Small Interfering RNAs as Critical Regulators of Plant Life Process: New Perspectives on Regulating the Transcriptomic Machinery
Abstract
:1. Introduction
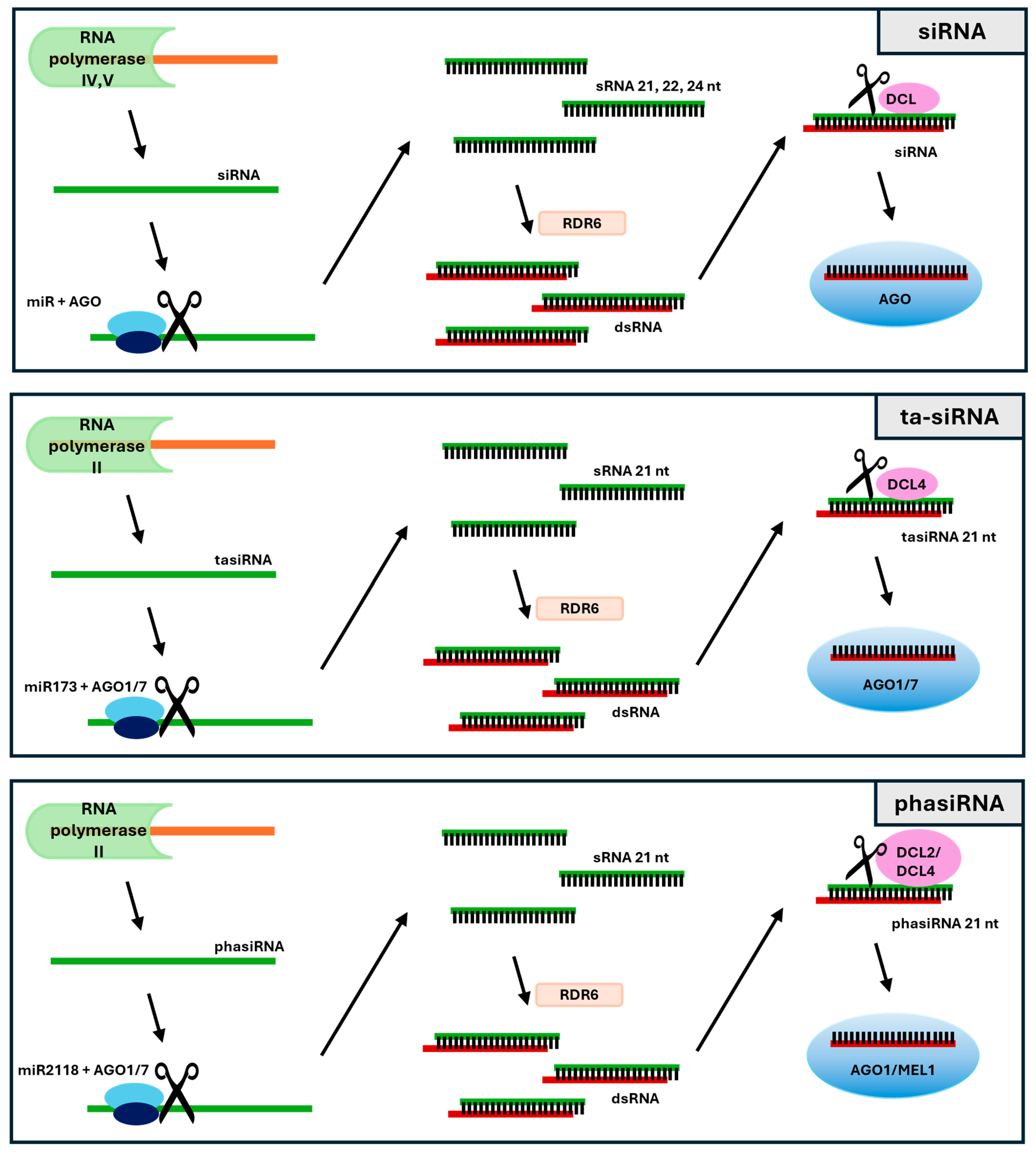
2. Small Interfering RNAs Biogenesis
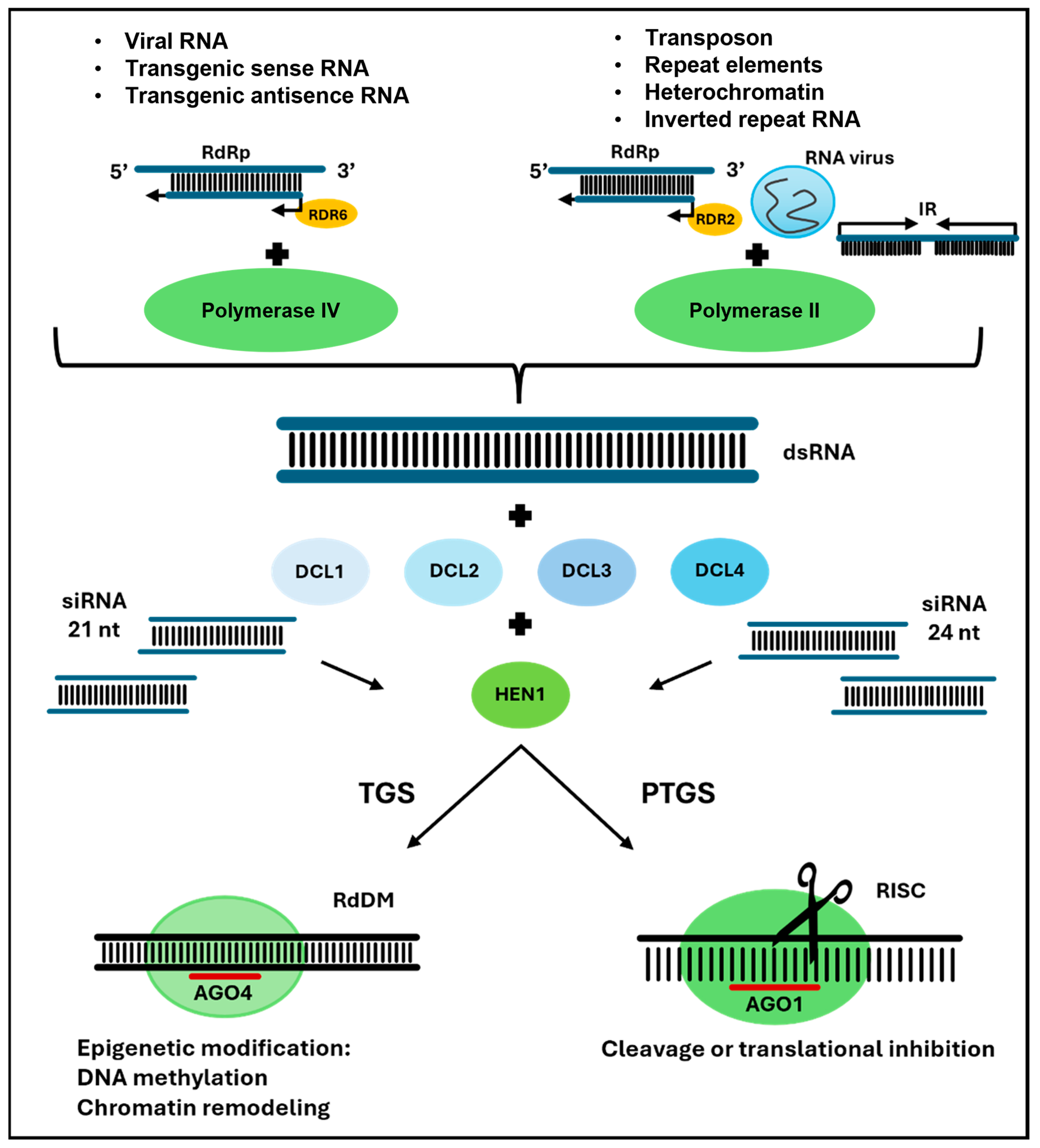
3. Mechanisms of siRNA Activity
4. Plant Development and Growth
5. siRNA Network Mechanisms in the Stress Response
5.1. siRNA-Induced Response to Abiotic Stress
5.2. siRNA-Induced Response to Biotic Stress
5.2.1. siRNAs in Pathogen Infection
5.2.2. siRNAs in Viral Infection
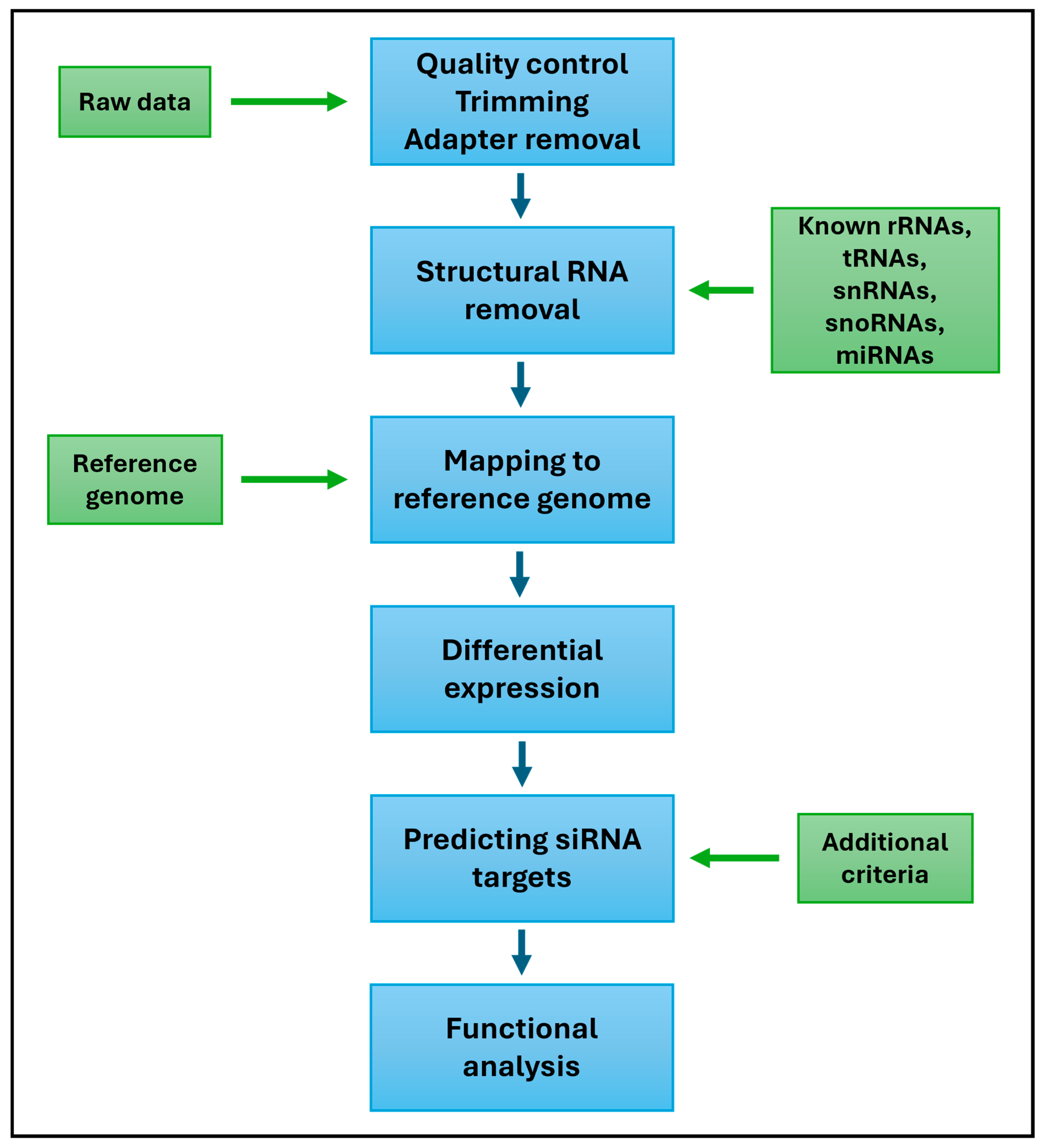
6. Methods of siRNA Identification and Analysis
7. Future Directions in Plant siRNA Research
8. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- An, Y.; Su, H.; Niu, Q.; Yin, S. Integrated Analysis of Coding and Non-coding RNAs Reveals the Molecular Mechanism Underlying Salt Stress Response in Medicago truncatula. Front. Plant Sci. 2022, 13, 891361. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.; Yang, L.; Zhuang, M.; Lv, H.; Wang, Y.; Zhang, Y.; Ji, J. Plant non-coding RNAs: The new frontier for the regulation of plant development and adaptation to stress. Plant Physiol. Biochem. 2024, 208, 108435. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Able, A.J.; Able, J.A. Integrated Analysis of Small RNA, Transcriptome, and Degradome Sequencing Reveals the Water-Deficit and Heat Stress Response Network in Durum Wheat. Int. J. Mol. Sci. 2020, 21, 6017. [Google Scholar] [CrossRef]
- Zhang, P.; Wu, W.; Chen, Q.; Chen, M. Non-Coding RNAs and their Integrated Networks. J. Integr. Bioinform. 2019, 16, 20190027. [Google Scholar] [CrossRef]
- Wang, Y.; Deng, X.W.; Zhu, D. From molecular basics to agronomic benefits: Insights into noncoding RNA-mediated gene regulation in plants. J. Integr. Plant Biol. 2022, 64, 2290–2308. [Google Scholar] [CrossRef]
- Sun, M. Advances in Plant Epigenetic Regulation of Abiotic Stress Response. Theor. Nat. Sci. 2025, 90, 81–87. [Google Scholar] [CrossRef]
- Fei, Q.; Xia, R.; Meyers, B.C. Phased, Secondary, Small Interfering RNAs in Posttranscriptional Regulatory Networks. Plant Cell 2013, 25, 2400–2415. [Google Scholar] [CrossRef]
- Tang, G. siRNA and miRNA: An insight into RISCs. Trends Biochem. Sci. 2005, 30, 106–114. [Google Scholar] [CrossRef]
- Miskiewicz, J.; Tomczyk, K.; Mickiewicz, A.; Sarzynska, J.; Szachniuk, M. Bioinformatics Study of Structural Patterns in Plant MicroRNA Precursors. BioMed Res. Int. 2017, 2017, 6783010. [Google Scholar] [CrossRef]
- Carthew, R.W.; Sontheimer, E.J. Origins and Mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642–655. [Google Scholar] [CrossRef]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.-L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A. Long non-coding RNAs: Definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef] [PubMed]
- Lam, J.K.W.; Chow, M.Y.T.; Zhang, Y.; Leung, S.W.S. siRNA Versus miRNA as Therapeutics for Gene Silencing. Mol. Ther. Nucleic Acids 2015, 4, e252. [Google Scholar] [CrossRef] [PubMed]
- Ranasinghe, P.; Addison, M.L.; Dear, J.W.; Webb, D.J. Small interfering RNA: Discovery, pharmacology and clinical development—An introductory review. Br. J. Pharmacol. 2023, 180, 2697–2720. [Google Scholar] [CrossRef]
- Zhu, J.-K. Reconstituting plant miRNA biogenesis. Proc. Natl. Acad. Sci. USA 2008, 105, 9851–9852. [Google Scholar] [CrossRef]
- Yang, S.; Kim, S.-H.; Yang, E.; Kang, M.; Joo, J.-Y. Molecular insights into regulatory RNAs in the cellular machinery. Exp. Mol. Med. 2024, 56, 1235–1249. [Google Scholar] [CrossRef]
- Filipowicz, W.; Jaskiewicz, L.; Kolb, F.A.; Pillai, R.S. Post-transcriptional gene silencing by siRNAs and miRNAs. Curr. Opin. Struct. Biol. 2005, 15, 331–341. [Google Scholar] [CrossRef]
- Vaucheret, H.; Voinnet, O. The plant siRNA landscape. Plant Cell 2023, 36, 246–275. [Google Scholar] [CrossRef]
- Phillips, J.R.; Dalmay, T.; Bartels, D. The role of small RNAs in abiotic stress. FEBS Lett. 2007, 581, 3592–3597. [Google Scholar] [CrossRef]
