Role of GmFRI-1 in Regulating Soybean Nodule Formation Under Cold Stress
Abstract
:1. Introduction
2. Results
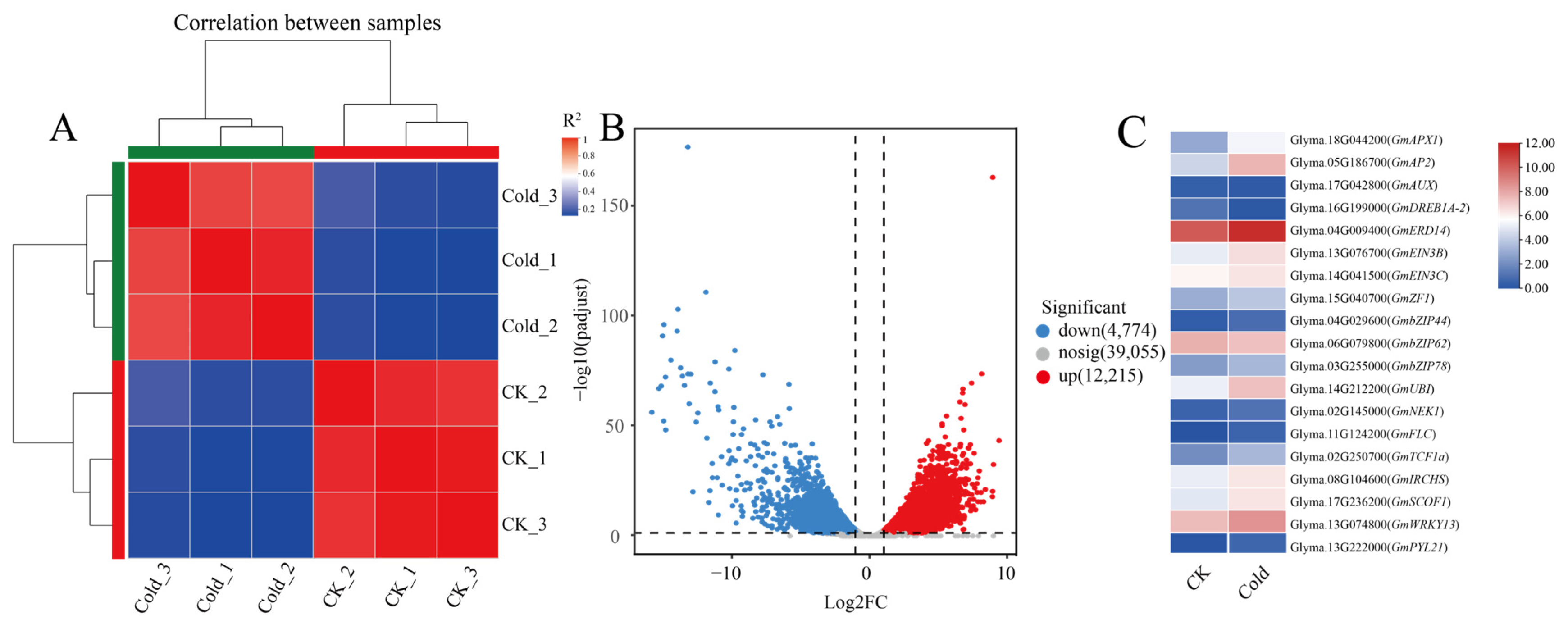
2.1. Comparative Transcriptomic Analysis of DEGs in Soybean Root Inoculated with Rhizobium Under Normal or Cold Conditions
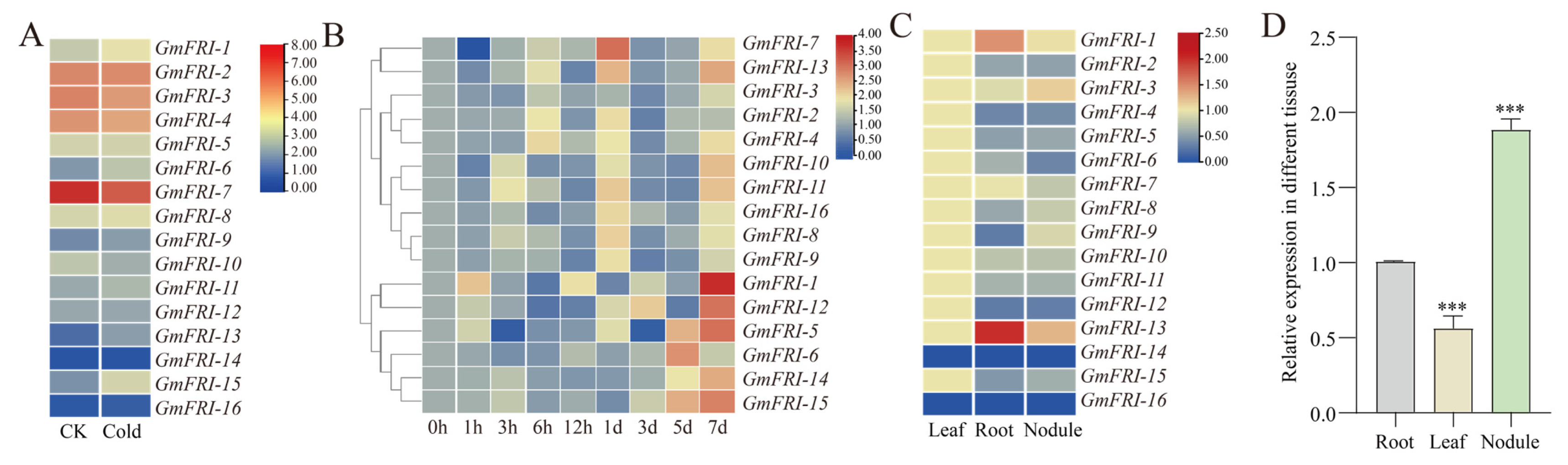
2.2. Expression Patterns of Soybean FRIGIDA Family Genes in Response to Rhizobium Inoculation
2.3. Physiological Changes Related to Overexpressing GmFRI-1 (OE-GmFRI-1) Under Low Temperature Conditions
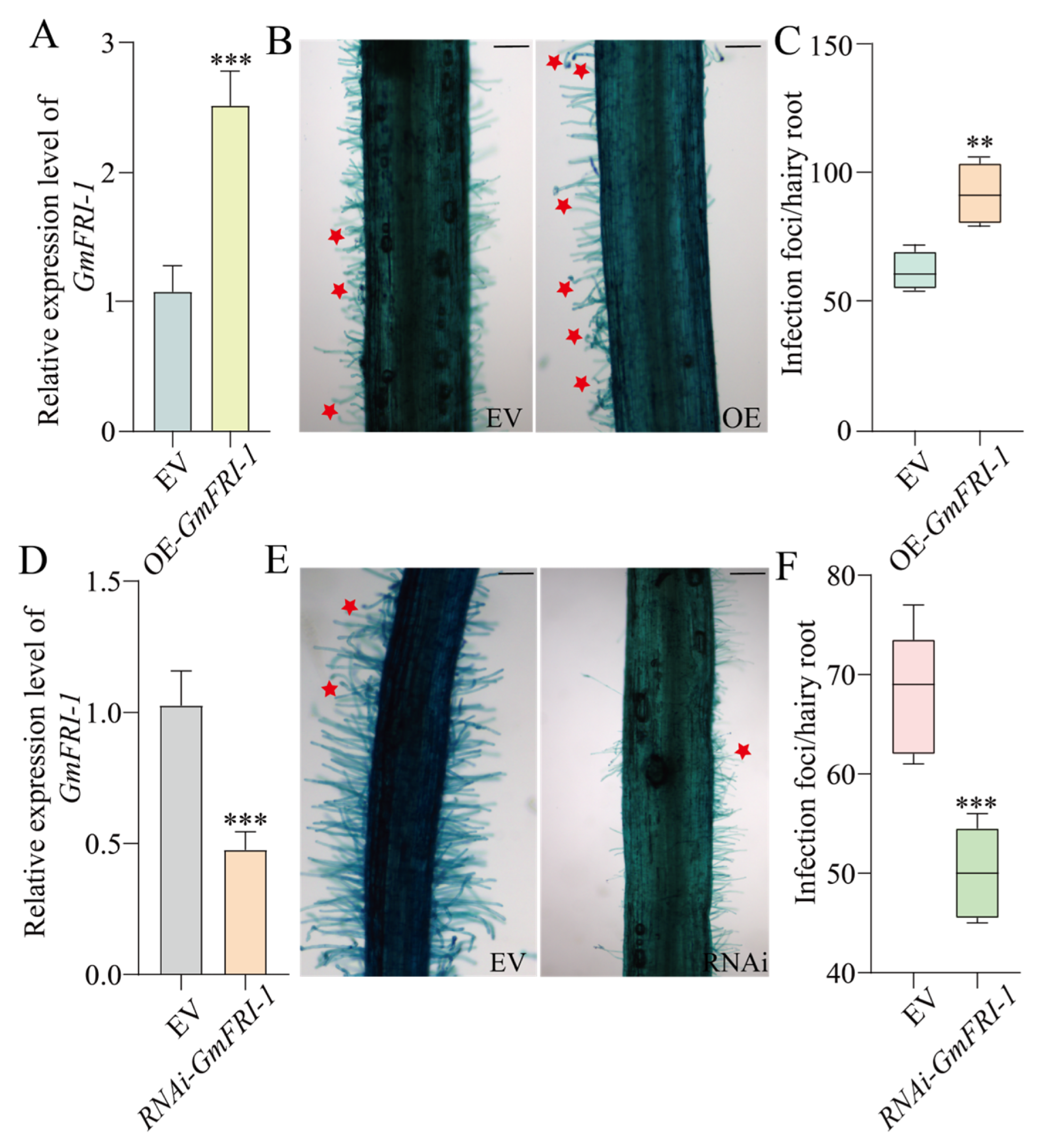
2.4. GmFRI-1 Positively Controls Soybean Nodulation
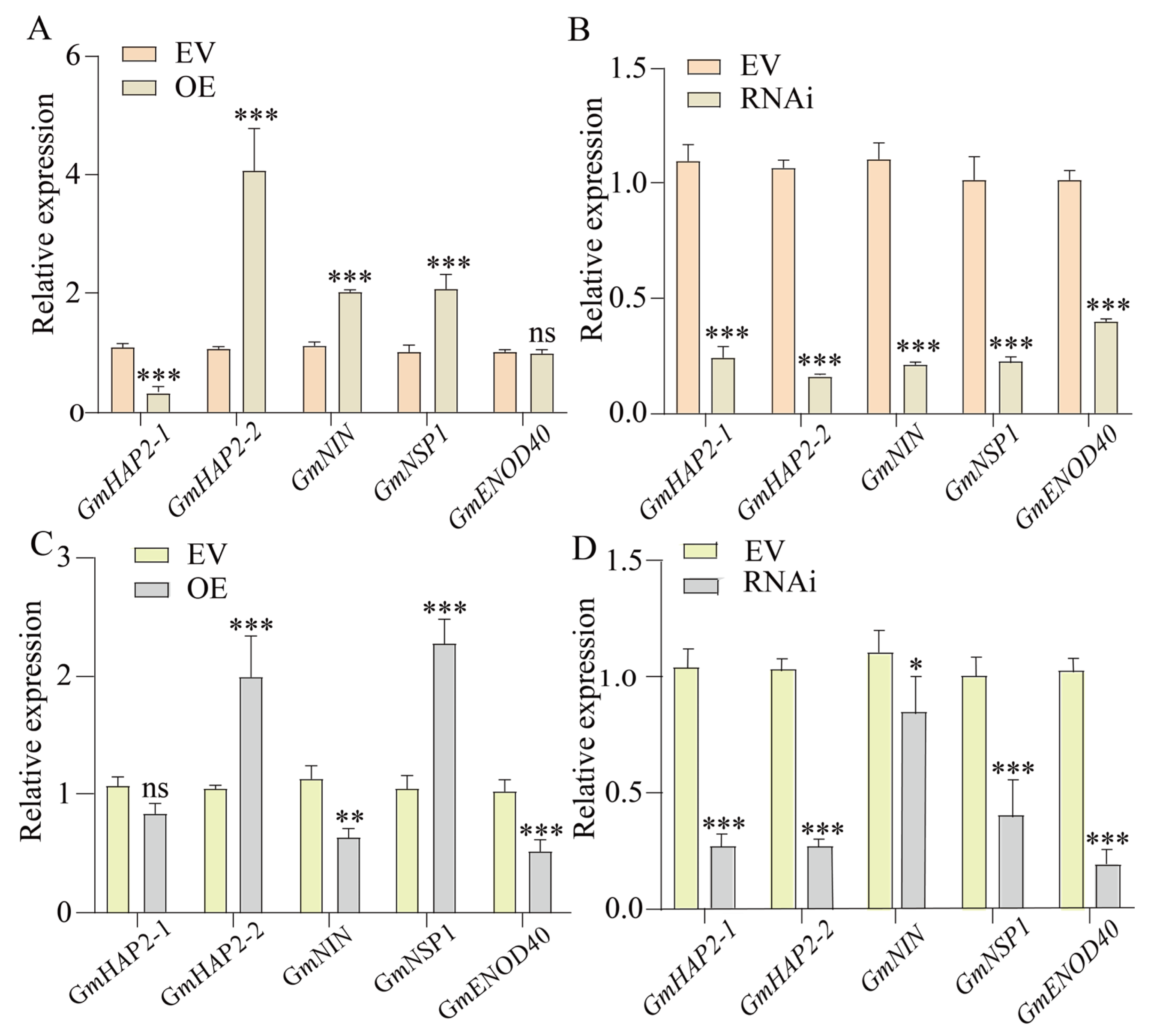
2.5. GmFRI-1 Controls Soybean Nodulation Through Regulating Nodule Factor Signaling Pathway Genes
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Transcriptomic Analysis
4.3. Gene Expression Analysis by Quantitative Real-Time PCR
4.4. Expression Analysis of Differentially Expressed Genes
4.5. Measurement and Analysis of Associated Physiological Parameters
4.6. Plasmid Construction and Genetic Transformation of Soybean Hairy Roots
4.7. Statistical Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hatfield, J.L.; Prueger, J.H. Temperature extremes: Effect on plant growth and development. Weather Clim. Extrem. 2015, 10, 4–10. [Google Scholar] [CrossRef]
