GDAP1 Is Dysregulated at DNA Methylation and H3K4me3 Levels in Alcohol Use Disorder
Abstract
1. Introduction
2. Results
2.1. Initial Analysis Revealed Altered Molecular Markers in AUD Patients, Independent of Sex, Age, or Smoking Behavior
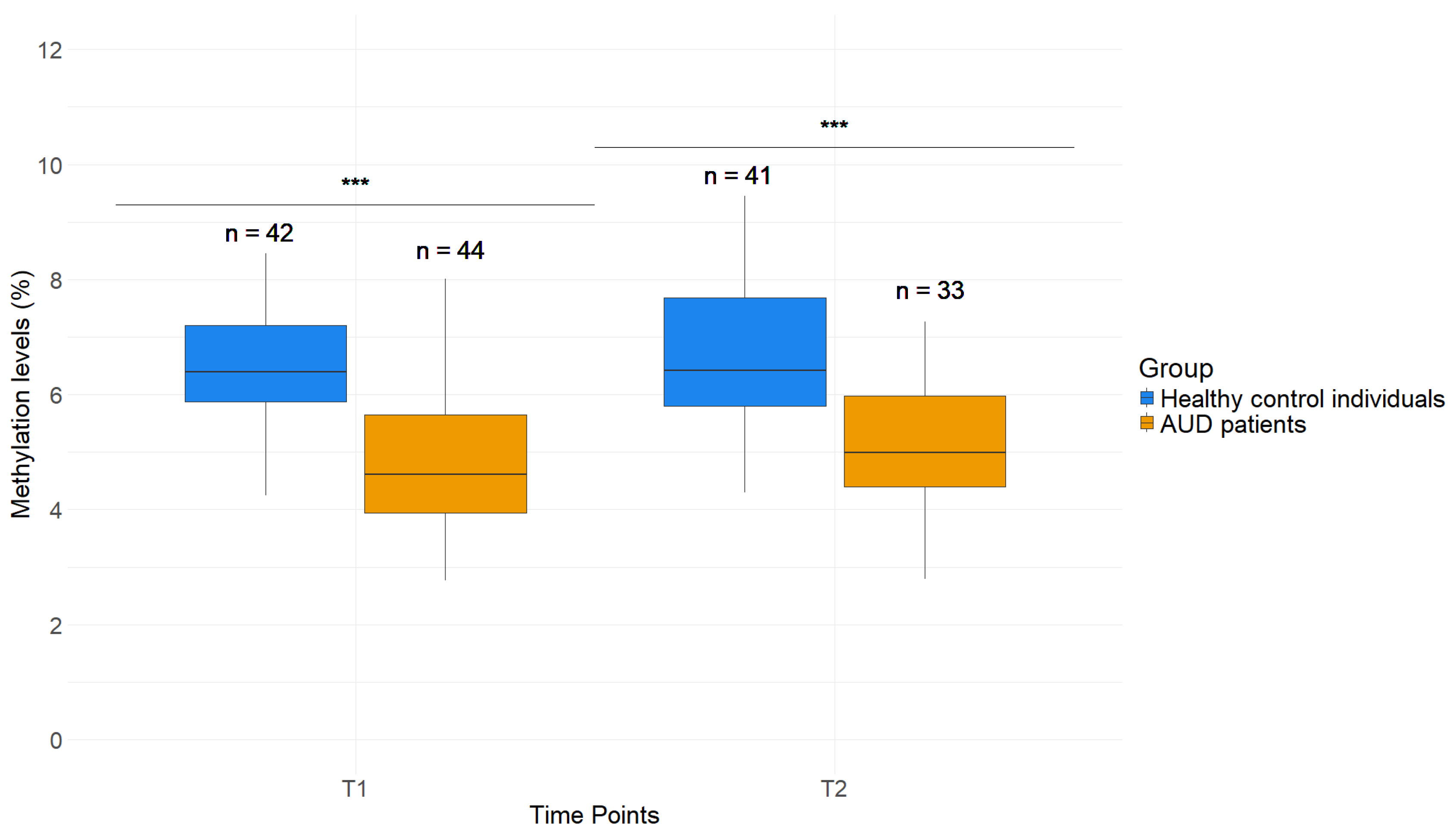
2.2. Lower DNAm Levels in GDAP1 Promoter in Whole Blood Samples of AUD Patients Compared to Healthy Control Individuals
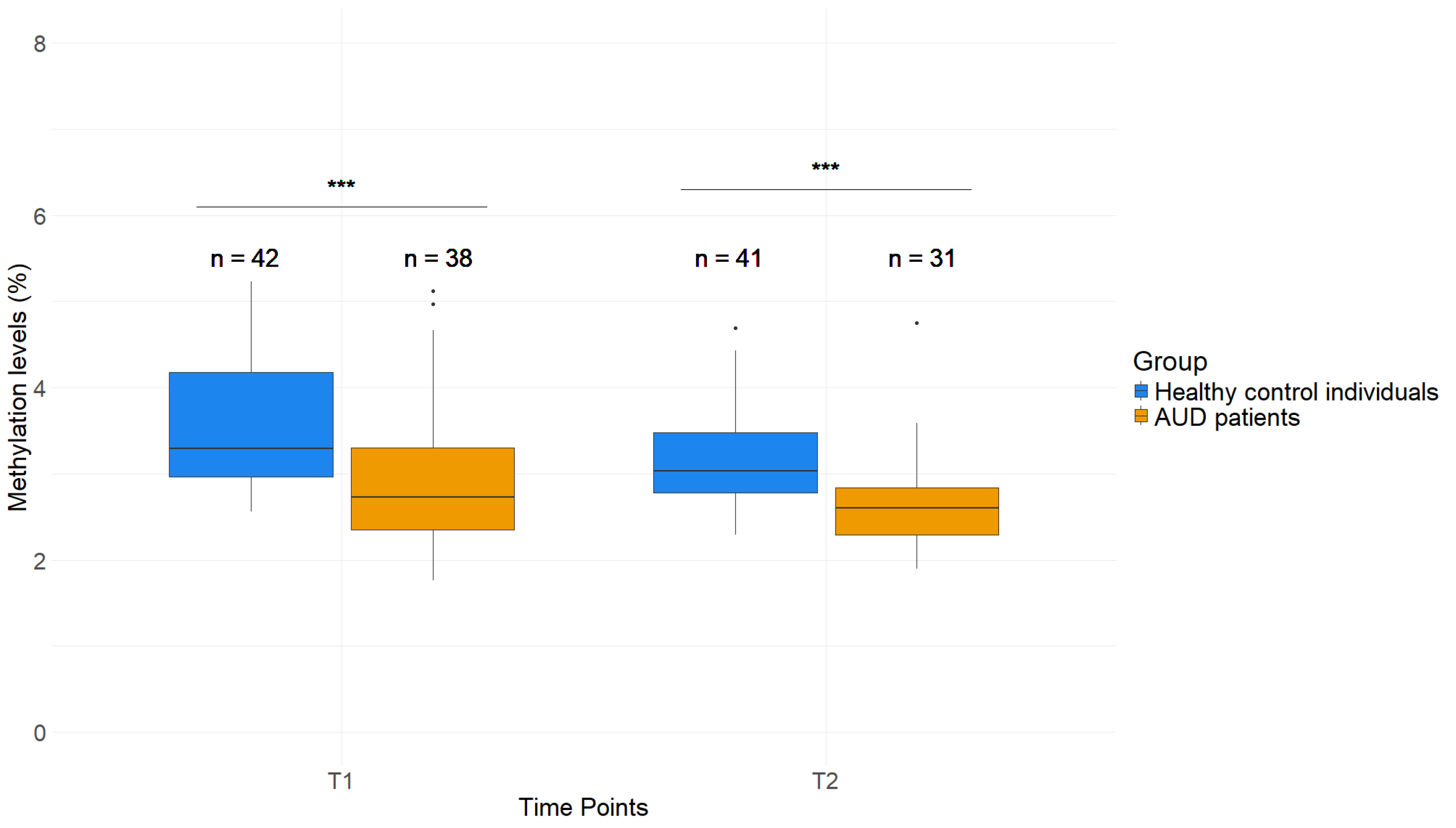
2.3. GDAP1 Promoters Display Lower DNAm Levels in Saliva of AUD Patients Compared to Healthy Control Individuals
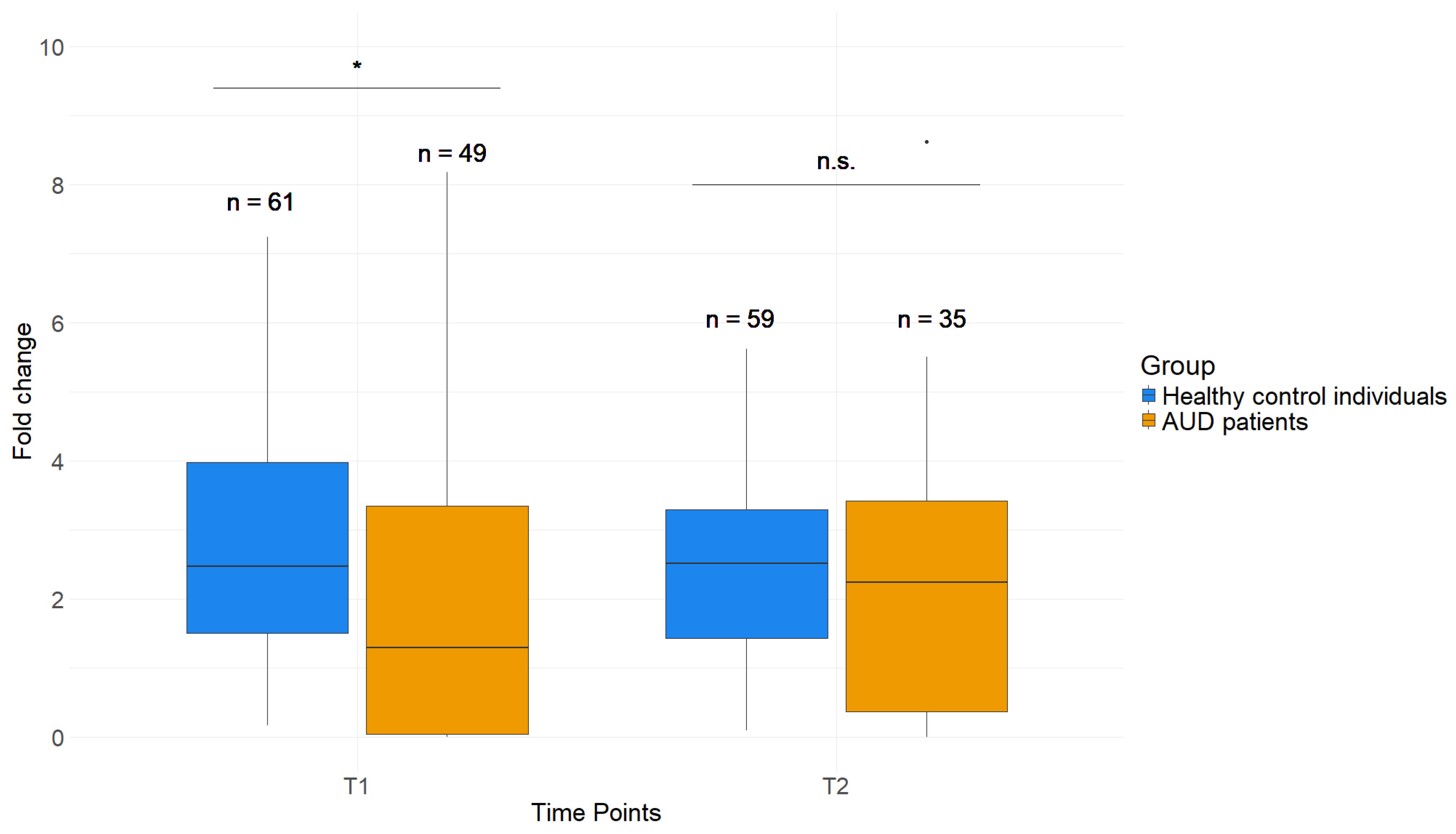
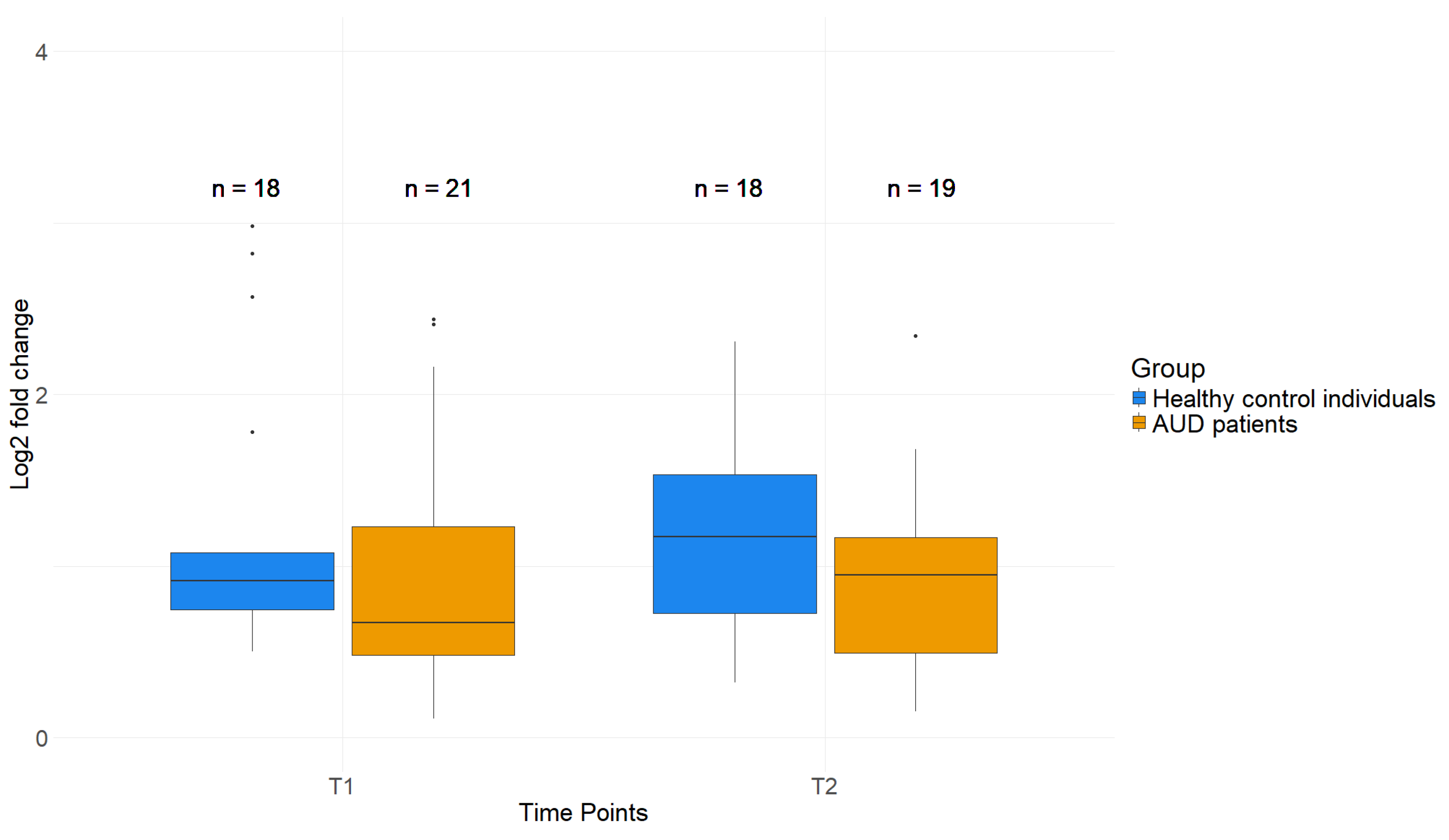
2.4. Reduced H3K4me3 Levels near TSS of AUD Patient PBMCs Compared to Healthy Control Individuals
2.5. Effects of AUD Therapy on H3K4me3 as Well as DNAm of Whole Blood and Saliva in GDAP1
2.6. No Difference in Gene Expression Levels of GDAP1 in Whole Blood of AUD Patients Compared to Healthy Control Individuals
3. Discussion
4. Materials and Methods
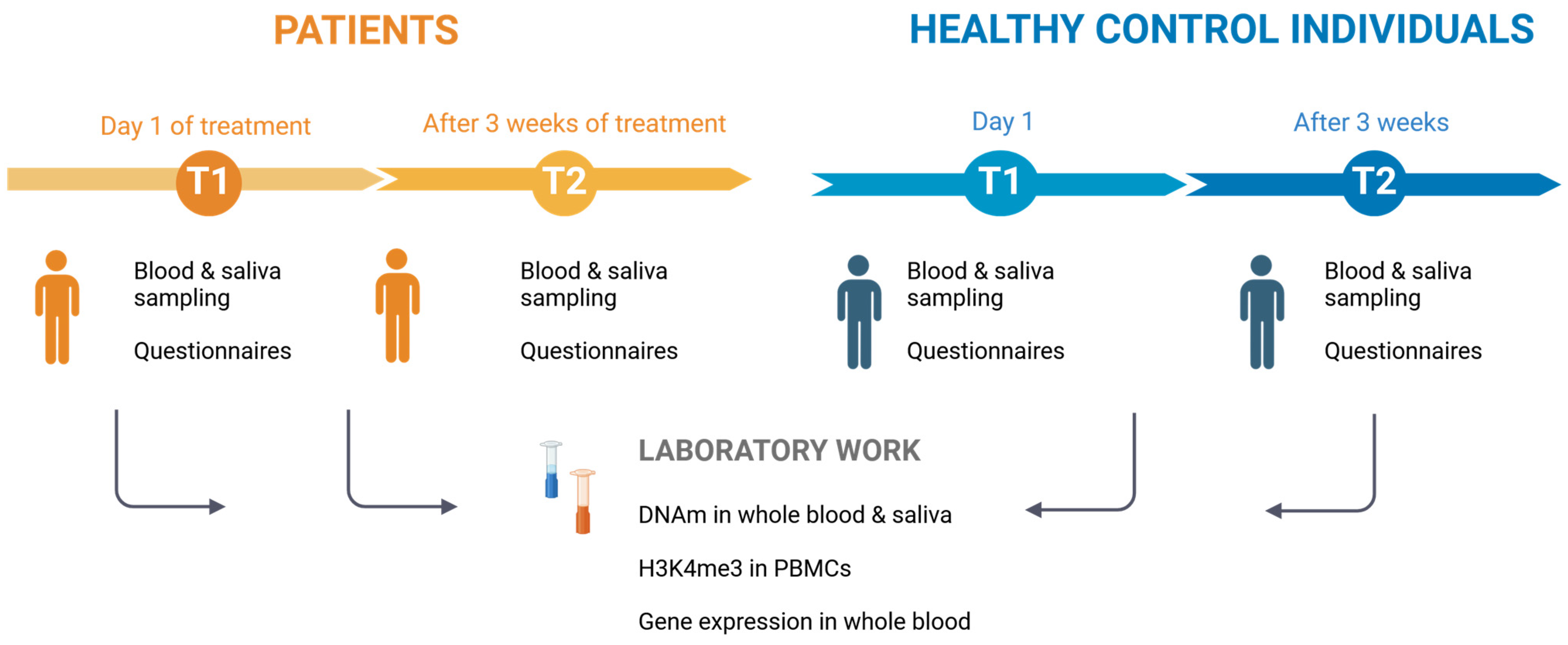
4.1. Study Sample
4.2. DNA-Methylation Analysis in Whole Blood and Saliva
4.3. H3K4-Trimethylation Analysis in Peripheral Blood Mononuclear Cells (PBMCs)
4.4. Gene Expression
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lim, S.S.; Vos, T.; Flaxman, A.D.; Danaei, G.; Shibuya, K.; Adair-Rohani, H.; Amann, M.; Anderson, H.R.; Andrews, K.G.; Aryee, M.; et al. A comparative risk assessment of burden of disease and injury attributable to 67 risk factors and risk factor clusters in 21 regions, 1990–2010: A systematic analysis for the Global Burden of Disease Study 2010. Lancet 2012, 380, 2224–2260. