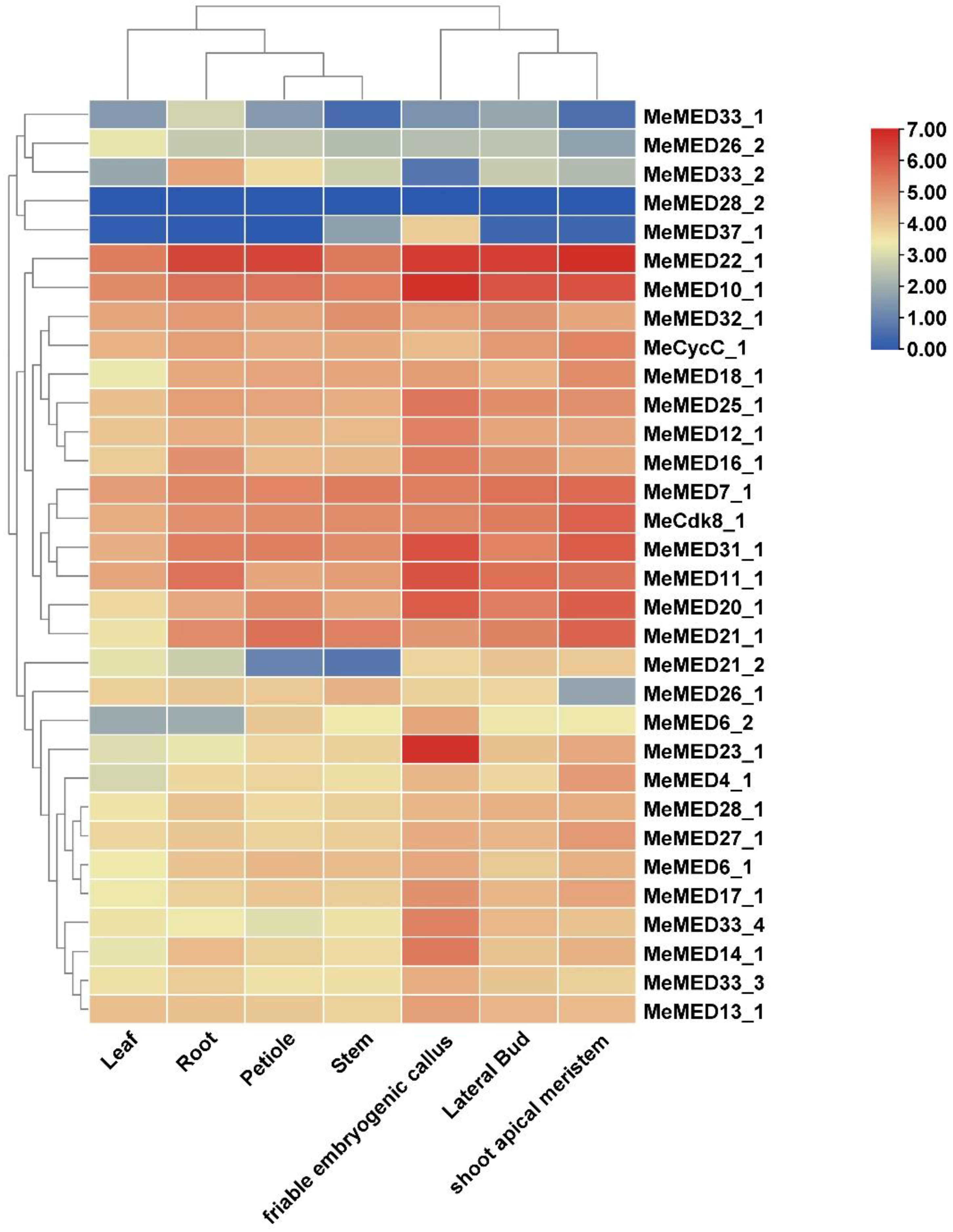
2.4. Expression Profiles of MeMED Genes in Different Tissues of Cassava Based on RNA-Seq
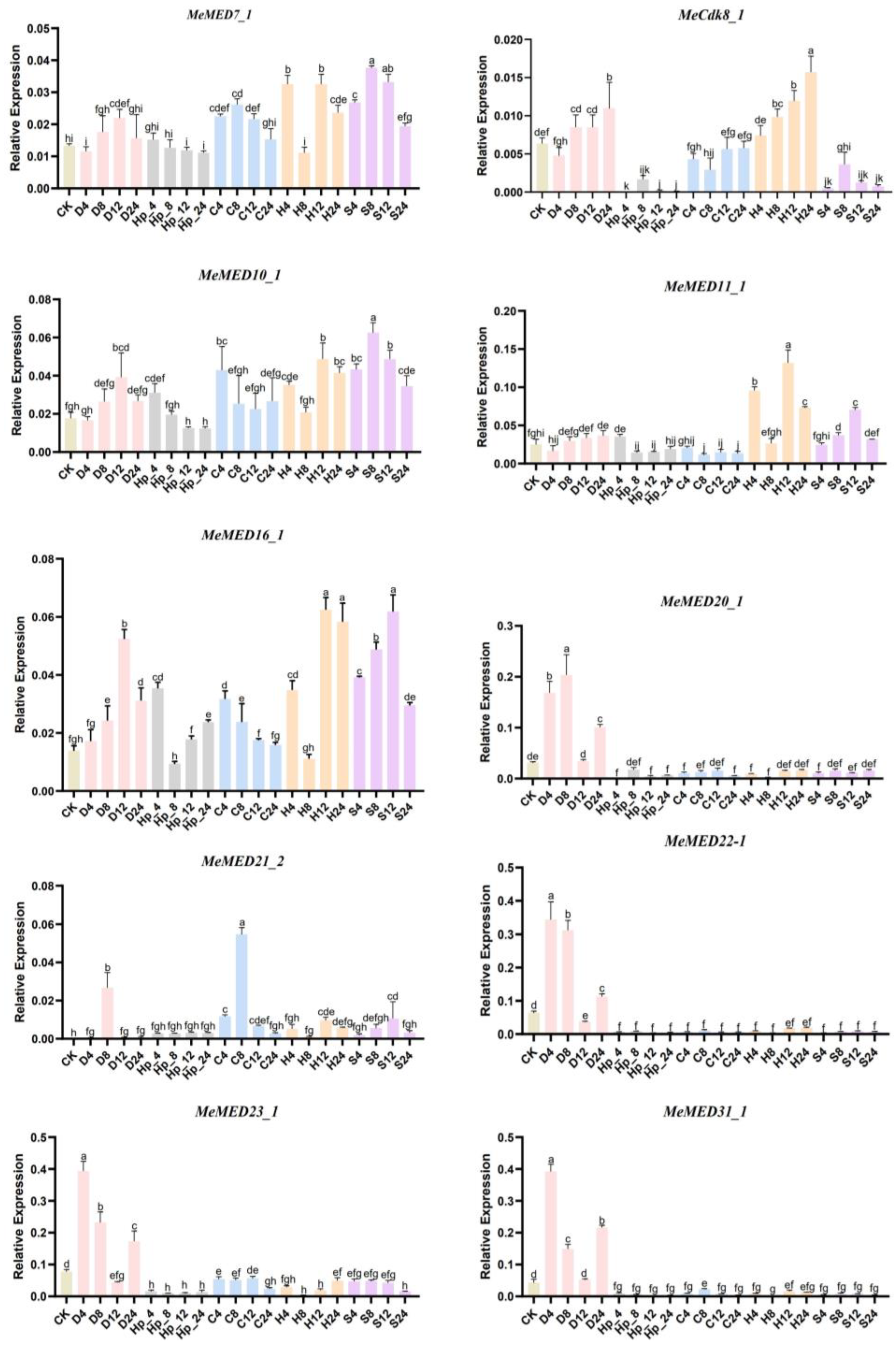
2.5. Expression Analysis of MeMED Genes in Response to Abiotic Stresses
3. Discussion
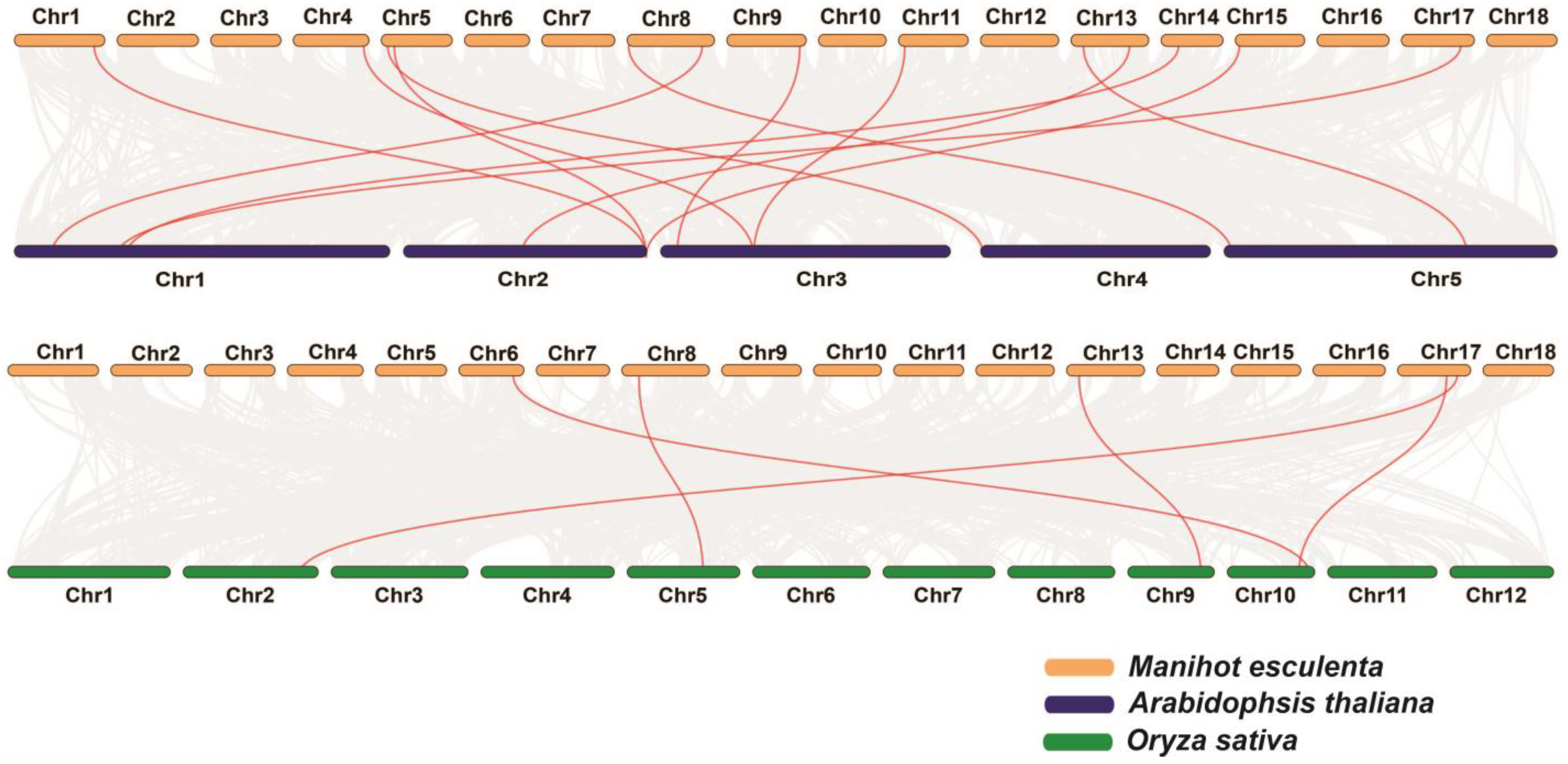
3.1. Phylogenetic Relationships and Evolution of the MeMED Gene Family
3.2. Regulation of MeMED Genes by Cis-Acting Elements
3.3. Expression Characteristics of MeMED Genes in Response to Stress
4. Material and Methods
4.1. Mining of MED Family in Manihot esculenta
4.2. Phylogeny of Cassava MED Proteins
4.3. Gene Structural Analysis of Cassava MED Proteins
4.4. Evolutionary Analysis of MeMED Genes
4.5. Expression Analyses of MeMED Genes in Different Tissues
4.6. RNA Extraction, cDNA Synthesis, and RT-qPCR Analysis
4.7. Plant Material and Stress Treatment Conditions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yang, Y.; Li, L.; Qu, L.J. Plant Mediator complex and its critical functions in transcription regulation. J. Integr. Plant Biol. 2016, 58, 106–118. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Q.Z.; Li, C.Y. The plant Mediator complex and its role in jasmonate signaling. J. Exp. Bot. 2019, 70, 3415–3424. [Google Scholar] [CrossRef] [PubMed]
- Schilbach, S.; Hantsche, M.; Tegunov, D.; Dienemann, C.; Wigge, C.; Urlaub, H.; Cramer, P. Structures of transcription pre-initiation complex with TFIIH and Mediator. Nature 2017, 551, 204. [Google Scholar] [CrossRef] [PubMed]
