Genome-Wide Identification and Functional Characterization of the Acyl-CoA Dehydrogenase (ACAD) Family in Fusarium sacchari
Abstract
1. Introduction
2. Results
2.1. Identification of Acyl-CoA Dehydrogenase Genes in F. sacchari
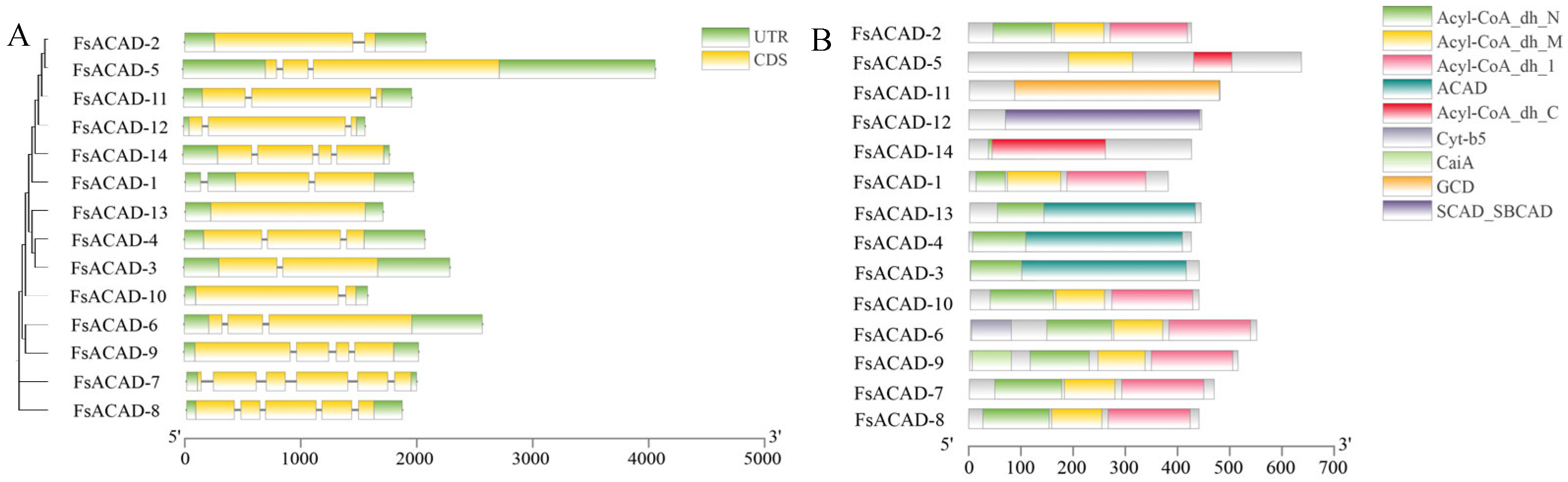
2.2. Gene Organization in the FsACAD Family
2.3. Phylogenetic Relationship Among ACAD Proteins
2.4. FsACAD Gene Expression Patterns During Sugarcane Infection
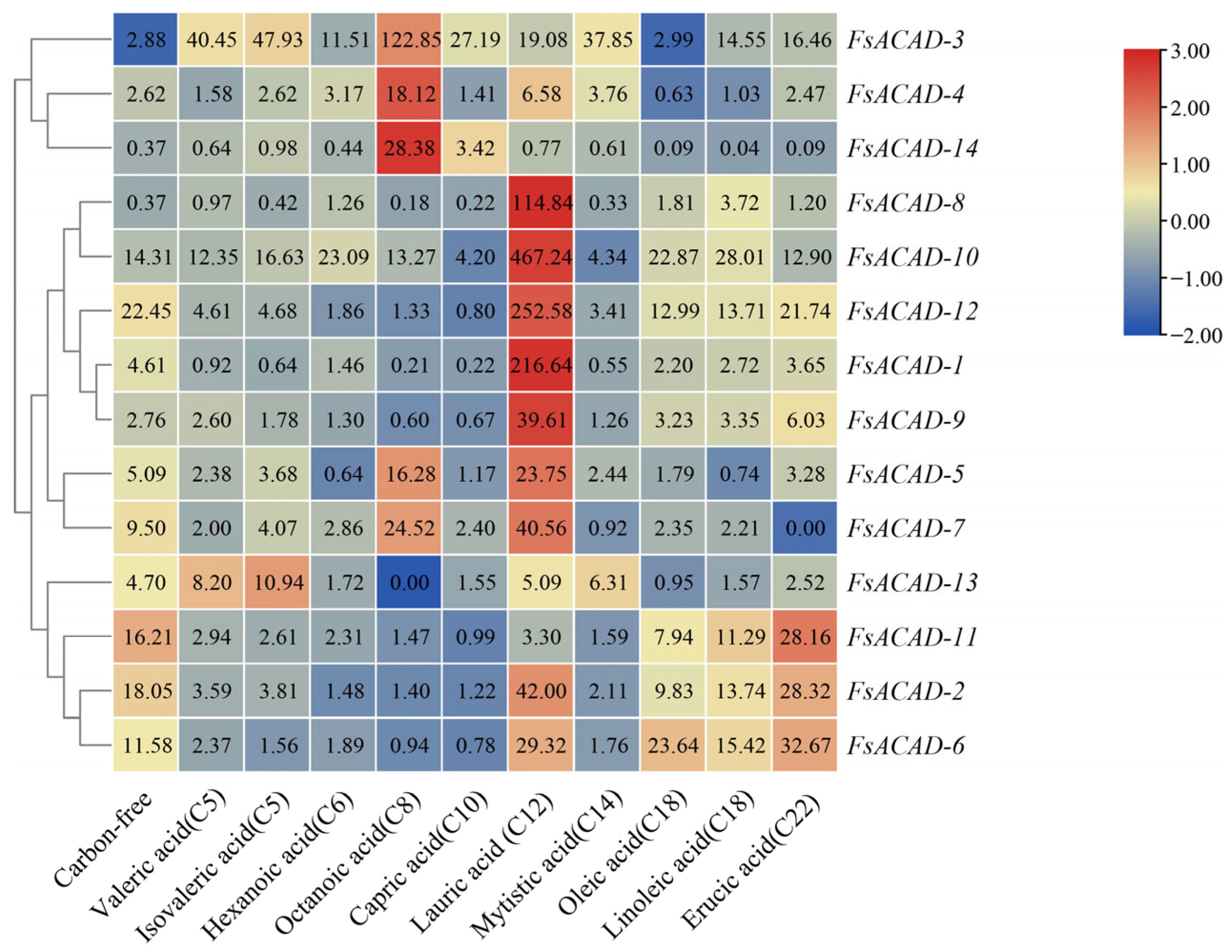
2.5. Expression of FsACADs Is Differentially Induced by Different Fatty Acids
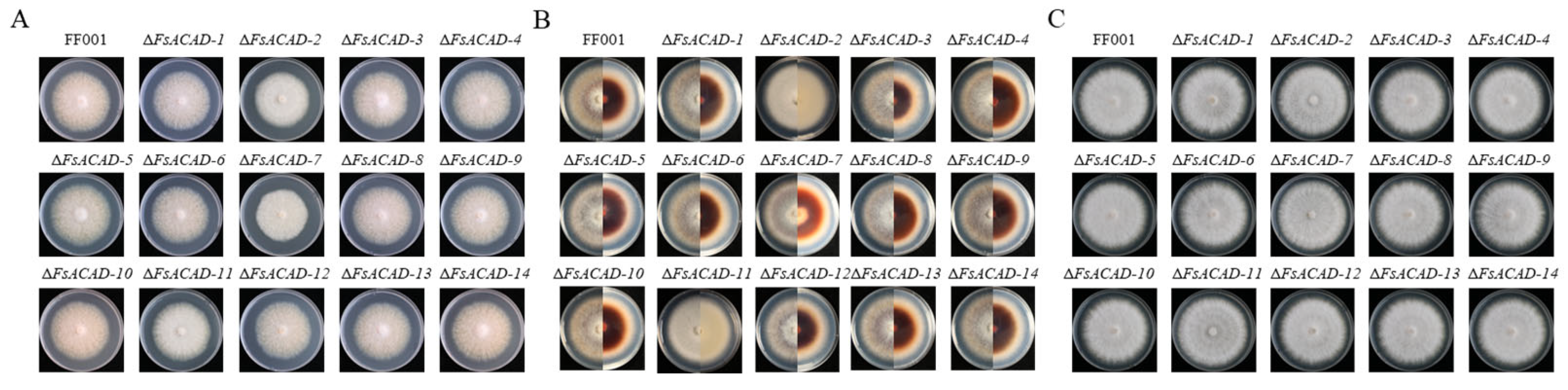
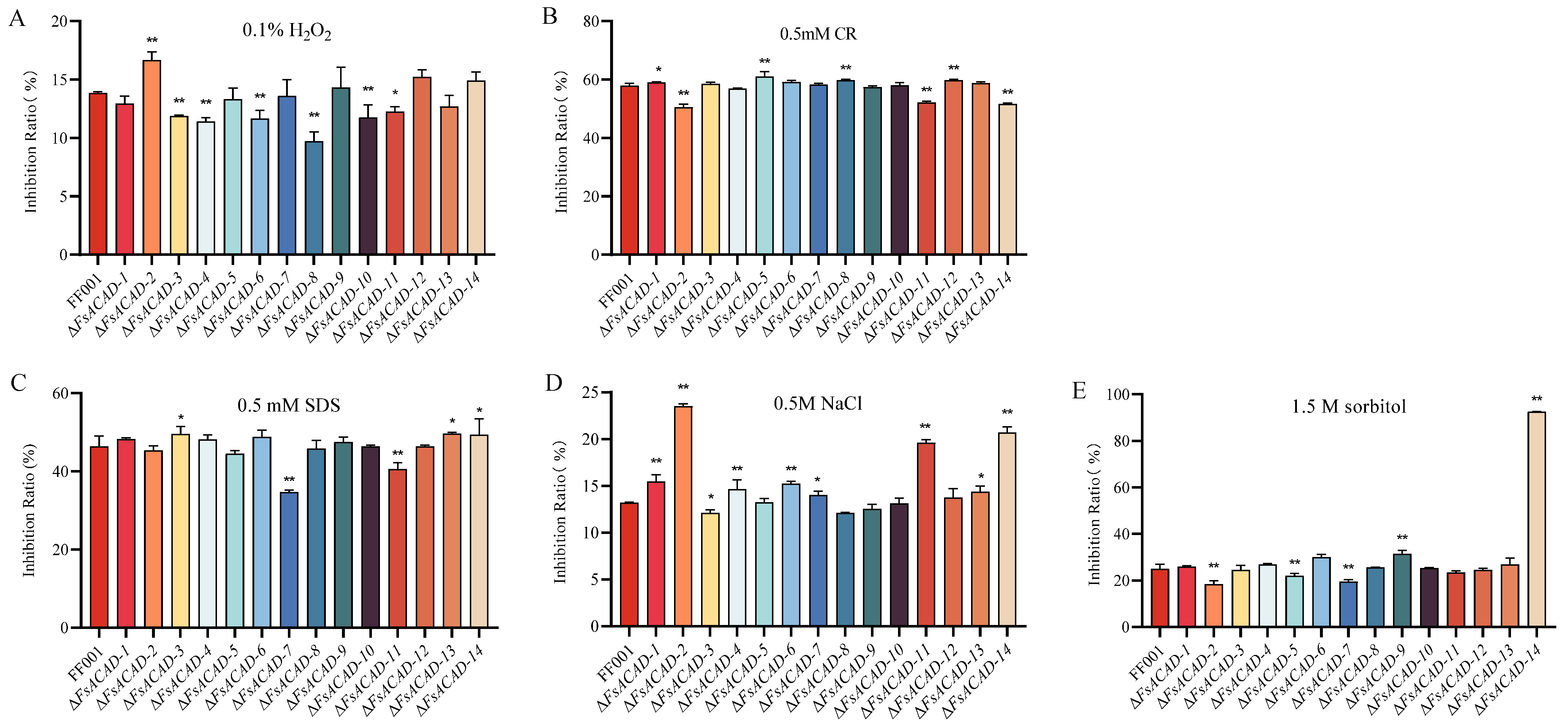
2.6. FsACADs Differentially Impact Hyphal Growth, Conidiation, and Stress Response
2.7. FsACADs Have Varied Impact on Fungal Utilization of Fatty Acids
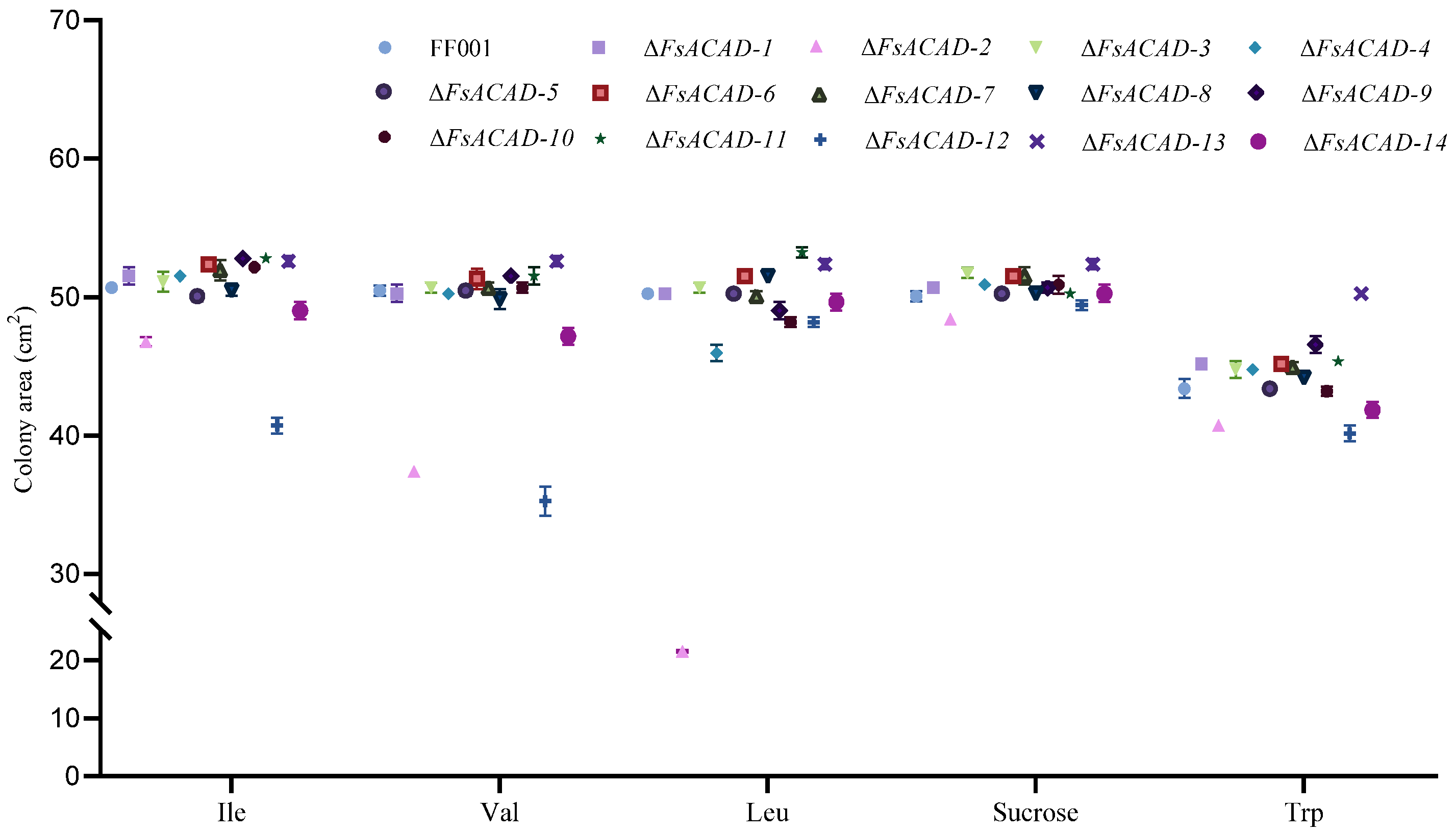
2.8. FsACADs Have Varied Impact on Utilization of Amino Acids by F. sacchari
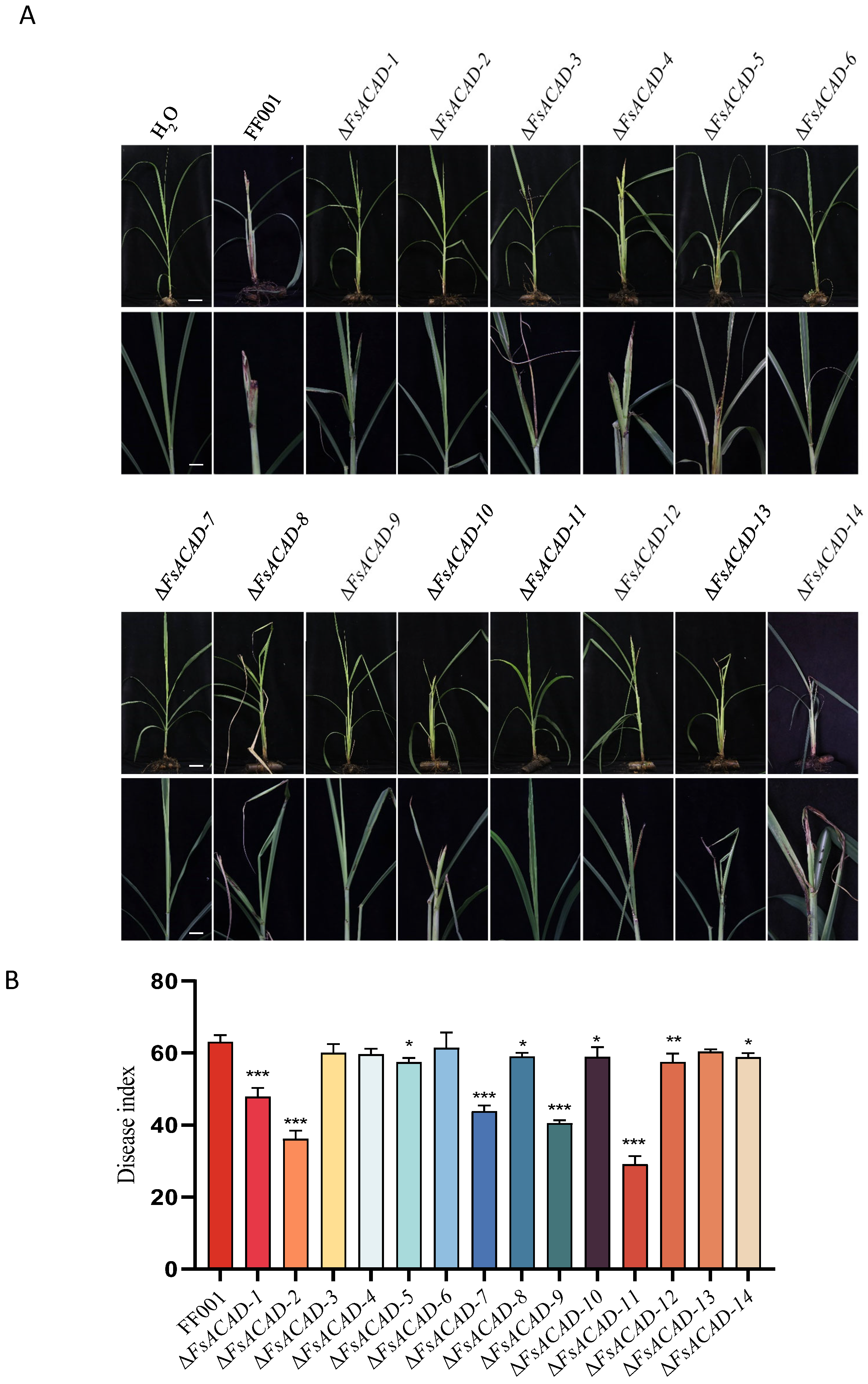
2.9. Contribution of FsACADs to Virulence
2.10. Phenotype of FsACAD-2/FsACAD-11 Double Knockout Mutants
3. Discussion
4. Materials and Methods
4.1. Fungal Strains and Their Culture Conditions
4.2. Bioinformatics Analysis
4.3. Gene Expression Profiling
4.4. Construction of FsACAD-Deletion Mutants
4.5. Stress Assay
4.6. Assays for Utilization of Fatty and Amino Acids as the Sole Carbon Source
4.7. Virulence Assays and Pathogenicity Analysis
4.8. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lund, J.; Rustan, A.C. Fatty acids: Structures and properties. In eLS; Wiley: Hoboken, NJ, USA, 2020; pp. 283–292. [Google Scholar]
- De Carvalho, C.C.C.R.; Caramujo, M.J. The various roles of fatty acids. Molecules 2018, 23, 2583. [Google Scholar] [CrossRef]
- Schulz, H. Chapter 3 Oxidation of fatty acids. In New Comprehensive Biochemistry; Elsevier: Amsterdam, The Netherlands, 1991; Volume 20, pp. 87–110. [Google Scholar]
- Goepfert, S.; Poirier, Y. β-Oxidation in fatty acid degradation and beyond. Curr. Opin. Plant Biol. 2007, 10, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Jallet, D.; Xing, D.; Hughes, A.; Moosburner, M.; Simmons, M.P.; Allen, A.E.; Peers, G. Mitochondrial fatty acid β-oxidation is required for storage-lipid catabolism in a marine diatom. New Phytol. 2020, 228, 946–958. [Google Scholar] [CrossRef] [PubMed]
- Maggio-Hall, L.A.; Lyne, P.; Wolff, J.A.; Keller, N.P. A single acyl-CoA dehydrogenase is required for catabolism of isoleucine, valine and short-chain fatty acids in Aspergillus nidulans. Fungal Genet. Biol. 2008, 45, 180–189. [Google Scholar] [CrossRef]