- Steponkus, P. Role of the Plasma Membrane in Freezing Injury and Cold Acclimation. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2003, 35, 543–584. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.K. Abiotic stress responses in plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef]
- Kocsy, G.; Galiba, G.; Brunold, C. Role of glutathione in adaptation and signalling during chilling and cold acclimation in plants. Physiol. Plant. 2001, 113, 158–164. [Google Scholar] [CrossRef]
- Balestrasse, K.B.; Tomaro, M.L.; Batlle, A.; Noriega, G.O. The role of 5-aminolevulinic acid in the response to cold stress in soybean plants. Phytochemistry 2010, 71, 2038–2045. [Google Scholar] [CrossRef]
- Xing, W.; Rajashekar, C.B. Glycine betaine involvement in freezing tolerance and water stress in Arabidopsis thaliana. Environ. Exp. Bot. 2001, 46, 21–28. [Google Scholar] [CrossRef]
- Miura, K.; Tada, Y. Regulation of water, salinity, and cold stress responses by salicylic acid. Front. Plant Sci. 2014, 5, 4. [Google Scholar] [CrossRef] [PubMed]
- Chinnusamy, V.; Zhu, J.; Zhu, J.K. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007, 12, 444–451. [Google Scholar] [CrossRef]
- Lee, T.-M.; Lur, H.-S.; Chu, C. Role of abscisic acid in chilling tolerance of rice (Oryza sativa L.) seedlings.: II. Modulation of free polyamine levels. Plant Sci. 1997, 126, 1–10. [Google Scholar] [CrossRef]
- Bhandari, K.; Nayyar, H. Low Temperature Stress in Plants: An Overview of Roles of Cryoprotectants in Defense. In Physiological Mechanisms and Adaptation Strategies in Plants Under Changing Environment; Springer: New York, NY, USA, 2014. [Google Scholar]
- Toorchi, M.; Yukawa, K.; Nouri, M.Z.; Komatsu, S. Proteomics approach for identifying osmotic-stress-related proteins in soybean roots. Peptides 2009, 30, 2108–2117. [Google Scholar] [CrossRef]
- Latef, A.; Hamed, A.A.; He, C. Arbuscular mycorrhizal influence on growth, photosynthetic pigments, osmotic adjustment and oxidative stress in tomato plants subjected to low temperature stress. Acta Physiol. Plant 2011, 33, 1217–1225. [Google Scholar] [CrossRef]
- Peleg, Z.; Blumwald, E. Hormone balance and abiotic stress tolerance in crop plants. Curr. Opin. Plant Biol. 2011, 14, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Raza, A.; Charagh, S.; Najafi-Kakavand, S.; Abbas, S.; Shoaib, Y.; Anwar, S.; Sharifi, S.; Lu, G.; Siddique, K.H.M. Role