- Kornberg, R.D. Mediator and the mechanism of transcriptional activation. Trends Biochem. Sci. 2005, 30, 235–239. [Google Scholar] [CrossRef]
- Plaschka, C.; Larivière, L.; Wenzeck, L.; Seizl, M.; Hemann, M.; Tegunov, D.; Petrotchenko, E.V.; Borchers, C.H.; Baumeister, W.; Herzog, F.; et al. Architecture of the RNA polymerase II-Mediator core initiation complex. Nature 2015, 518, 376–380. [Google Scholar] [CrossRef]
- Robinson, P.J.; Trnka, M.J.; Bushnell, D.A.; Davis, R.E.; Mattei, P.J.; Burlingame, A.L.; Kornberg, R.D. Structure of a Complete Mediator-RNA Polymerase II Pre-Initiation Complex. Cell 2016, 166, 1411–1422. [Google Scholar] [CrossRef] [PubMed]
- Mathur, S.; Vyas, S.; Kapoor, S.; Tyagi, A.K. The Mediator Complex in Plants: Structure, Phylogeny, and Expression Profiling of Representative Genes in a Dicot (Arabidopsis) and a Monocot (Rice) during Reproduction and Abiotic Stress. Plant Physiol. 2011, 157, 1609–1627. [Google Scholar] [CrossRef] [PubMed]
- Chadick, J.Z.; Asturias, F.J. Structure of eukaryotic Mediator complexes. Trends Biochem. Sci. 2005, 30, 264–271. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Yang, S.; Fan, B.; Zhu, C.; Chen, Z. The Mediator Complex: A Central Coordinator of Plant Adaptive Responses to Environmental Stresses. Int. J. Mol. Sci. 2022, 23, 6170. [Google Scholar] [CrossRef] [PubMed]
- Chong, L.; Guo, P.; Zhu, Y. Mediator Complex: A Pivotal Regulator of ABA Signaling Pathway and Abiotic Stress Response in Plants. Int. J. Mol. Sci. 2020, 21, 7755. [Google Scholar] [CrossRef]