- Zhan, J.; Meyers, B.C. Plant Small RNAs: Their Biogenesis, Regulatory Roles, and Functions. Annu. Rev. Plant Biol. 2023, 74, 21–51. [Google Scholar] [CrossRef]
- Bielewicz, D.; Kalak, M.; Kalyna, M.; Windels, D.; Barta, A.; Vazquez, F.; Szweykowska-Kulinska, Z.; Jarmolowski, A. Introns of plant pri-miRNAs enhance miRNA biogenesis. Embo Rep. 2013, 14, 622–628. [Google Scholar] [CrossRef]
- Qin, C.; Li, B.; Fan, Y.; Zhang, X.; Yu, Z.; Ryabov, E.; Zhao, M.; Wang, H.; Shi, N.; Zhang, P.; et al. Roles of Dicer-Like Proteins 2 and 4 in Intra- and Intercellular Antiviral Silencing. Plant Physiol. 2017, 174, 1067–1081. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Xue, Y.; Zhang, L.; Zhong, Z.; Feng, S.; Wang, C.; Xiao, L.; Yang, Z.; Harris, C.J.; Wu, Z.; et al. Mechanism of siRNA production by a plant Dicer-RNA complex in dicing-competent conformation. Science 2021, 374, 1152–1157. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Feng, Y.; Zhu, Z. Dicer-like (DCL) proteins in plants. Funct. Integr. Genom. 2009, 9, 277–286. [Google Scholar] [CrossRef]
- Nielsen, C.P.S.; Arribas-Hernández, L.; Han, L.; Reichel, M.; Woessmann, J.; Daucke, R.; Bressendorff, S.; López-Márquez, D.; Andersen, S.U.; Pumplin, N.; et al. Evidence for an RNAi-independent role of Arabidopsis DICER-LIKE2 in growth inhibition and basal antiviral resistance. Plant Cell 2024, 36, 2289–2309. [Google Scholar] [CrossRef]
- Akbar, S.; Wei, Y.; Zhang, M.-Q. RNA Interference: Promising Approach to Combat Plant Viruses. Int. J. Mol. Sci. 2022, 23, 5312. [Google Scholar] [CrossRef]
- Sanan-Mishra, N.; Jailani, A.A.K.; Mandal, B.; Mukherjee, S.K. Secondary siRNAs in Plants: Biosynthesis, Various Functions, and Applications in Virology. Front. Plant Sci. 2021, 12, 610283. [Google Scholar] [CrossRef]
- Parent, J.; Bouteiller, N.; Elmayan, T.; Vaucheret, H. Respective contributions of Arabidopsis DCL2 and DCL4 to RNA silencing. Plant J. 2014, 81, 223–232. [Google Scholar] [CrossRef]
- Leonetti, P.; Ghasemzadeh, A.; Consiglio, A.; Gursinsky, T.; Behrens, S.; Pantaleo, V. Endogenous activated small interfering RNAs in virus-infected Brassicaceae crops show a common host gene-silencing pattern affecting photosynthesis and stress response. New Phytol. 2020, 229, 1650–1664. [Google Scholar] [CrossRef]
- Mlotshwa, S.; Pruss, G.J.; Peragine, A.; Endres, M.W.; Li, J.; Chen, X.; Poethig, R.S.; Bowman, L.H.; Vance, V. DICER-LIKE2 Plays a Primary Role in Transitive Silencing of Transgenes in Arabidopsis. PLoS ONE 2008, 3, e1755. [Google Scholar] [CrossRef]
- Holoch, D.; Moazed, D. RNA-mediated epigenetic regulation of gene expression. Nat. Rev. Genet. 2015, 16, 71–84. [Google Scholar] [CrossRef]
- Hamar, É.; Szaker, H.M.; Kis, A.; Dalmadi, Á.; Miloro, F.; Szittya, G.; Taller, J.; Gyula, P.; Csorba, T.; Havelda, Z. Genome-Wide Identification of RNA Silencing-Related Genes and Their Expressional Analysis in Response to Heat Stress in Barley (Hordeum vulgare L.). Biomolecules 2020, 10, 929. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Allen, E.; Wilken, A.; Carrington, J.C. DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2005, 102, 12984–12989. [Google Scholar] [CrossRef] [PubMed]
- Kakiyama, S.; Tabara, M.; Nishibori, Y.; Moriyama, H.; Fukuhara, T. Long DCL4-substrate dsRNAs efficiently induce RNA interference in plant cells. Sci. Rep. 2019, 9, 6920. [Google Scholar] [CrossRef]
- Montavon, T.; Kwon, Y.; Zimmermann, A.; Hammann, P.; Vincent, T.; Cognat, V.; Bergdoll, M.; Michel, F.; Dunoyer, P. Characterization of DCL4 missense alleles provides insights into its ability to process distinct classes of dsRNA substrates. Plant J. 2018, 95, 204–218. [Google Scholar] [CrossRef]
- Devers, E.A.; Brosnan, C.A.; Sarazin, A.; Schott, G.; Lim, P.; Lehesranta, S.; Helariutta, Y.; Voinnet, O. In planta dynamics, transport biases, and endogenous functions of mobile siRNAs in Arabidopsis. Plant J. 2023, 115, 1377–1393. [Google Scholar] [CrossRef]