- Tahri-Joutey, M.; Andreoletti, P.; Surapureddi, S.; Nasser, B.; Cherkaoui-Malki, M.; Latruffe, N. Mechanisms mediating the regulation of peroxisomal fatty acid beta-oxidation by PPARα. Int. J. Mol. Sci. 2021, 22, 8969. [Google Scholar] [CrossRef] [PubMed]
- Kunau, W.-H.; Dommes, V.; Schulz, H. β-Oxidation of fatty acids in mitochondria, peroxisomes, and bacteria: A century of continued progress. Prog. Lipid Res. 1995, 34, 267–342. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.J.; Brown, T.W.; Eitzen, G.A.; Rachubinski, R.A. Regulation of peroxisome size and number by fatty acid β-oxidation in the yeast Yarrowia lipolytica. J. Biol. Chem. 2000, 275, 20168–20178. [Google Scholar] [CrossRef] [PubMed]
- Maggio-Hall, L.A.; Keller, N.P. Mitochondrial β-oxidation in Aspergillus nidulans. Mol. Microbiol. 2004, 54, 1173–1185. [Google Scholar] [CrossRef] [PubMed]
- Bartlett, K.; Eaton, S. Mitochondrial β-oxidation. Eur. J. Biochem. 2004, 271, 462–469. [Google Scholar] [CrossRef] [PubMed]
- Lakshmanan, P.; Geijskes, R.J.; Aitken, K.S.; Grof, C.L.P.; Bonnett, G.D.; Smith, G.R. Sugarcane biotechnology: The challenges and opportunities. Vitr. Cell. Dev. Biol. Plant 2005, 41, 345–363. [Google Scholar] [CrossRef]
- Qi, Y.; Gao, X.; Zeng, Q.; Zheng, Z.; Wu, C.; Yang, R.; Feng, X.; Wu, Z.; Fan, L.; Huang, Z. Sugarcane breeding, Germplasm development and related Molecular research in China. Sugar Tech 2022, 24, 73–85. [Google Scholar] [CrossRef]
- Vishwakarma, S.K.; Kumar, P.; Nigam, A.; Singh, A.; Kumar, A. Pokkah boeng: An emerging disease of sugarcane. J. Plant Pathol. Microbiol. 2013, 4, 170. [Google Scholar] [CrossRef]
- Mohammadi, A.; Nejad, R.F.; Mofrad, N.N. Fusarium verticillioides from sugarcane, vegetative compatibility groups and pathogenicity. Plant Prot. Sci. 2012, 48, 80–84. [Google Scholar] [CrossRef]
- Khani, K.; Alizadeh, A.; Nejad, R.F.; Tehrani, A.S. Pathogenicity of Fusarium proliferatum, a new causal agent of pokkah boeng in sugarcane. In Proceedings of the International Society of Sugar Cane Technologists, São Paulo, Brazil, 24–28 June 2013. [Google Scholar]
- Bao, Y.; Akbar, S.; Yao, W.; Xu, Y.; Xu, J.; Powell, C.A.; Chen, B.; Zhang, M. Genetic diversity and pathogenicity of Fusarium fujikuroi species complex (FFSC) causing sugarcane pokkah boeng disease (PBD) in China. Plant Dis. 2023, 107, 1299–1309. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.R.; Huang, H.J.; Li, Y.X.; Li, Y.J.; Li, J.Q.; Chen, B.S. First report of Fusarium sacchari causing sugarcane pokkah boeng in China. Plant Dis. 2020, 104, 1553–1554. [Google Scholar] [CrossRef]