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Global Status Report on Alcohol and Health 2018; Vladimir Poznyak, D.R., Ed.; Alcohol, Drugs and Addictive Behaviors (ADA): Geneva, Switzerland, 2018; p. 450. ISBN 978-92-4-156563-9. [Google Scholar]
- World Health Organization (WHO). Global Status Report on Alcohol and Health and Treatment of Substance Use Disorders; World Health Organization: Geneva, Switzerland, 2024. [Google Scholar]
- Deutsches Institut für Medizinische Dokumentation und Information (DIMDI) im Auftrag des Bundesministeriums für Gesundheit (BMG) unter Beteiligung der Arbeitsgruppe ICD des Kuratoriums für Fragen der Klassifikation im Gesundheitswesen (KKG). Internationale statistische Klassifikation der Krankheiten und 524 verwandter Gesundheitsprobleme, 10. Revision, German Modification; KKG: Köln, Germany, 2023. (In German) [Google Scholar]
- Karg, K.; Sen, S. Gene x environment interaction models in psychiatric genetics. Curr. Top. Behav. Neurosci. 2012, 12, 441–462. [Google Scholar] [CrossRef]
- Liu, L.; Li, Y.; Tollefsbol, T.O. Gene-environment interactions and epigenetic basis of human diseases. Curr. Issues Mol. Biol. 2008, 10, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, D.W. Alcoholism and heredity. A review and hypothesis. Arch. Gen. Psychiatry 1979, 36, 57–61. [Google Scholar] [CrossRef]