- Chen, X. Small RNAs and Their Roles in Plant Development. Annu. Rev. Cell Dev. Biol. 2009, 25, 21–44. [Google Scholar] [CrossRef]
- Halder, K.; Chaudhuri, A.; Abdin, M.Z.; Datta, A. Tweaking the Small Non-Coding RNAs to Improve Desirable Traits in Plant. Int. J. Mol. Sci. 2023, 24, 3143. [Google Scholar] [CrossRef]
- Allen, E.; Xie, Z.; Gustafson, A.M.; Carrington, J.C. microRNA-Directed Phasing during Trans-Acting siRNA Biogenesis in Plants. Cell 2005, 121, 207–221. [Google Scholar] [CrossRef]
- Kunej, U.; Jakše, J.; Radišek, S.; Štajner, N. Core RNA interference genes involved in miRNA and ta-siRNA biogenesis in hops and their expression analysis after challenging with verticillium nonalfalfae. Int. J. Mol. Sci. 2021, 22, 4224. [Google Scholar] [CrossRef]
- Chen, X.; Rechavi, O. Plant and animal small RNA communications between cells and organisms. Nat. Rev. Mol. Cell Biol. 2021, 23, 185–203. [Google Scholar] [CrossRef]
- Hollick, J.B. Paramutation: A trans-homolog interaction affecting heritable gene regulation. Curr. Opin. Plant Biol. 2012, 15, 536–543. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Yan, X.; Gu, C.; Yuan, X. Biogenesis, Trafficking, and Function of Small RNAs in Plants. Front. Plant Sci. 2022, 13, 825477. [Google Scholar] [CrossRef] [PubMed]
- Pikaard, C.S.; Haag, J.R.; Ream, T.; Wierzbicki, A.T. Roles of RNA polymerase IV in gene silencing. Trends Plant Sci. 2008, 13, 390–397. [Google Scholar] [CrossRef]
- Shabalina, S.; Koonin, E. Origins and evolution of eukaryotic RNA interference. Trends Ecol. Evol. 2008, 23, 578–587. [Google Scholar] [CrossRef]
- Kryovrysanaki, N.; James, A.; Tselika, M.; Bardani, E.; Kalantidis, K. RNA silencing pathways in plant development and defense. Int. J. Dev. Biol. 2022, 66, 163–175. [Google Scholar] [CrossRef]
- Baumann, K. How plants silence stress. Nat. Rev. Mol. Cell Biol. 2020, 21, 303. [Google Scholar] [CrossRef]
- Guleria, P.; Mahajan, M.; Bhardwaj, J.; Yadav, S.K. Plant Small RNAs: Biogenesis, Mode of Action and Their Roles in Abiotic Stresses. Genom. Proteom. Bioinform. 2011, 9, 183–199. [Google Scholar] [CrossRef]
- Dana, H.; Chalbatani, G.M.; Mahmoodzadeh, H.; Gharagouzlo, E.; Karimloo, R.; Rezaiean, O.; Moradzadeh, A.; Mazraeh, A.; Marmari, V.; Rashno, M.M.; et al. Molecular Mechanisms and Biological Functions of siRNA. Int. J. Biomed. Sci. 2017, 13, 48–57. [Google Scholar] [CrossRef]
- Orban, T.I.; Izaurralde, E. Decay of mRNAs targeted by RISC requires XRN1, the Ski complex, and the exosome. RNA 2005, 11, 459–469. [Google Scholar] [CrossRef]
- Tomari, Y.; Zamore, P.D. Perspective: Machines for RNAi. Genes Dev. 2005, 19, 517–529. [Google Scholar] [CrossRef]
- Liu, W.; Shoji, K.; Naganuma, M.; Tomari, Y.; Iwakawa, H.-O. The mechanisms of siRNA selection by plant Argonaute proteins triggering DNA methylation. Nucleic Acids Res. 2022, 50, 12997–13010. [Google Scholar] [CrossRef] [PubMed]
- Wierzbicki, A.T.; Cocklin, R.; Mayampurath, A.; Lister, R.; Rowley, M.J.; Gregory, B.D.; Ecker, J.R.; Tang, H.; Pikaard, C.S. Spatial and functional relationships among Pol V-associated loci, Pol IV-dependent siRNAs, and cytosine methylation in the Arabidopsis epigenome. Genes Dev. 2012, 26, 1825–1836. [Google Scholar] [CrossRef] [PubMed]
- Erdmann, R.M.; Picard, C.L. RNA-directed DNA methylation. PLoS Genet. 2020, 16, e1009034. [Google Scholar] [CrossRef]
- Ashapkin, V.V.; Kutueva, L.I.; Aleksandrushkina, N.I.; Vanyushin, B.F. Epigenetic Regulation of Plant Gametophyte Development. Int. J. Mol. Sci. 2019, 20, 3051. [Google Scholar] [CrossRef]
- Palauqui, J.C.; Elmayan, T.; de Borne, F.D.; Crete, P.; Charles, C.; Vaucheret, H. Frequencies, Timing, and Spatial Patterns of Co-Suppression of Nitrate Reductase and Nitrite Reductase in Transgenic Tobacco Plants. Plant Physiol. 1996, 112, 1447–1456. [Google Scholar] [CrossRef]
- Yoo, B.-C.; Kragler, F.; Varkonyi-Gasic, E.; Haywood, V.; Archer-Evans, S.; Lee, Y.M.; Lough, T.J.; Lucas, W.J. A Systemic Small RNA Signaling System in Plants. Plant Cell 2004, 16, 1979–2000. [Google Scholar] [CrossRef]