- Lin, Z.; Xu, S.; Que, Y.; Wang, J.; Comstock, J.C.; Wei, J.; McCord, P.H.; Chen, B.; Chen, R.; Zhang, M. Species-specific detection and identification of Fusarium species complex, the causal agent of sugarcane pokkah boeng in China. PLoS ONE 2014, 9, e104195. [Google Scholar] [CrossRef]
- Liu, S.; Ruan, W.; Li, J.; Xu, H.; Wang, J.; Gao, Y.; Wang, J. Biological control of phytopathogenic fungi by fatty acids. Mycopathologia 2008, 166, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Trépanier, M.; Bécard, G.; Moutoglis, P.; Willemot, C.; Gagné, S.; Avis Tyler, J.; Rioux, J.-A. Dependence of arbuscular-mycorrhizal fungi on their plant host for palmitic acid synthesis. Appl. Environ. Microbiol. 2005, 71, 5341–5347. [Google Scholar] [CrossRef]
- Wipperman, M.F.; Sampson, N.S.; Thomas, S.T. Pathogen roid rage: Cholesterol utilization by Mycobacterium tuberculosis. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 269–293. [Google Scholar] [CrossRef]
- Kim, J.J.P.; Miura, R. Acyl-CoA dehydrogenases and acyl-CoA oxidases. Structural basis for mechanistic similarities and differences. Eur. J. Biochem. 2004, 271, 483–493. [Google Scholar] [CrossRef] [PubMed]
- Swigoňová, Z.; Mohsen, A.-W.; Vockley, J. Acyl-CoA dehydrogenases: Dynamic history of protein family evolution. J. Mol. Evol. 2009, 69, 176–193. [Google Scholar] [CrossRef] [PubMed]
- Beites, T.; Jansen, R.S.; Wang, R.; Jinich, A.; Rhee, K.Y.; Schnappinger, D.; Ehrt, S. Multiple acyl-CoA dehydrogenase deficiency kills Mycobacterium tuberculosis in vitro and during infection. Nat. Commun. 2021, 12, 6593. [Google Scholar] [CrossRef]
- Aliyu, S.R.; Lin, L.; Chen, X.; Abdul, W.; Lin, Y.; Otieno, F.J.; Shabbir, A.; Batool, W.; Zhang, Y.; Tang, W.; et al. Disruption of putative short-chain acyl-CoA dehydrogenases compromised free radical scavenging, conidiogenesis, and pathogenesis of Magnaporthe oryzae. Fungal Genet. Biol. 2019, 127, 23–34. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Misheva, M.; Kotzamanis, K.; Davies, L.C.; Tyrrell, V.J.; Rodrigues, P.; Benavides, G.A.; Hinz, C.; Murphy, R.C.; Kennedy, P.D.; Taylor, P.R.; et al. Oxylipin metabolism is controlled by mitochondrial β-oxidation during bacterial inflammation. Nat. Commun. 2022, 13, 139. [Google Scholar] [CrossRef] [PubMed]
- McMullen, E.; Hertenstein, H.; Strassburger, K.; Deharde, L.; Brankatschk, M.; Schirmeier, S. Glycolytically impaired Drosophila glial cells fuel neural metabolism via β-oxidation. Nat. Commun. 2023, 14, 2996. [Google Scholar] [CrossRef]