- Liu, I.C.; Blacker, D.L.; Xu, R.; Fitzmaurice, G.; Lyons, M.J.; Tsuang, M.T. Genetic and environmental contributions to the development of alcohol dependence in male twins. Arch. Gen. Psychiatry 2004, 61, 897–903. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kendler, K.S.; Neale, M.C.; Heath, A.C.; Kessler, R.C.; Eaves, L.J. A twin-family study of alcoholism in women. Am. J. Psychiatry 1994, 151, 707–715. [Google Scholar] [CrossRef]
- Prescott, C.A.; Kendler, K.S. Genetic and environmental contributions to alcohol abuse and dependence in a population-based sample of male twins. Am. J. Psychiatry 1999, 156, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Jirtle, R. Epigenetics: How Genes and Environment Interact—Randy Jirtle. Ph.D. Thesis, Duke University, Durham, NC, USA, 2012. [Google Scholar]
- Weinhold, B. Epigenetics: The science of change. Environ. Health Perspect. 2006, 114, A160–A167. [Google Scholar] [CrossRef] [PubMed]
- Gordon, L.; Joo, J.E.; Powell, J.E.; Ollikainen, M.; Novakovic, B.; Li, X.; Andronikos, R.; Cruickshank, M.N.; Conneely, K.N.; Smith, A.K.; et al. Neonatal DNA methylation profile in human twins is specified by a complex interplay between intrauterine environmental and genetic factors, subject to tissue-specific influence. Genome Res. 2012, 22, 1395–1406. [Google Scholar] [CrossRef] [PubMed]