- Zhai, J.; Zhang, H.; Arikit, S.; Huang, K.; Nan, G.-L.; Walbot, V.; Meyers, B.C. Spatiotemporally dynamic, cell-type–dependent premeiotic and meiotic phasiRNAs in maize anthers. Proc. Natl. Acad. Sci. USA 2015, 112, 3146–3151. [Google Scholar] [CrossRef]
- Zhang, Y.-C.; Lei, M.-Q.; Zhou, Y.-F.; Yang, Y.-W.; Lian, J.-P.; Feng, Y.-Z.; Zhou, K.-R.; He, R.-R.; He, H.; Zhang, Z.; et al. Reproductive phasiRNAs regulate reprogramming of gene expression and meiotic progression in rice. Nat. Commun. 2020, 11, 6031. [Google Scholar] [CrossRef]
- Chow, H.T.; Mosher, R.A. Small RNA-mediated DNA methylation during plant reproduction. Plant Cell 2023, 35, 1787–1800. [Google Scholar] [CrossRef]
- Wu, H.; Li, B.; Iwakawa, H.-O.; Pan, Y.; Tang, X.; Ling-Hu, Q.; Liu, Y.; Sheng, S.; Feng, L.; Zhang, H.; et al. Plant 22-nt siRNAs mediate translational repression and stress adaptation. Nature 2020, 581, 89–93. [Google Scholar] [CrossRef]
- Wu, J.; Huang, W.; He, Z. Dendrimers as Carriers for siRNA Delivery and Gene Silencing: A Review. Sci. World J. 2013, 2013, 630654. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, S.; Han, S.; Li, X.; Tong, Z.; Qi, L. Deciphering Small Noncoding RNAs during the Transition from Dormant Embryo to Germinated Embryo in Larches (Larix leptolepis). PLoS ONE 2013, 8, e81452. [Google Scholar] [CrossRef] [PubMed]
- Chitwood, D.H.; Nogueira, F.T.; Howell, M.D.; Montgomery, T.A.; Carrington, J.C.; Timmermans, M.C. Pattern formation via small RNA mobility. Genes Dev. 2009, 23, 549–554. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ke, L.; Wu, G.; Xu, Y.; Wu, X.; Xia, R.; Deng, X.; Xu, Q. miR3954 is a trigger of phasiRNAs that affects flowering time in citrus. Plant J. 2017, 92, 263–275. [Google Scholar] [CrossRef] [PubMed]
- Tucker, M.; Okada, T.; Hu, Y.; Scholefield, A.; Taylor, J.M.; Koltunow, A.M.G. Somatic small RNA pathways promote the mitotic events of megagametogenesis during female reproductive development in Arabidopsis. Development 2012, 139, 1399–1404. [Google Scholar] [CrossRef]
- Montgomery, T.A.; Howell, M.D.; Cuperus, J.T.; Li, D.; Hansen, J.E.; Alexander, A.L.; Chapman, E.J.; Fahlgren, N.; Allen, E.; Carrington, J.C. Specificity of ARGONAUTE7-miR390 Interaction and Dual Functionality in TAS3 Trans-Acting siRNA Formation. Cell 2008, 133, 128–141. [Google Scholar] [CrossRef]
- Xia, R.; Xu, J.; Meyers, B.C. The emergence, evolution, and diversification of the miR390-TAS3-ARF pathway in land plants. Plant Cell 2017, 29, 1232–1247. [Google Scholar] [CrossRef]
- Hunter, C.; Willmann, M.R.; Wu, G.; Yoshikawa, M.; de la Luz Gutiérrez-Nava, M.; Poethig, S.R. Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development 2006, 133, 2973–2981. [Google Scholar] [CrossRef]
- Luo, L.; Yang, X.; Guo, M.; Lan, T.; Yu, Y.; Mo, B.; Chen, X.; Gao, L.; Liu, L. TRANS-ACTING SIRNA3-derived short interfering RNAs confer cleavage of mRNAs in rice. Plant Physiol. 2021, 188, 347–362. [Google Scholar] [CrossRef]
- Ellis, C.M.; Nagpal, P.; Young, J.C.; Hagen, G.; Guilfoyle, T.J.; Reed, J.W. AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development 2005, 132, 4563–4574. [Google Scholar] [CrossRef]
- Glazińska, P.; Kulasek, M.; Glinkowski, W.; Wojciechowski, W.; Kosiński, J. Integrated Analysis of Small RNA, Transcriptome and Degradome Sequencing Provides New Insights into Floral Development and Abscission in Yellow Lupine (Lupinus luteus L.). Int. J. Mol. Sci. 2019, 20, 5122. [Google Scholar] [CrossRef] [PubMed]
- Marin, E.; Jouannet, V.; Herz, A.; Lokerse, A.S.; Weijers, D.; Vaucheret, H.; Nussaume, L.; Crespi, M.D.; Maizel, A. miR390, Arabidopsis TAS3 tasiRNAs, and Their AUXIN RESPONSE FACTOR Targets Define an Autoregulatory Network Quantitatively Regulating Lateral Root Growth. Plant Cell 2010, 22, 1104–1117. [Google Scholar] [CrossRef] [PubMed]