- Guerra, I.; Ferreira, H.B.; Melo, T.; Rocha, H.; Moreira, S.; Diogo, L.; Domingues, M.R.; Moreira, A. Mitochondrial Fatty Acid β-Oxidation Disorders: From Disease to Lipidomic Studies—A Critical Review. Int. J. Mol. Sci. 2022, 23, 13933. [Google Scholar] [CrossRef]
- Clifford, M.N.; King, L.J.; Kerimi, A.; Pereira-Caro, M.G.; Williamson, G. Metabolism of phenolics in coffee and plant-based foods by canonical pathways: An assessment of the role of fatty acid β-oxidation to generate biologically-active and -inactive intermediates. Crit. Rev. Food Sci. Nutr. 2022, 64, 3326–3383. [Google Scholar] [CrossRef] [PubMed]
- Santos Souza, H.F.; Marsiccobetre, S.; Souza, R.O.O.; Luévano-Martínez, L.A.; Silber, A.M. Branched chain amino acids catabolism as a source of new drug targets in pathogenic protists. Exp. Parasitol. 2023, 249, 108499. [Google Scholar] [CrossRef] [PubMed]
- Sauer, S.W.; Okun, J.G.; Schwab, M.A.; Crnic, L.R.; Hoffmann, G.F.; Goodman, S.I.; Koeller, D.M.; Kölker, S. Bioenergetics in glutaryl-Coenzyme A dehydrogenase deficiency: A role for glutaryl-Coenzyme A. J. Biol. Chem. 2005, 280, 21830–21836. [Google Scholar] [CrossRef] [PubMed]
- Rashan, E.H.; Bartlett, A.K.; Khana, D.B.; Zhang, J.; Jain, R.; Smith, A.J.; Baker, Z.N.; Cook, T.; Caldwell, A.; Chevalier, A.R.; et al. ACAD10 and ACAD11 enable mammalian 4-hydroxy acid lipid catabolism. bioRxiv 2024. [Google Scholar] [CrossRef]
- Kim, J.J.; Wang, M.; Paschke, R. Crystal structures of medium-chain acyl-CoA dehydrogenase from pig liver mitochondria with and without substrate. Proc. Natl. Acad. Sci. USA 1993, 90, 7523–7527. [Google Scholar] [CrossRef]
- Toogood, H.S.; van Thiel, A.; Basran, J.; Sutcliffe, M.J.; Scrutton, N.S.; Leys, D. Extensive domain motion and electron transfer in the human electron transferring flavoprotein·medium chain acyl-CoA dehydrogenase complex. J. Biol. Chem. 2004, 279, 32904–32912. [Google Scholar] [CrossRef] [PubMed]
- Tiffany, K.A.; Roberts, D.L.; Wang, M.; Paschke, R.; Mohsen, A.-W.A.; Vockley, J.; Kim, J.-J.P. Structure of human isovaleryl-CoA dehydrogenase at 2.6 Å resolution: structural basis for substrate specificity. Biochemistry 1997, 36, 8455–8464. [Google Scholar] [CrossRef] [PubMed]
- Djordjevic, S.; Pace, C.P.; Stankovich, M.T.; Kim, J.-J.P. Three-dimensional structure of butyryl-CoA dehydrogenase from Megasphaera elsdenii. Biochemistry 1995, 34, 2163–2171. [Google Scholar] [CrossRef] [PubMed]