- Kidd, B.N.; Cahill, D.M.; Manners, J.M.; Schenk, P.M.; Kazan, K. Diverse roles of the Mediator complex in plants. Semin. Cell Dev. Biol. 2011, 22, 741–748. [Google Scholar] [CrossRef] [PubMed]
- Elfving, N.; Davoine, C.; Benlloch, R.; Blomberg, J.; Brännström, K.; Müller, D.; Nilsson, A.; Ulfstedt, M.; Ronne, H.; Wingsle, G.; et al. The Arabidopsis thaliana Med25 mediator subunit integrates environmental cues to control plant development. Proc. Natl. Acad. Sci. USA 2011, 108, 8245–8250. [Google Scholar] [CrossRef]
- Pei, Z.-M.; Zhu, Y.; Wang, B.; Tang, K.; Hsu, C.-C.; Xie, S.; Du, H.; Yang, Y.; Tao, W.A.; Zhu, J.-K. An Arabidopsis Nucleoporin NUP85 modulates plant responses to ABA and salt stress. PLoS Genet. 2017, 13, e1007124. [Google Scholar]
- Crawford, T.; Karamat, F.; Lehotai, N.; Rentoft, M.; Blomberg, J.; Strand, Å.; Björklund, S. Specific functions for Mediator complex subunits from different modules in the transcriptional response of Arabidopsis thaliana to abiotic stress. Sci. Rep. 2020, 10, 5073. [Google Scholar] [CrossRef] [PubMed]
- Li, X.H.; Yang, R.; Gong, Y.F.; Chen, H.M. The Arabidopsis Mediator Complex Subunit MED19a is Involved in ABI5-mediated ABA Responses. J. Plant Biol. 2018, 61, 97–110. [Google Scholar] [CrossRef]
- Lee, M.; Dominguez-Ferreras, A.; Kaliyadasa, E.; Huang, W.J.; Antony, E.; Stevenson, T.; Lehmann, S.; Schäfer, P.; Knight, M.R.; Ntoukakis, V.; et al. Mediator Subunits MED16, MED14, and MED2 Are Required for Activation of ABRE-Dependent Transcription in Arabidopsis. Front. Plant Sci. 2021, 12, 649720. [Google Scholar] [CrossRef]