- Alhassan, A.R.; Dzinyela, R.; Dargiri, S.A.; Suglo, P.; Yang, L.; Movahedi, A. Regulatory roles of small RNAs in plant growth, breeding, and stress adaptation. Plant Growth Regul. 2025, 1–19. [Google Scholar] [CrossRef]
- Kirolinko, C.; Hobecker, K.; Wen, J.; Mysore, K.S.; Niebel, A.; Blanco, F.A.; Zanetti, M.E. Auxin Response Factor 2 (ARF2), ARF3, and ARF4 Mediate Both Lateral Root and Nitrogen Fixing Nodule Development in Medicago truncatula. Front. Plant Sci. 2021, 12, 659061. [Google Scholar] [CrossRef]
- Molinier, J.; Ries, G.; Zipfel, C.; Hohn, B. Transgeneration memory of stress in plants. Nature 2006, 442, 1046–1049. [Google Scholar] [CrossRef]
- Surdonja, K.; Eggert, K.; Hajirezaei, M.-R.; Harshavardhan, V.T.; Seiler, C.; Von Wirén, N.; Sreenivasulu, N.; Kuhlmann, M. Increase of DNA Methylation at the HvCKX2.1 Promoter by Terminal Drought Stress in Barley. Epigenomes 2017, 1, 9. [Google Scholar] [CrossRef]
- Bélanger, S.; Pokhrel, S.; Czymmek, K.J.; Meyers, B.C. Pre-meiotic, 24-nt reproductive phasiRNAs are abundant in anthers of wheat and barley but not rice and maize. Plant Physiol. 2020, 184, 1407–1423. [Google Scholar] [CrossRef]
- Borsani, O.; Zhu, J.; Verslues, P.E.; Sunkar, R.; Zhu, J.-K. Endogenous siRNAs Derived from a Pair of Natural cis-Antisense Transcripts Regulate Salt Tolerance in Arabidopsis. Cell 2005, 123, 1279–1291. [Google Scholar] [CrossRef]
- Yao, Y.; Ni, Z.; Peng, H.; Sun, F.; Xin, M.; Sunkar, R.; Zhu, J.-K.; Sun, Q. Non-coding small RNAs responsive to abiotic stress in wheat (Triticum aestivum L.). Funct. Integr. Genom. 2010, 10, 187–190. [Google Scholar] [CrossRef]
- Moldovan, D.; Spriggs, A.; Yang, J.; Pogson, B.J.; Dennis, E.S.; Wilson, I.W. Hypoxia-responsive microRNAs and trans-acting small interfering RNAs in Arabidopsis. J. Exp. Bot. 2009, 61, 165–177. [Google Scholar] [CrossRef]
- Liu, Y.; Teng, C.; Xia, R.; Meyers, B.C. PhasiRNAs in Plants: Their Biogenesis, Genic Sources, and Roles in Stress Responses, Development, and Reproduction. Plant Cell 2020, 32, 3059–3080. [Google Scholar] [CrossRef] [PubMed]
- Hilbricht, T.; Varotto, S.; Sgaramella, V.; Bartels, D.; Salamini, F.; Furini, A. Retrotransposons and siRNA have a role in the evolution of desiccation tolerance leading to resurrection of the plant Craterostigma plantagineum. New Phytol. 2008, 179, 877–887. [Google Scholar] [CrossRef]
- Katiyar-Agarwal, S.; Morgan, R.; Dahlbeck, D.; Borsani, O.; Villegas, A.; Zhu, J.-K.; Staskawicz, B.J.; Jin, H. A pathogen-inducible endogenous siRNA in plant immunity. Proc. Natl. Acad. Sci. USA 2006, 103, 18002–18007. [Google Scholar] [CrossRef] [PubMed]
- Hewezi, T.; Howe, P.; Maier, T.R.; Baum, T.J. Arabidopsis Small RNAs and Their Targets During Cyst Nematode Parasitism. Mol. Plant-Microbe Interact. 2008, 21, 1622–1634. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Dodds, P.N.; Chen, J.; Outram, M.A. Pathogen perception and signaling in plant immunity. Plant Cell 2024, 36, 1465–1481. [Google Scholar] [CrossRef]
- Qiao, Y.; Xia, R.; Zhai, J.; Hou, Y.; Feng, L.; Zhai, Y.; Ma, W. Small RNAs in Plant Immunity and Virulence of Filamentous Pathogens. Annu. Rev. Phytopathol. 2021, 59, 265–288. [Google Scholar] [CrossRef]
- Mann, C.W.; Sawyer, A.; Gardiner, D.M.; Mitter, N.; Carroll, B.J.; Eamens, A.L. RNA-based control of fungal pathogens in plants. Int. J. Mol. Sci. 2023, 24, 12391. [Google Scholar] [CrossRef]
- Khraiwesh, B.; Zhu, J.-K.; Zhu, J. Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim. Biophys. Acta 2012, 1819, 137–148. [Google Scholar] [CrossRef]
- Ding, T.; Li, W.; Li, F.; Ren, M.; Wang, W. microRNAs: Key Regulators in Plant Responses to Abiotic and Biotic Stresses via Endogenous and Cross-Kingdom Mechanisms. Int. J. Mol. Sci. 2024, 25, 1154. [Google Scholar] [CrossRef]
- Jyothsna, S.; Alagu, M. Role of phasiRNAs in plant-pathogen interactions: Molecular perspectives and bioinformatics tools. Physiol. Mol. Biol. Plants 2022, 28, 947–961. [Google Scholar] [CrossRef] [PubMed]