- Hoang, T.T.; Lee, Y.; McCartney, D.L.; Kersten, E.T.G.; Page, C.M.; Hulls, P.M.; Lee, M.; Walker, R.M.; Breeze, C.E.; Bennett, B.D.; et al. Comprehensive evaluation of smoking exposures and their interactions on DNA methylation. EBioMedicine 2024, 100, 104956. [Google Scholar] [CrossRef] [PubMed]
- Jin, B.; Li, Y.; Robertson, K.D. DNA methylation: Superior or subordinate in the epigenetic hierarchy? Genes. Cancer 2011, 2, 607–617. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.S.; Shin, W.J.; Lee, J.E.; Do, J.T. CpG and Non-CpG Methylation in Epigenetic Gene Regulation and Brain Function. Genes 2017, 8, 148. [Google Scholar] [CrossRef] [PubMed]
- Deaton, A.M.; Bird, A. CpG islands and the regulation of transcription. Genes. Dev. 2011, 25, 1010–1022. [Google Scholar] [CrossRef]
- Greer, E.L.; Shi, Y. Histone methylation: A dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 2012, 13, 343–357. [Google Scholar] [CrossRef]
- Eissenberg, J.C.; Shilatifard, A. Histone H3 lysine 4 (H3K4) methylation in development and differentiation. Dev. Biol. 2010, 339, 240–249. [Google Scholar] [CrossRef]
- Shilatifard, A. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr. Opin. Cell Biol. 2008, 20, 341–348. [Google Scholar] [CrossRef]
- Longley, M.J.; Lee, J.; Jung, J.; Lohoff, F.W. Epigenetics of alcohol use disorder-A review of recent advances in DNA methylation profiling. Addict. Biol. 2021, 26, e13006. [Google Scholar] [CrossRef] [PubMed]