- Battaile, K.P.; Molin-Case, J.; Paschke, R.; Wang, M.; Bennett, D.; Vockley, J.; Kim, J.-J.P. Crystal structure of rat short chain acyl-CoA dehydrogenase complexed with acetoacetyl-CoA: Comparison with other acyl-CoA dehydrogenases. J. Biol. Chem. 2002, 277, 12200–12207. [Google Scholar] [CrossRef] [PubMed]
- Battaile, K.P.; Nguyen, T.V.; Vockley, J.; Kim, J.-J.P. Structures of Isobutyryl-CoA dehydrogenase and enzyme-product complex: Comparison with isovaleryl—and short-chain acyl-CoA dehydrogenases. J. Biol. Chem. 2004, 279, 16526–16534. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.; Wang, M.; Paschke, R.; Rao, K.S.; Frerman, F.E.; Kim, J.-J.P. Crystal structures of human glutaryl-CoA dehydrogenase with and without an alternate substrate: Structural bases of dehydrogenation and decarboxylation reactions. Biochemistry 2004, 43, 9674–9684. [Google Scholar] [CrossRef]
- Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A.; et al. The conserved domain database in 2023. Nucleic Acids Res. 2023, 51, D384–D388. [Google Scholar] [CrossRef] [PubMed]
- Ruprecht, A.; Maddox, J.; Stirling, A.J.; Visaggio, N.; Seah, S.Y.K. Characterization of Novel Acyl Coenzyme A Dehydrogenases Involved in Bacterial Steroid Degradation. J. Bacteriol. 2015, 197, 1360–1367. [Google Scholar] [CrossRef]
- Gerstner, B.; Gratopp, A.; Marcinkowski, M.; Sifringer, M.; Obladen, M.; Bührer, C. Glutaric acid and its metabolites cause apoptosis in immature oligodendrocytes: A novel mechanism of white matter degeneration in glutaryl-CoA dehydrogenase deficiency. Pediatr. Res. 2005, 57, 771–776. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Schuck, P.F.; Busanello, E.N.B.; Tonin, A.M.; Viegas, C.M.; Ferreira, G.C. Neurotoxic effects of trans-glutaconic acid in Rats. Oxidative Med. Cell. Longev. 2013, 2013, 607610. [Google Scholar] [CrossRef] [PubMed]
- Barroso, M.; Gertzen, M.; Puchwein-Schwepcke, A.F.; Preisler, H.; Sturm, A.; Reiss, D.D.; Danecka, M.K.; Muntau, A.C.; Gersting, S.W. Glutaryl-CoA dehydrogenase misfolding in glutaric acidemia type 1. Int. J. Mol. Sci. 2023, 24, 13158. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zheng, X.; Zhu, M.; Chen, M.; Zhang, S.; He, F.; Chen, X.; Lv, J.; Pei, M.; Zhang, Y.; et al. MoIVD-mediated leucine catabolism is required for vegetative growth, conidiation and full virulence of the rice blast fungus Magnaporthe oryzae. Front. Microbiol. 2019, 10, 444. [Google Scholar] [CrossRef] [PubMed]
- John, F.; Leslie, B.A.S. Media—recipes and preparation. In The Fusarium Laboratory Manual; Wiley: Hoboken, NJ, USA, 2006; pp. 3–14. [Google Scholar]