- Shahid, S.; Kim, G.; Johnson, N.R.; Wafula, E.; Wang, F.; Coruh, C.; Bernal-Galeano, V.; Phifer, T.; Depamphilis, C.W.; Westwood, J.H.; et al. MicroRNAs from the parasitic plant Cuscuta campestris target host messenger RNAs. Nature 2018, 553, 82–85. [Google Scholar] [CrossRef] [PubMed]
- Ellendorff, U.; Fradin, E.F.; de Jonge, R.; Thomma, B.P.H.J. RNA silencing is required for Arabidopsis defence against Verticillium wilt disease. J. Exp. Bot. 2008, 60, 591–602. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, Q.; Zhang, J.; Wu, L.; Qi, Y.; Zhou, J.-M. Identification of MicroRNAs Involved in Pathogen-Associated Molecular Pattern-Triggered Plant Innate Immunity. Plant Physiol. 2010, 152, 2222–2231. [Google Scholar] [CrossRef]
- Wagh, S.G.; Alam, M.M.; Kobayashi, K.; Yaeno, T.; Yamaoka, N.; Toriba, T.; Hirano, H.-Y.; Nishiguchi, M. Analysis of rice RNA-dependent RNA polymerase 6 (OsRDR6) gene in response to viral, bacterial and fungal pathogens. J. Gen. Plant Pathol. 2015, 82, 12–17. [Google Scholar] [CrossRef]
- Canto-Pastor, A.; Santos, B.A.M.C.; Valli, A.A.; Summers, W.; Schornack, S.; Baulcombe, D.C. Enhanced resistance to bacterial and oomycete pathogens by short tandem target mimic RNAs in tomato. Proc. Natl. Acad. Sci. USA 2019, 116, 2755–2760. [Google Scholar] [CrossRef]
- Boccara, M.; Sarazin, A.; Thiébeauld, O.; Jay, F.; Voinnet, O.; Navarro, L.; Colot, V. The Arabidopsis miR472-RDR6 Silencing Pathway Modulates PAMP- and Effector-Triggered Immunity through the Post-transcriptional Control of Disease Resistance Genes. PLoS Pathog. 2014, 10, e1003883. [Google Scholar] [CrossRef]
- Cai, Q.; Liang, C.; Wang, S.; Hou, Y.; Gao, L.; Liu, L.; He, W.; Ma, W.; Mo, B.; Chen, X. The disease resistance protein SNC1 represses the biogenesis of microRNAs and phased siRNAs. Nat. Commun. 2018, 9, 5080. [Google Scholar] [CrossRef]
- Kong, X.; Yang, M.; Le, B.H.; He, W.; Hou, Y. The master role of siRNAs in plant immunity. Mol. Plant Pathol. 2022, 23, 1565–1574. [Google Scholar] [CrossRef]
- Ding, S.-W.; Voinnet, O. Antiviral Immunity Directed by Small RNAs. Cell 2007, 130, 413–426. [Google Scholar] [CrossRef]
- Weiberg, A.; Jin, H. Small RNAs—The secret agents in the plant–pathogen interactions. Curr. Opin. Plant Biol. 2015, 26, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, A.J.; Baulcombe, D.C. A Species of Small Antisense RNA in Posttranscriptional Gene Silencing in Plants. Science 1999, 286, 950–952. [Google Scholar] [CrossRef] [PubMed]
- Septiani, P.; Pramesti, Y.; Ghildan, M.; Aprilia, K.Z.; Awaludin, R.; Medina, S.; Subandiyah, S.; Meitha, K. RNAi-based biocontrol for crops: A revised expectation for a non-recent technology. Planta 2025, 261, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Soto, A.; Chacón-Cerdas, R. RNAi crop protection advances. Int. J. Mol. Sci. 2021, 22, 12148. [Google Scholar] [CrossRef] [PubMed]
- Bakhat, N.; Jiménez-Sánchez, A.; Ruiz-Jiménez, L.; Padilla-Roji, I.; Velasco, L.; Pérez-García, A.; Fernández-Ortuño, D. Fungal effector genes involved in the suppression of chitin signaling as novel targets for the control of powdery mildew disease via a nontransgenic RNA interference approach. Pest Manag. Sci. 2025. [Google Scholar] [CrossRef]
- Guo, Z.; Li, Y.; Ding, S.-W. Small RNA-based antimicrobial immunity. Nat. Rev. Immunol. 2018, 19, 31–44. [Google Scholar] [CrossRef]
- Brodersen, P.; Voinnet, O. The diversity of RNA silencing pathways in plants. Trend Genet. 2006, 22, 268–280. [Google Scholar] [CrossRef]
- Garcia-Ruiz, H.; Takeda, A.; Chapman, E.J.; Sullivan, C.M.; Fahlgren, N.; Brempelis, K.J.; Carrington, J.C. Arabidopsis RNA-Dependent RNA Polymerases and Dicer-Like Proteins in Antiviral Defense and Small Interfering RNA Biogenesis during Turnip Mosaic Virus Infection. Plant Cell 2010, 22, 481–496. [Google Scholar] [CrossRef]