- Zhu, Y.F.; Huang, P.C.; Guo, P.C.; Chong, L.; Yu, G.B.; Sun, X.L.; Hu, T.; Li, Y.; Hsu, C.C.; Tang, K.; et al. CDK8 is associated with RAP2.6 and SnRK2.6 and positively modulates abscisic acid signaling and drought response in Arabidopsis. New Phytol. 2020, 228, 1573–1590. [Google Scholar] [CrossRef] [PubMed]
- Knight, H.; Veale, E.L.; Warren, G.J.; Knight, M.R. The sfr6 mutation in Arabidopsis suppresses low-temperature induction of genes dependent on the CRT/DRE sequence motif. Plant Cell 1999, 11, 875–886. [Google Scholar] [CrossRef]
- Liang, L.; Wang, D.; Xu, D.; Xiao, J.; Tang, W.; Song, X.; Yu, G.; Liang, Z.; Xie, M.; Xu, Z.; et al. Comparative phylogenetic analysis of the mediator complex subunit in asparagus bean (Vigna unguiculata ssp. sesquipedialis) and its expression profile under cold stress. BMC Genom. 2024, 25, 149. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.; Giraud, E.; Duncan, O.; Law, S.R.; Wang, Y.; Xu, L.; Narsai, R.; Carrie, C.; Walker, H.; Day, D.A.; et al. Cyclin-dependent Kinase E1 (CDKE1) Provides a Cellular Switch in Plants between Growth and Stress Responses. J. Biol. Chem. 2013, 288, 3449–3459. [Google Scholar] [CrossRef] [PubMed]
- Ohama, N.; Moo, T.L.; Chua, N.H. Differential requirement of MED14/17 recruitment for activation of heat inducible genes. New Phytol. 2021, 229, 3360–3376. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.J.; Liu, P.; Shao, J.F.; Li, C.Q.; Wang, B.; Guo, X.; Yan, B.; Xia, Y.J.; Peng, M. Analysis of different strategies adapted by two cassava cultivars in response to drought stress: Ensuring survival or continuing growth. J. Exp. Bot. 2015, 66, 1477–1488. [Google Scholar] [CrossRef] [PubMed]