- Philibert, R.A.; Penaluna, B.; White, T.; Shires, S.; Gunter, T.; Liesveld, J.; Erwin, C.; Hollenbeck, N.; Osborn, T. A pilot examination of the genome-wide DNA methylation signatures of subjects entering and exiting short-term alcohol dependence treatment programs. Epigenetics 2014, 9, 1212–1219. [Google Scholar] [CrossRef] [PubMed]
- Bruckmann, C.; Di Santo, A.; Karle, K.N.; Batra, A.; Nieratschker, V. Validation of differential GDAP1 DNA methylation in alcohol dependence and its potential function as a biomarker for disease severity and therapy outcome. Epigenetics 2016, 11, 456–463. [Google Scholar] [CrossRef]
- Atlas, T.H.P. GDAP1. 2024. Available online: https://www.proteinatlas.org/ENSG00000104381-GDAP1/tissue (accessed on 23 February 2024).
- Cantarero, L.; Juarez-Escoto, E.; Civera-Tregon, A.; Rodriguez-Sanz, M.; Roldan, M.; Benitez, R.; Hoenicka, J.; Palau, F. Mitochondria-lysosome membrane contacts are defective in GDAP1-related Charcot-Marie-Tooth disease. Hum. Mol. Genet. 2021, 29, 3589–3605. [Google Scholar] [CrossRef]
- Marco, A.; Cuesta, A.; Pedrola, L.; Palau, F.; Marin, I. Evolutionary and structural analyses of GDAP1, involved in Charcot-Marie-Tooth disease, characterize a novel class of glutathione transferase-related genes. Mol. Biol. Evol. 2004, 21, 176–187. [Google Scholar] [CrossRef]
- NiH. Charcot-Marie-Tooth Disease. 2022. Available online: https://www.ninds.nih.gov/health-information/disorders/charcot-marie-tooth-disease (accessed on 1 December 2024).