- Wang, X.-B.; Wu, Q.; Ito, T.; Cillo, F.; Li, W.-X.; Chen, X.; Yu, J.-L.; Ding, S.-W. RNAi-mediated viral immunity requires amplification of virus-derived siRNAs in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2009, 107, 484–489. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, T.; Li, J.; Wu, N.; Wu, G.; Chen, X.; He, L.; Chen, J. Chinese wheat mosaic virus-derived vsiRNA-20 can regulate virus infection in wheat through inhibition of vacuolar- (H+)-PPase induced cell death. New Phytol. 2019, 226, 205–220. [Google Scholar] [CrossRef]
- Liu, P.; Zhang, X.; Zhang, F.; Xu, M.; Ye, Z.; Wang, K.; Liu, S.; Han, X.; Cheng, Y.; Zhong, K.; et al. A virus-derived siRNA activates plant immunity by interfering with ROS scavenging. Mol. Plant 2021, 14, 1088–1103. [Google Scholar] [CrossRef]
- Cao, M.; Du, P.; Wang, X.; Yu, Y.-Q.; Qiu, Y.-H.; Li, W.; Gal-On, A.; Zhou, C.; Li, Y.; Ding, S.-W. Virus infection triggers widespread silencing of host genes by a distinct class of endogenous siRNAs in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 14613–14618. [Google Scholar] [CrossRef]
- Wang, M.; Dean, R.A. Movement of small RNAs in and between plants and fungi. Mol. Plant Pathol. 2020, 21, 589–601. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Pignatta, D.; Bendix, C.; Brunkard, J.O.; Cohn, M.M.; Tung, J.; Sun, H.; Kumar, P.; Baker, B. MicroRNA regulation of plant innate immune receptors. Proc. Natl. Acad. Sci. USA 2012, 109, 1790–1795. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Gomollon, S. Detecting sRNAs by Northern blotting. Methods Mol. Biol. 2011, 732, 25–38. [Google Scholar] [CrossRef] [PubMed]
- Koeppe, S.; Kawchuk, L.; Kalischuk, M. RNA Interference Past and Future Applications in Plants. Int. J. Mol. Sci. 2023, 24, 9755. [Google Scholar] [CrossRef] [PubMed]
- Barrera-Figueroa, B.E.; Gao, L.; Wu, Z.; Zhou, X.; Zhu, J.; Jin, H.; Liu, R.; Zhu, J.-K. High throughput sequencing reveals novel and abiotic stress-regulated microRNAs in the inflorescences of rice. BMC Plant Biol. 2012, 12, 132. [Google Scholar] [CrossRef]
- Shivaprasad, P.V.; Chen, H.-M.; Patel, K.; Bond, D.M.; Santos, B.A.; Baulcombe, D.C. A MicroRNA Superfamily Regulates Nucleotide Binding Site–Leucine-Rich Repeats and Other mRNAs. Plant Cell 2012, 24, 859–874. [Google Scholar] [CrossRef]
- Morgado, L.; Johannes, F. Computational tools for plant small RNA detection and categorization. Briefings Bioinform. 2017, 20, 1181–1192. [Google Scholar] [CrossRef]
- Thiebaut, F.; A Rojas, C.; Grativol, C.; Motta, M.R.; Vieira, T.; Regulski, M.; Martienssen, R.A.; Farinelli, L.; Hemerly, A.S.; Ferreira, P.C. Genome-wide identification of microRNA and siRNA responsive to endophytic beneficial diazotrophic bacteria in maize. BMC Genom. 2014, 15, 776. [Google Scholar] [CrossRef]
- Medina, C.; da Rocha, M.; Magliano, M.; Raptopoulo, A.; Marteu, N.; Lebrigand, K.; Abad, P.; Favery, B.; Jaubert-Possamai, S. Characterization of siRNAs clusters in Arabidopsis thaliana galls induced by the root-knot nematode Meloidogyne incognita. BMC Genom. 2018, 19, 943. [Google Scholar] [CrossRef]
- Ge, F.; Huang, X.; Hu, H.; Zhang, Y.; Li, Z.; Zou, C.; Peng, H.; Li, L.; Gao, S.; Pan, G.; et al. Endogenous small interfering RNAs associated with maize embryonic callus formation. PLoS ONE 2017, 12, e0180567. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Xia, Y.; Li, R.; Zhang, H.; Siddique, K.H.; Guo, P. Identification and Analysis of Small Interfering RNAs Associated With Heat Stress in Flowering Chinese Cabbage Using High-Throughput Sequencing. Front. Genet. 2021, 12, 746816. [Google Scholar] [CrossRef]
- Yu, D.; Meng, Y.; Zuo, Z.; Xue, J.; Wang, H. NATpipe: An integrative pipeline for systematical discovery of natural antisense transcripts (NATs) and phase-distributed nat-siRNAs from de novo assembled transcriptomes. Sci. Rep. 2016, 6, 21666. [Google Scholar] [CrossRef]
- Lei, P.; Qi, N.; Yan, J.; Zhu, X.; Liu, X.; Xuan, Y.; Fan, H.; Chen, L.; Duan, Y.; Wang, Y. Genome-wide identification of small interfering RNAs from sRNA libraries constructed from soybean cyst nematode resistant and susceptible cultivars. Gene 2022, 832, 146557. [Google Scholar] [CrossRef]