- Amir, S.; Sutar, S.; Singh, S.K.; Baghela, A. A rapid and efficient method of fungal genomic DNA extraction, suitable for PCR based molecular methods. Plant Pathol. Quar. 2015, 5, 74–81. [Google Scholar] [CrossRef]
- Lan, X.; Yao, Z.; Zhou, Y.; Shang, J.; Lin, H.; Nuss, D.L.; Chen, B. Deletion of cpku80 gene in the chestnut blight fungus, Cryphonectria parasitica, enhanced gene target-ing efficiency, Cryphonectria parasitica, enhances gene disruption efficiency. Curr. Genet. 2008, 53, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yao, Z.; Zhao, L.; Yu, M.; Chen, B.; Zou, C. Redundant and Distinct Roles of Two 14-3-3 Proteins in Fusarium sacchari, Pathogen of Sugarcane Pokkah Boeng Disease. J. Fungi 2024, 10, 257. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Li, F.; Huang, Y.; Yu, Q.; Huang, Z.; Zeng, Q.; Chen, B.; Meng, J. FsCGBP, a Cutinase G-Box Binding Protein, Regulates the Growth, Development, and Virulence of Fusarium sacchari, the Pathogen of Sugarcane Pokkah Boeng Disease. J. Fungi 2024, 10, 246. [Google Scholar] [CrossRef]
- Cai, X.H.; Chen, T.; Wang, R.Y.; Fan, Y.J.; Li, Y.; Hu, S.N.; Yuan, Z.M.; Li, H.G.; Li, X.Y.; Zhao, S.Y.; et al. Forecasting wildfire disease on tobacco: Toward developing a high-accuracy prediction model for disease index using local climate factors and support vector regression. Theor. Appl. Climatol. 2019, 137, 2139–2149. [Google Scholar] [CrossRef]
- Wang, Z.-P.; Sun, H.-J.; Guo, Q.; Xu, S.-Q.; Wang, J.-H.; Lin, S.-H.; Zhang, M.-Q. Artificial inoculation method of pokkah boeng disease of sugarcane and screening of resistant germplasm resources in subtropical China. Sugar Tech 2017, 19, 283–292. [Google Scholar] [CrossRef]



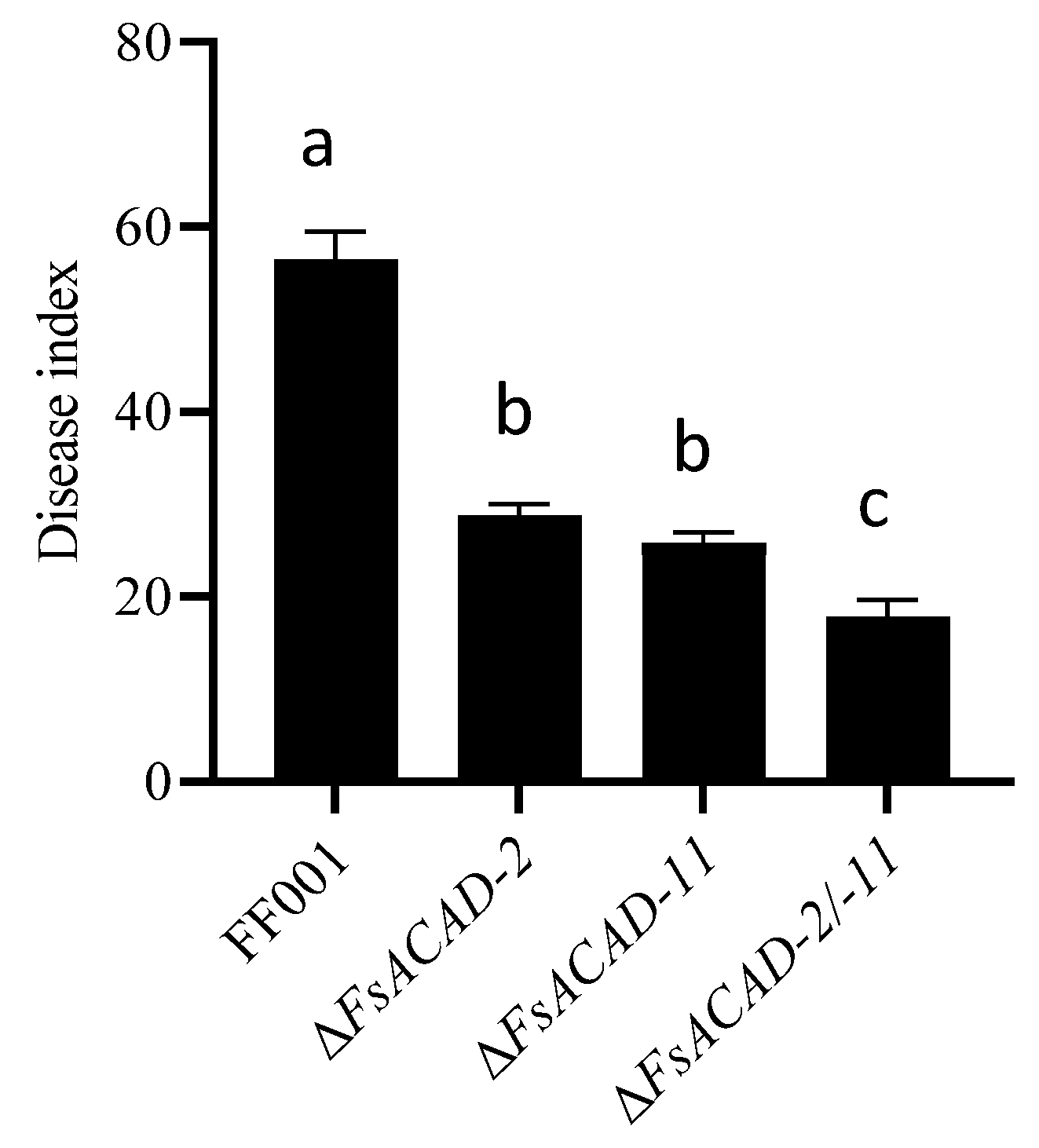
| Gene | Accession No. | Chromosome a | Protein Size (aa) | Subcellular Localization | Annotation |
|---|---|---|---|---|---|
| FsACAD-1 | PP466006 | Chrom5(+) | 381 | mitochondria | Acyl-CoA dehydrogenase medium-chain specific mitochondrial precursor |
| FsACAD-2 | PP466007 | Chrom6(−) | 427 | mitochondria | Isovaleryl-CoA dehydrogenase |
| FsACAD-3 | PP466008 | Chrom3(+) | 439 | mitochondria | Acyl dehydrogenase medium-chain specific mitochondrial precursor |
| FsACAD-4 | PP466009 | Chrom3(+) | 426 | mitochondria | Acyl-CoA dehydrogenase medium-chain specific mitochondrial precursor |
| FsACAD-5 | PP466010 | Chrom4(+) | 638 | mitochondria | Acyl-CoA dehydrogenase family member 11 |
| FsACAD-6 | PP466011 | Chrom4(+) | 548 | mitochondria | Acyl-CoA dehydrogenase |