- Baxter, R.V.; Ben Othmane, K.; Rochelle, J.M.; Stajich, J.E.; Hulette, C.; Dew-Knight, S.; Hentati, F.; Ben Hamida, M.; Bel, S.; Stenger, J.E.; et al. Ganglioside-induced differentiation-associated protein-1 is mutant in Charcot-Marie-Tooth disease type 4A/8q21. Nat. Genet. 2002, 30, 21–22. [Google Scholar] [CrossRef] [PubMed]
- Rzepnikowska, W.; Kochanski, A. A role for the GDAP1 gene in the molecular pathogenesis of Charcot-Marie-Tooth disease. Acta Neurobiol. Exp. (Wars) 2018, 78, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Nieratschker, V. Created in BioRender. Available online: https://BioRender.com/h20l580 (accessed on 23 December 2024).
- Balasubramanian, D.; Akhtar-Zaidi, B.; Song, L.; Bartels, C.F.; Veigl, M.; Beard, L.; Myeroff, L.; Guda, K.; Lutterbaugh, J.; Willis, J.; et al. H3K4me3 inversely correlates with DNA methylation at a large class of non-CpG-island-containing start sites. Genome Med. 2012, 4, 47. [Google Scholar] [CrossRef] [PubMed]
- Weigert, R.; Hetzel, S.; Bailly, N.; Haggerty, C.; Ilik, I.A.; Yung, P.Y.K.; Navarro, C.; Bolondi, A.; Kumar, A.S.; Anania, C.; et al. Dynamic antagonism between key repressive pathways maintains the placental epigenome. Nat. Cell Biol. 2023, 25, 579–591. [Google Scholar] [CrossRef] [PubMed]
- Sharifi-Zarchi, A.; Gerovska, D.; Adachi, K.; Totonchi, M.; Pezeshk, H.; Taft, R.J.; Scholer, H.R.; Chitsaz, H.; Sadeghi, M.; Baharvand, H.; et al. DNA methylation regulates discrimination of enhancers from promoters through a H3K4me1-H3K4me3 seesaw mechanism. BMC Genom. 2017, 18, 964. [Google Scholar] [CrossRef] [PubMed]
- Cruz, C.; Della Rosa, M.; Krueger, C.; Gao, Q.; Horkai, D.; King, M.; Field, L.; Houseley, J. Tri-methylation of histone H3 lysine 4 facilitates gene expression in ageing cells. Elife 2018, 7, e34081. [Google Scholar] [CrossRef]
- Wang, P.; Lin, C.; Smith, E.R.; Guo, H.; Sanderson, B.W.; Wu, M.; Gogol, M.; Alexander, T.; Seidel, C.; Wiedemann, L.M.; et al. Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II. Mol. Cell Biol. 2009, 29, 6074–6085. [Google Scholar] [CrossRef]
- LaMere, S.A.; Thompson, R.C.; Komori, H.K.; Mark, A.; Salomon, D.R. Promoter H3K4 methylation dynamically reinforces activation-induced pathways in human CD4 T cells. Genes. Immun. 2016, 17, 283–297. [Google Scholar] [CrossRef] [PubMed]
- Santos-Rosa, H.; Schneider, R.; Bannister, A.J.; Sherriff, J.; Bernstein, B.E.; Emre, N.C.; Schreiber, S.L.; Mellor, J.; Kouzarides, T. Active genes are tri-methylated at K4 of histone H3. Nature 2002, 419, 407–411. [Google Scholar] [CrossRef] [PubMed]
- Ninova, M.; Fejes Toth, K.; Aravin, A.A. The control of gene expression and cell identity by H3K9 trimethylation. Development 2019, 146, dev181180. [Google Scholar] [CrossRef]
- Cai, Y.; Zhang, Y.; Loh, Y.P.; Tng, J.Q.; Lim, M.C.; Cao, Z.; Raju, A.; Lieberman Aiden, E.; Li, S.; Manikandan, L.; et al. H3K27me3-rich genomic regions can function as silencers to repress gene expression via chromatin interactions. Nat. Commun. 2021, 12, 719. [Google Scholar] [CrossRef] [PubMed]
- Bu, G.; Zhu, W.; Liu, X.; Zhang, J.; Yu, L.; Zhou, K.; Wang, S.; Li, Z.; Fan, Z.; Wang, T.; et al. Coordination of zygotic genome activation entry and exit by H3K4me3 and H3K27me3 in porcine early embryos. Genome Res. 2022, 32, 1487–1501. [Google Scholar] [CrossRef] [PubMed]
- de Mendoza, A.; Nguyen, T.V.; Ford, E.; Poppe, D.; Buckberry, S.; Pflueger, J.; Grimmer, M.R.; Stolzenburg, S.; Bogdanovic, O.; Oshlack, A.; et al. Large-scale manipulation of promoter DNA methylation reveals context-specific transcriptional responses and stability. Genome Biol. 2022, 23, 163. [Google Scholar] [CrossRef] [PubMed]
- Crews, F.T.; Coleman, L.G., Jr.; Macht, V.A.; Vetreno, R.P. Alcohol, HMGB1, and Innate Immune Signaling in the Brain. Alcohol. Res. 2024, 44, 04. [Google Scholar] [CrossRef] [PubMed]