- Guo, Q.; Qu, X.; Jin, W. PhaseTank: Genome-wide computational identification of phasiRNAs and their regulatory cascades. Bioinformatics 2014, 31, 284–286. [Google Scholar] [CrossRef]
- Fu, K.; Wu, Q.; Jiang, N.; Hu, S.; Ye, H.; Hu, Y.; Li, L.; Li, T.; Sun, Z. Identification and Expressional Analysis of siRNAs Responsive to Fusarium graminearum Infection in Wheat. Int. J. Mol. Sci. 2023, 24, 16005. [Google Scholar] [CrossRef]
- Xu, W.-B.; Cao, F.; Liu, P.; Yan, K.; Guo, Q.-H. The multifaceted role of RNA-based regulation in plant stress memory. Front. Plant Sci. 2024, 15, 1387575. [Google Scholar] [CrossRef]
- Mierziak, J.; Wojtasik, W. Epigenetic weapons of plants against fungal pathogens. BMC Plant Biol. 2024, 24, 1–23. [Google Scholar] [CrossRef]
- Jain, H.; Kaur, R.; Sain, S.K.; Siwach, P. Development, Design, and Application of Efficient siRNAs Against Cotton Leaf Curl Virus-Betasatellite Complex to Mediate Resistance Against Cotton Leaf Curl Disease. Indian J. Microbiol. 2024, 64, 558–571. [Google Scholar] [CrossRef]
- Ali, A.; Shahbaz, M.; Ölmez, F.; Fatima, N.; Umar, U.U.D.; Ali, A.; Akram, M.; Seelan, J.S.S.; Baloch, F.S. RNA interference: A promising biotechnological approach to combat plant pathogens, mechanism and future prospects. World J. Microbiol. Biotechnol. 2024, 40, 339. [Google Scholar] [CrossRef]
- Deng, P.; Muhammad, S.; Cao, M.; Wu, L. Biogenesis and regulatory hierarchy of phased small interfering RNAs in plants. Plant Biotechnol. J. 2018, 16, 965–975. [Google Scholar] [CrossRef] [PubMed]
- Curaba, J.; Singh, M.B.; Bhalla, P.L. miRNAs in the crosstalk between phytohormone signalling pathways. J. Exp. Bot. 2014, 65, 1425–1438. [Google Scholar] [CrossRef] [PubMed]


| Category | siRNAs | miRNAs |
|---|---|---|
| Origins | Encoded by transposons, virus, heterochromatin | Distinct genomic loci encoded by their genes |
| Biogenesis, precursor | Long bimolecular RNA | sRNA molecules that include an imperfect stem-loop secondary structure |
| Processing enzyme | Dicer-like enzymes | |
| Accessory proteins | Different cofactors depending on the siRNA class, e.g., RDRs | SE and HYL1 proteins |
| Length | 21, 23, 24 nt | 18–24 nt |
| Mechanism of action | Associate with RISC to mediate mRNA TGS and PTGS mechanisms | Mediate the silencing of the same genes from which they originate |
| Working principle | Function through RISC by complementarity targeting mRNA | |
| Complementarity to targets | Fully complementary to mRNA | Partially complementary to the mRNA targets |
| Sequence conservation | Low conservation, depending on the dsRNA source | Highly conserved across plant species |
| Epigenetic effects | Induce TGS via DNA methylation | Typically absent |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Puchta-Jasińska, M.; Bolc, P.; Pietrusińska-Radzio, A.; Motor, A.; Boczkowska, M. Small Interfering RNAs as Critical Regulators of Plant Life Process: New Perspectives on Regulating the Transcriptomic Machinery. Int. J. Mol. Sci. 2025, 26, 1624. https://doi.org/10.3390/ijms26041624
Puchta-Jasińska M, Bolc P, Pietrusińska-Radzio A, Motor A, Boczkowska M. Small Interfering RNAs as Critical Regulators of Plant Life Process: New Perspectives on Regulating the Transcriptomic Machinery. International Journal of Molecular Sciences. 2025; 26(4):1624. https://doi.org/10.3390/ijms26041624
Chicago/Turabian StylePuchta-Jasińska, Marta, Paulina Bolc, Aleksandra Pietrusińska-Radzio, Adrian Motor, and Maja Boczkowska. 2025. "Small Interfering RNAs as Critical Regulators of Plant Life Process: New Perspectives on Regulating the Transcriptomic Machinery" International Journal of Molecular Sciences 26, no. 4: 1624. https://doi.org/10.3390/ijms26041624
APA StylePuchta-Jasińska, M., Bolc, P., Pietrusińska-Radzio, A., Motor, A., & Boczkowska, M. (2025). Small Interfering RNAs as Critical Regulators of Plant Life Process: New Perspectives on Regulating the Transcriptomic Machinery. International Journal of Molecular Sciences, 26(4), 1624. https://doi.org/10.3390/ijms26041624