- Li, S.B.; Cui, Y.Y.; Zhou, Y.; Luo, Z.T.; Liu, J.D.; Zhao, M.M. The industrial applications of cassava: Current status, opportunities and prospects. J. Sci. Food Agric. 2017, 97, 2282–2290. [Google Scholar] [CrossRef] [PubMed]
- Marand, A.P.; Eveland, A.L.; Kaufmann, K.; Springer, N.M. cis-Regulatory Elements in Plant Development, Adaptation, and Evolution. Annu. Rev. Plant Biol. 2023, 74, 111–137. [Google Scholar] [CrossRef] [PubMed]
- Knight, H.; Mugford, S.G.; Ülker, B.; Gao, D.H.; Thorlby, G.; Knight, M.R. Identification of SFR6, a key component in cold acclimation acting post-translationally on CBF function. Plant J. 2009, 58, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Li, G.J.; Bressan, R.A.; Song, C.P.; Zhu, J.K.; Zhao, Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.P.; Chen, G.; Gao, F.; Xu, R.; Li, N.; Zhang, Y.Y.; Li, Y.H. Transcriptional Repression of the APC/C Activator Genes CCS52A1/A2 by the Mediator Complex Subunit MED16 Controls Endoreduplication and Cell Growth in Arabidopsis. Plant Cell 2019, 31, 1899–1912. [Google Scholar] [CrossRef] [PubMed]
- Rhee, S.Y.; Beavis, W.; Berardini, T.Z.; Chen, G.; Dixon, D.; Doyle, A.; Garcia-Hernandez, M.; Huala, E.; Lander, G.; Montoya, M.; et al. The Arabidopsis Information Resource (TAIR): A model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res. 2003, 31, 224–228. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.N.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I.; et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015, 43, D222–D226. [Google Scholar] [CrossRef] [PubMed]