- Bruckmann, C.; Islam, S.A.; MacIsaac, J.L.; Morin, A.M.; Karle, K.N.; Di Santo, A.; Wust, R.; Lang, I.; Batra, A.; Kobor, M.S.; et al. DNA methylation signatures of chronic alcohol dependence in purified CD3(+) T-cells of patients undergoing alcohol treatment. Sci. Rep. 2017, 7, 6605. [Google Scholar] [CrossRef] [PubMed]
- Schlaepfer, I.R.; Hoft, N.R.; Ehringer, M.A. The genetic components of alcohol and nicotine co-addiction: From genes to behavior. Curr. Drug Abuse Rev. 2008, 1, 124–134. [Google Scholar] [CrossRef]
- True, W.R.; Xian, H.; Scherrer, J.F.; Madden, P.A.; Bucholz, K.K.; Heath, A.C.; Eisen, S.A.; Lyons, M.J.; Goldberg, J.; Tsuang, M. Common genetic vulnerability for nicotine and alcohol dependence in men. Arch. Gen. Psychiatry 1999, 56, 655–661. [Google Scholar] [CrossRef]
- Swan, G.E.; Carmelli, D.; Cardon, L.R. The consumption of tobacco, alcohol, and coffee in Caucasian male twins: A multivariate genetic analysis. J. Subst. Abuse 1996, 8, 19–31. [Google Scholar] [CrossRef] [PubMed]
- Mundorf, A.; Rommel, S.; Verheyen, M.; Mergia, E.; Peters, M.; Freund, N. Cigarette smoke exposure has region-specific effects on GDAP1 expression in mouse hippocampus. Psychiatry Res. 2020, 289, 112979. [Google Scholar] [CrossRef] [PubMed]
- Benayoun, B.A.; Pollina, E.A.; Ucar, D.; Mahmoudi, S.; Karra, K.; Wong, E.D.; Devarajan, K.; Daugherty, A.C.; Kundaje, A.B.; Mancini, E.; et al. H3K4me3 breadth is linked to cell identity and transcriptional consistency. Cell 2014, 158, 673–688. [Google Scholar] [CrossRef] [PubMed]
- Klein, J.A.; Ackerman, S.L. Oxidative stress, cell cycle, and neurodegeneration. J. Clin. Investig. 2003, 111, 785–793. [Google Scholar] [CrossRef] [PubMed]
- GeneCards. GDAP1 Gene—Ganglioside Induced Differentiation Associated Protein 1. 2024. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=GDAP1 (accessed on 12 January 2024).
- Li, J.; O, W.; Li, W.; Jiang, Z.G.; Ghanbari, H.A. Oxidative stress and neurodegenerative disorders. Int. J. Mol. Sci. 2013, 14, 24438–24475. [Google Scholar] [CrossRef] [PubMed]
- Chopra, K.; Tiwari, V. Alcoholic neuropathy: Possible mechanisms and future treatment possibilities. Br. J. Clin. Pharmacol. 2012, 73, 348–362. [Google Scholar] [CrossRef]
- Julian, T.; Glascow, N.; Syeed, R.; Zis, P. Alcohol-related peripheral neuropathy: A systematic review and meta-analysis. J. Neurol. 2019, 266, 2907–2919. [Google Scholar] [CrossRef] [PubMed]
- Hawley, R.J.; Kurtzke, J.F.; Armbrustmacher, V.W.; Saini, N.; Manz, H. The course of alcoholic-nutritional peripheral neuropathy. Acta Neurol. Scand. 1982, 66, 582–589. [Google Scholar] [CrossRef] [PubMed]
- Uzar, E.; Tamam, Y.; Evliyaoglu, O.; Tuzcu, A.; Beyaz, C.; Acar, A.; Aydin, B.; Tasdemir, N. Serum prolidase activity and oxidative status in patients with diabetic neuropathy. Neurol. Sci. 2012, 33, 875–880. [Google Scholar] [CrossRef] [PubMed]
- Mallet, M.L.; Hadjivassiliou, M.; Sarrigiannis, P.G.; Zis, P. The Role of Oxidative Stress in Peripheral Neuropathy. J. Mol. Neurosci. 2020, 70, 1009–1017. [Google Scholar] [CrossRef] [PubMed]
- Eftekharpour, E.; Fernyhough, P. Oxidative Stress and Mitochondrial Dysfunction Associated with Peripheral Neuropathy in Type 1 Diabetes. Antioxid. Redox Signal 2022, 37, 578–596. [Google Scholar] [CrossRef] [PubMed]
- Areti, A.; Yerra, V.G.; Naidu, V.; Kumar, A. Oxidative stress and nerve damage: Role in chemotherapy induced peripheral neuropathy. Redox Biol. 2014, 2, 289–295. [Google Scholar] [CrossRef]
- Monforte, R.; Estruch, R.; Valls-Sole, J.; Nicolas, J.; Villalta, J.; Urbano-Marquez, A. Autonomic and peripheral neuropathies in patients with chronic alcoholism. A dose-related toxic effect of alcohol. Arch. Neurol. 1995, 52, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Tagliaferro, S.S. Comparison of DNA extraction methods for samples from old blood collections. Biotechniques 2021, 70, 243–250. [Google Scholar] [CrossRef]
- Lapaire, O.; Johnson, K.L.; Bianchi, D.W. Method for extraction of high-quantity and -quality cell-free DNA from amniotic fluid. Methods Mol. Biol. 2008, 444, 303–309. [Google Scholar] [PubMed]