| FsACAD-7 | PP466012 | Chrom4(−) | 469 | mitochondria | Long-chain specific acyl-CoA dehydrogenase |
| FsACAD-8 | PP466013 | Chrom4(−) | 441 | mitochondria | Long-chain specific acyl-CoA dehydrogenase |
| FsACAD-9 | PP466014 | Chrom7(−) | 514 | mitochondria | Very long-chain specific acyl-CoA dehydrogenase |
| FsACAD-10 | PP466015 | Chrom7(+) | 438 | mitochondria | Acyl-CoA dehydrogenase related to the alkylation response protein AidB |
| FsACAD-11 | PP466016 | Chrom7(+) | 480 | mitochondria | Glutaryl-CoA dehydrogenase |
| FsACAD-12 | PP466017 | Chrom7(+) | 446 | mitochondria | Short/branched chain-specific acyl-CoA dehydrogenase |
| FsACAD-13 | PP466018 | Chrom8(+) | 443 | mitochondria | Acyl-CoA dehydrogenase fadE12 |
| FsACAD-14 | PP466019 | Chrom2(+) | 426 | mitochondria | Acyl-CoA dehydrogenase |
| Strain | Average Colony Size (cm2) | ||
|---|---|---|---|
| PDA | MM | CM | |
| FF001 | 36.14 ± 0.312 bc | 48.81 ± 0.358 c | 46.37 ± 0.352 a |
| ΔFsACAD-1 | 33.70 ± 1.372 ef | 47.58 ± 0.352 d | 46.37 ± 0.352 a |
| ΔFsACAD-2 | 29.39 ± 0.283 g | 45.96 ± 0.605 ef | 32.68 ± 0.883 i |
| ΔFsACAD-3 | 32.68 ± 0.505 f | 43.59 ± 0.585 g | 44.58 ± 1.484 bc |
| ΔFsACAD-4 | 35.43 ± 0.300 cd | 47.58 ± 0.352 d | 43.20 ± 0.335 de |
| ΔFsACAD-5 | 38.85 ± 0.323 a | 46.77 ± 0.699 de | 45.57 ± 0.687 ab |
| ΔFsACAD-6 | 33.35 ± 0.300 ef | 46.57 ± 0.605 e | 45.37 ± 0.595 ab |
| ΔFsACAD-7 | 24.34 ± 0.254 h | 50.06 ± 0.364 b | 35.26 ± 0.525 h |
| ΔFsACAD-8 | 33.87 ± 0.789 ef | 45.56 ± 0.346 f | 42.24 ± 0.335 e |
| ΔFsACAD-9 | 34.39 ± 0.300 de | 47.58 ± 0.352 d | 43.99 ± 1.224 cd |
| ΔFsACAD-10 | 36.14 ± 0.312 bc | 46.37 ± 0.352 ef | 42.62 ± 0.335 e |
| ΔFsACAD-11 | 33.19 ± 1.357 ef | 45.96 ± 0.605 ef | 38.85 ± 0.641 g |
| ΔFsACAD-12 | 33.01 ± 0.294 f | 47.58 ± 0.352 d | 42.62 ± 0.335 e |
| ΔFsACAD-13 | 33.35 ± 0.300 ef | 47.58 ± 0.352 d | 45.56 ± 0.346 ab |
| ΔFsACAD-14 | 37.22 ± 0.824 b | 51.11 ± 0.364 a | 40.16 ± 0.976 f |
| Strains | PDA Medium | MM | CM | |||
|---|---|---|---|---|---|---|
| Average (Spore/cm2) | Fold Change | Average (Spore/cm2) | Fold Change | Average (Spore/cm2) | Fold Change | |
| FF001 | 3.95 × 106 ± 0.69 × 106 | 1.00 bcd | 3.69 × 106 ± 0.59 × 106 | 1.00 d | 5.81 × 106 ± 1.52 × 106 | 1.00 a |
| ΔFsACAD-1 | 3.57 × 106 ± 0.90 × 106 | 0.90 bcd | 7.38 × 106 ± 1.43 × 106 | 2.00 b | 6.30 × 106 ± 0.58 × 106 | 1.09 a |
| ΔFsACAD-2 | 0.60 × 106 ± 0.06 × 106 | 0.15 e | 1.59 × 106 ± 0.30 × 106 | 0.43 e | 1.42 × 106 ± 0.14 × 106 | 0.24 d |
| ΔFsACAD-3 | 2.80 × 106 ± 1.29 × 106 | 0.71 cd | 4.67 × 106 ± 0.79 × 106 | 1.27 d | 4.07 × 106 ± 0.37 × 106 | 0.70 bc |