- Garg, V.K.; Avashthi, H.; Tiwari, A.; Jain, P.A.; Ramkete, P.W.; Kayastha, A.M.; Singh, V.K. MFPPI—Multi FASTA ProtParam Interface. Bioinformation 2016, 12, 74–77. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11 Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Goodstein, D.M.; Shu, S.Q.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Koch, M.A.; Haubold, B.; Mitchell-Olds, T. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Mol. Biol. Evol. 2000, 17, 1483–1498. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
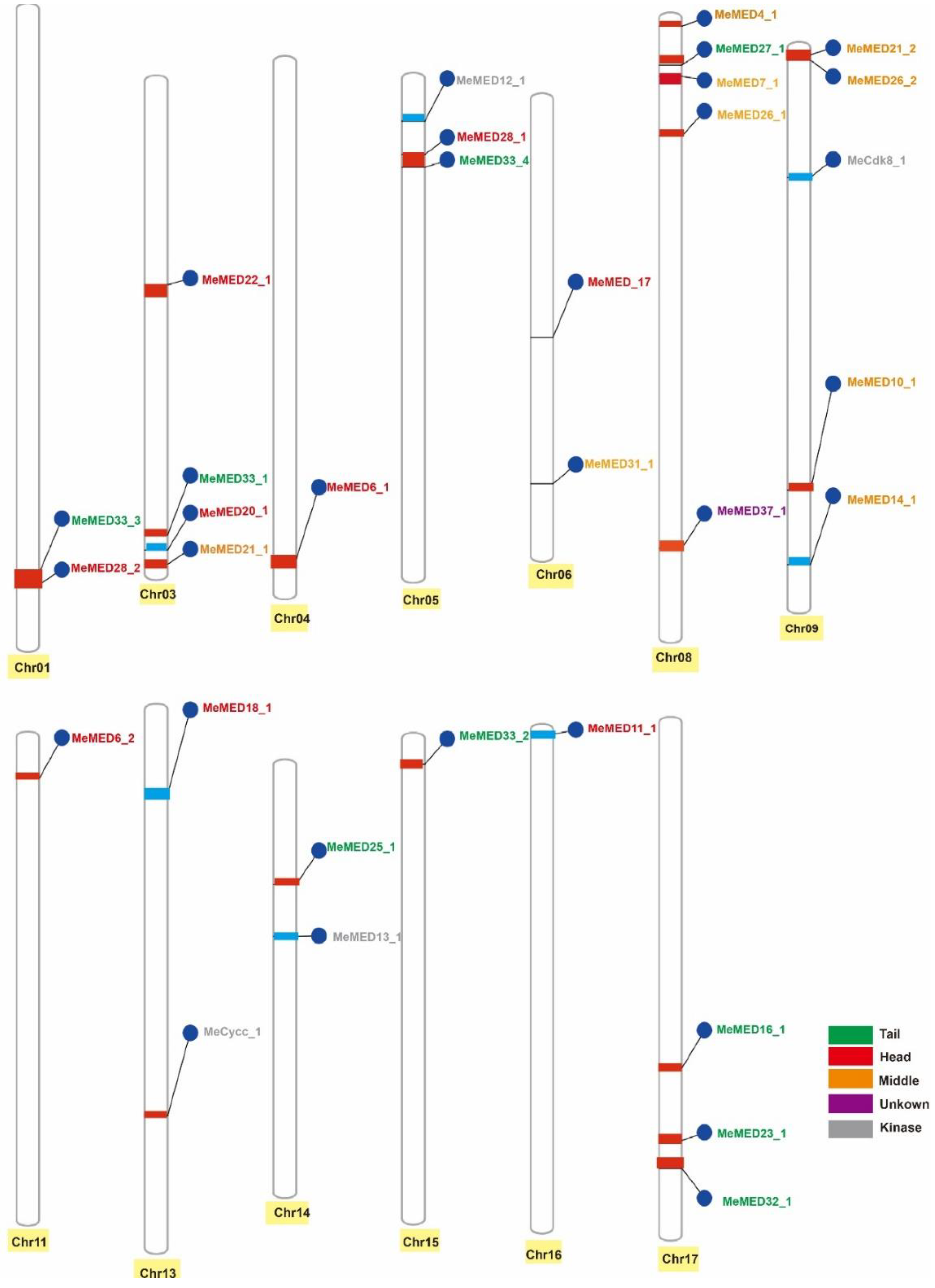
| Gene Name | Chr | CDS (bp) | Protein Length (A.A) | Protein Molecular Weight (KDa) | pI | Gravy | Mediator Module | |
|---|---|---|---|---|---|---|---|---|
| Manes.01G210100.1 | MeMED33_3 | 1 | 3987 | 1329 | 144.55 | 6.71 | 0.169 | Tail |
| Manes.01G220201.1 | MeMED28_2 | 1 | 318 | 106 | 12.41 | 6.13 | −0.185 | Head |
| Manes.03G077300.1 | MeMED22_1 | 3 | 477 | 159 | 17.00 | 6.12 | −0.226 | Head |
| Manes.03G185600.9 | MeMED33_1 | 3 | 3969 | 1323 | 144.45 | 6.68 | 0.227 | Tail |
| Manes.03G199800.1 | MeMED20_1 | 3 | 666 | 222 | 25.36 | 6.08 | −0.266 | Head |
| Manes.03G210700.11 | MeMED21_1 | 3 | 405 | 135 | 14.69 | 4.4 | −0.37 | Middle |
| Manes.04G136400.1 | MeMED6_1 | 4 | 741 | 247 | 27.51 | 5.18 | −0.378 | Head |
| Manes.05G034300.10 | MeMED12_1 | 5 | 6798 | 2266 | 250.60 | 8.92 | −0.27 | Kinase |