- Rotondo, J.C.; Borghi, A.; Selvatici, R.; Magri, E.; Bianchini, E.; Montinari, E.; Corazza, M.; Virgili, A.; Tognon, M.; Martini, F. Hypermethylation-Induced Inactivation of the IRF6 Gene as a Possible Early Event in Progression of Vulvar Squamous Cell Carcinoma Associated With Lichen Sclerosus. JAMA Dermatol. 2016, 152, 928–933. [Google Scholar] [CrossRef] [PubMed]
- Leontiou, C.A.; Hadjidaniel, M.D.; Mina, P.; Antoniou, P.; Ioannides, M.; Patsalis, P.C. Bisulfite Conversion of DNA: Performance Comparison of Different Kits and Methylation Quantitation of Epigenetic Biomarkers that Have the Potential to Be Used in Non-Invasive Prenatal Testing. PLoS ONE 2015, 10, e0135058. [Google Scholar] [CrossRef] [PubMed]
- Kottaridi, C.; Koureas, N.; Margari, N.; Terzakis, E.; Bilirakis, E.; Pappas, A.; Chrelias, C.; Spathis, A.; Aga, E.; Pouliakis, A.; et al. A Pyrosequencing Assay for the Quantitative Methylation Analysis of GALR1 in Endometrial Samples: Preliminary Results. Biomed. Res. Int. 2015, 2015, 756359. [Google Scholar] [CrossRef] [PubMed]
- Fusco, G.; Amoroso, M.G.; Montesano, N.G.; Viscardi, M. Development of a pyrosequencing assay for the typing of alphaherpesviruses. MethodsX 2015, 2, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Kassambara. ggpubr: ’ggplot2’ Based Publication Ready Plots. R Package Version 0.6.0. 2023. Available online: https://cran.r-project.org/web/packages/ggpubr/index.html (accessed on 21 December 2024).
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
| Patients | Healthy Control Individuals | p-Values | ||
|---|---|---|---|---|
| Sample Size (n) | 74 | 86 | ||
| Sex | Female | 26 (35.135%) | 55 (63.953%) | p < 0.001 |
| Male | 48 (64.865%) | 31 (36.047%) | p < 0.001 | |
| Age | All | 49.9 ± 11.5 | 39.1 ± 13.8 | p < 0.001 |
| Female | 49.4 ± 13.3 | 37.7 ± 12.7 | p < 0.001 | |
| Male | 50.1 ± 10.5 | 41.6 ± 15.4 | p = 0.006 | |
| Smokers | 58 (79.378%) | 4 (4.651%) | p < 0.001 | |
| AUDIT | T1 | 27.1 (±7.5) | 2.6 (±1.9) | p < 0.001 |
| T2 | 22.6 (±9.0) | 2.9 (±2.0) | p < 0.001 | |
| OCDS | T1 | 21.5 (±7.3) | 1.7 (±1.8) | p < 0.001 |
| T2 | 13.4 (±6.3) | 1.5 (±1.9) | p < 0.001 | |
| GSI | T1 | 65.9 (±9.5) | 45.7 (±8.9) | p < 0.001 |
| T2 | 55.63 (±11.2) | 44.5 (±9.1) | p < 0.001 | |
| a. Blood DNA Methylation | b. Saliva DNA Methylation | c. H3K4me3 Fold Change | |
|---|---|---|---|
| Group (patients/healthy controls) | p < 0.001 (***) | p < 0.001 (***) | p = 0.015 (**) |
| Sex | p = 0.05 (*) | n.s. | n.s. |
| Age | n.s. | p = 0.003 (**) | n.s. |
| Smoking | n.s. | n.s. | n.s. |
| Timepoint | n.s. | p = 0.01 (**) | n.s. |
| Time × timepoint | n.s. | n.s. | n.s. |
| Random effect standard deviation | 0.1699 | 0.1124 | 1.030 |
| Residual standard deviation | 0.1297 | 0.1731 | 1.315 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kawecka, E.; Plättner, H.; Ederer, L.; Niemann, K.; Pasche, S.; Zimmermann, M.; Edelmann, S.; Nieratschker, V. GDAP1 Is Dysregulated at DNA Methylation and H3K4me3 Levels in Alcohol Use Disorder. Int. J. Mol. Sci. 2025, 26, 1623. https://doi.org/10.3390/ijms26041623
Kawecka E, Plättner H, Ederer L, Niemann K, Pasche S, Zimmermann M, Edelmann S, Nieratschker V. GDAP1 Is Dysregulated at DNA Methylation and H3K4me3 Levels in Alcohol Use Disorder. International Journal of Molecular Sciences. 2025; 26(4):1623. https://doi.org/10.3390/ijms26041623
Chicago/Turabian StyleKawecka, Emilia, Henning Plättner, Lena Ederer, Kilian Niemann, Sarah Pasche, Milan Zimmermann, Susanne Edelmann, and Vanessa Nieratschker. 2025. "GDAP1 Is Dysregulated at DNA Methylation and H3K4me3 Levels in Alcohol Use Disorder" International Journal of Molecular Sciences 26, no. 4: 1623. https://doi.org/10.3390/ijms26041623
APA StyleKawecka, E., Plättner, H., Ederer, L., Niemann, K., Pasche, S., Zimmermann, M., Edelmann, S., & Nieratschker, V. (2025). GDAP1 Is Dysregulated at DNA Methylation and H3K4me3 Levels in Alcohol Use Disorder. International Journal of Molecular Sciences, 26(4), 1623. https://doi.org/10.3390/ijms26041623