| ΔFsACAD-4 | 3.10 × 106 ± 1.50 × 106 | 0.78 cd | 6.81 × 106 ± 0.86 × 106 | 1.85 b | 4.14 × 106 ± 0.37 × 106 | 0.71 bc |
| ΔFsACAD-5 | 6.96 × 106 ± 0.62 × 106 | 1.76 a | 7.27 × 106 ± 0.90 × 106 | 1.97 b | 5.50 × 106 ± 1.14 × 106 | 0.95 ab |
| ΔFsACAD-6 | 3.87 × 106 ± 0.66 × 106 | 0.98 bcd | 4.35 × 106 ± 0.32 × 106 | 1.18 d | 4.09 × 106 ± 0.05 × 106 | 0.70 bc |
| ΔFsACAD-7 | 2.28 × 106 ± 0.45 × 106 | 0.58 de | 10.11 × 106 ± 0.70 × 106 | 2.74 a | 3.55 × 106 ± 0.53 × 106 | 0.61 c |
| ΔFsACAD-8 | 3.95 × 106 ± 0.42 × 106 | 1.00 bcd | 4.57 × 106 ± 0.24 × 106 | 1.24 d | 5.20 × 106 ± 0.29 × 106 | 0.9 ab |
| ΔFsACAD-9 | 5.04 × 106 ± 0.58 × 106 | 1.28 b | 4.51 × 106 ± 0.15 × 106 | 1.22 d | 3.11 × 106 ± 0.31 × 106 | 0.54 c |
| ΔFsACAD-10 | 4.39 × 106 ± 1.09 × 106 | 1.11 bc | 6.35 × 106 ± 0.41 × 106 | 1.72 bc | 3.37 × 106 ± 0.29 × 106 | 0.58 c |
| ΔFsACAD-11 | 0.73 × 106 ± 0.03 × 106 | 0.19 e | 1.56 × 106 ± 0.19 × 106 | 0.42 e | 0.77 × 106 ± 0.03 × 106 | 0.13 d |
| ΔFsACAD-12 | 2.70 × 106 ± 0.43 × 106 | 0.69 cd | 4.99 × 106 ± 0.63 × 106 | 1.35 cd | 5.51 × 106 ± 0.15 × 106 | 0.95 ab |
| ΔFsACAD-13 | 3.63 × 106 ± 0.57 × 106 | 0.92 bcd | 1.98 × 106 ± 0.53 × 106 | 0.54 e | 3.65 × 106 ± 0.53 × 106 | 0.63 c |
| ΔFsACAD-14 | 5.42 × 106 ± 0.31 × 106 | 1.37 ab | 4.63 × 106 ± 0.46 × 106 | 1.26 d | 5.53 × 106 ± 0.32 × 106 | 0.95 ab |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeng, Q.; Yu, Q.; Mo, Y.; Liang, H.; Chen, B.; Meng, J. Genome-Wide Identification and Functional Characterization of the Acyl-CoA Dehydrogenase (ACAD) Family in Fusarium sacchari. Int. J. Mol. Sci. 2025, 26, 973. https://doi.org/10.3390/ijms26030973
Zeng Q, Yu Q, Mo Y, Liang H, Chen B, Meng J. Genome-Wide Identification and Functional Characterization of the Acyl-CoA Dehydrogenase (ACAD) Family in Fusarium sacchari. International Journal of Molecular Sciences. 2025; 26(3):973. https://doi.org/10.3390/ijms26030973
Chicago/Turabian StyleZeng, Quan, Quan Yu, Yingxi Mo, Haoming Liang, Baoshan Chen, and Jiaorong Meng. 2025. "Genome-Wide Identification and Functional Characterization of the Acyl-CoA Dehydrogenase (ACAD) Family in Fusarium sacchari" International Journal of Molecular Sciences 26, no. 3: 973. https://doi.org/10.3390/ijms26030973
APA StyleZeng, Q., Yu, Q., Mo, Y., Liang, H., Chen, B., & Meng, J. (2025). Genome-Wide Identification and Functional Characterization of the Acyl-CoA Dehydrogenase (ACAD) Family in Fusarium sacchari. International Journal of Molecular Sciences, 26(3), 973. https://doi.org/10.3390/ijms26030973