| Manes.05G061600.1 | MeMED28_1 | 5 | 438 | 146 | 17.12 | 5.78 | −0.886 | Head |
| Manes.05G073300.1 | MeMED33_4 | 5 | 3993 | 1331 | 143.11 | 7.22 | 0.185 | Tail |
| Manes.06G049700.1 | MeMED17_1 | 6 | 1998 | 666 | 74.30 | 5.65 | −0.3 | Head |
| Manes.06G129600.1 | MeMED31_1 | 6 | 621 | 207 | 23.70 | 9.28 | −0.637 | Middle |
| Manes.08G002200.1 | MeMED4_1 | 8 | 1239 | 413 | 44.99 | 4.97 | −0.542 | Middle |
| Manes.08G033200.3 | MeMED27_1 | 8 | 1263 | 421 | 46.41 | 8.76 | −0.292 | Tail |
| Manes.08G039900.1 | MeMED7_1 | 8 | 507 | 169 | 19.64 | 9.16 | −0.675 | Middle |
| Manes.08G061400.1 | MeMED26_1 | 8 | 1431 | 477 | 53.66 | 5.56 | −0.948 | Middle |
| Manes.08G118700.1 | MeMED37_1 | 8 | 1983 | 661 | 73.14 | 5.28 | −0.463 | Unknow |
| Manes.09G000400.6 | MeMED21_2 | 9 | 405 | 135 | 14.74 | 4.42 | −0.407 | Middle |
| Manes.09G002800.2 | MeMED26_2 | 9 | 1443 | 481 | 54.11 | 5.94 | −0.844 | Middle |
| Manes.09G101000.3 | MeMED10_1 | 9 | 564 | 188 | 20.41 | 5.23 | −0.384 | Middle |
| Manes.09G152800.6 | MeMED14_1 | 9 | 5478 | 1826 | 197.87 | 7.96 | −0.202 | Middle |
| Manes.11G030300.3 | MeMED6_2 | 11 | 744 | 248 | 27.74 | 5.2 | −0.468 | Head |
| Manes.13G095300.2 | MeMED18_1 | 13 | 690 | 230 | 24.59 | 6.71 | 0.137 | Head |
| Manes.14G098600.1 | MeMED25_1 | 14 | 2511 | 837 | 89.26 | 8.79 | −0.343 | Tail |
| Manes.14G121800.7 | MeMED13_1 | 14 | 5925 | 1975 | 213.55 | 5.64 | −0.208 | Kinase |
| Manes.15G022100.1 | MeMED33_2 | 15 | 3972 | 1324 | 143.98 | 6.29 | 0.185 | Tail |
| Manes.16G001300.1 | MeMED11_1 | 16 | 351 | 117 | 13.26 | 5.78 | −0.521 | Head |
| Manes.17G043400.1 | MeMED16_1 | 17 | 3756 | 1252 | 135.29 | 6.1 | −0.232 | Tail |
| Manes.17G082000.1 | MeMED23_1 | 17 | 4839 | 1613 | 180.90 | 6.64 | −0.08 | Tail |
| Manes.17G099700.2 | MeMED32_1 | 17 | 441 | 147 | 15.40 | 4.69 | −0.098 | Tail |
| Manes.13G050900.1 | MeCycC_1 | 13 | 762 | 254 | 29.76 | 6.54 | −0.098 | Kinase |
| Manes.09G052700.5 | MeCdk8_1 | 9 | 1434 | 478 | 53.00 | 9.24 | −0.491 | Kinase |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, L.; Sun, S.; Zhu, L.; Chen, X.; Xu, R.; Wu, L.; Gu, S. Genome-Wide Identification and Expression Analysis of the Mediator Complex Subunit Gene Family in Cassava. Int. J. Mol. Sci. 2025, 26, 1666. https://doi.org/10.3390/ijms26041666
Zhou L, Sun S, Zhu L, Chen X, Xu R, Wu L, Gu S. Genome-Wide Identification and Expression Analysis of the Mediator Complex Subunit Gene Family in Cassava. International Journal of Molecular Sciences. 2025; 26(4):1666. https://doi.org/10.3390/ijms26041666
Chicago/Turabian StyleZhou, Lingling, Shuhui Sun, Linlong Zhu, Xian Chen, Ran Xu, Lian Wu, and Shuang Gu. 2025. "Genome-Wide Identification and Expression Analysis of the Mediator Complex Subunit Gene Family in Cassava" International Journal of Molecular Sciences 26, no. 4: 1666. https://doi.org/10.3390/ijms26041666
APA StyleZhou, L., Sun, S., Zhu, L., Chen, X., Xu, R., Wu, L., & Gu, S. (2025). Genome-Wide Identification and Expression Analysis of the Mediator Complex Subunit Gene Family in Cassava. International Journal of Molecular Sciences, 26(4), 1666. https://doi.org/10.3390/ijms26041666