of phytohormones in regulating cold stress tolerance: Physiological and molecular approaches for developing cold-smart crop plants. Plant Stress 2023, 8, 100152. [Google Scholar] [CrossRef]
- Lewis, B.D.; Karlin-Neumann, G.; Davis, R.W.; Spalding, E.P. Ca(2+)-activated anion channels and membrane depolarizations induced by blue light and cold in Arabidopsis seedlings. Plant Physiol. 1997, 114, 1327–1334. [Google Scholar] [CrossRef]
- Wang, D.Z.; Jin, Y.N.; Ding, X.H.; Wang, W.J.; Zhai, S.S.; Bai, L.P.; Guo, Z.F. Gene Regulation and Signal Transduction in the ICE-CBF-COR Signaling Pathway during Cold Stress in Plants. Biochem. Biokhimiia 2017, 82, 1103–1117. [Google Scholar] [CrossRef]
- Thalhammer, A.; Hincha, D.K. A mechanistic model of COR15 protein function in plant freezing tolerance: Integration of structural and functional characteristics. Plant Signal. Behav. 2014, 9, e977722. [Google Scholar] [CrossRef]
- Steponkus, P.L.; Uemura, M.; Joseph, R.A.; Gilmour, S.J.; Thomashow, M.F. Mode of action of the COR15a gene on the freezing tolerance of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 1998, 95, 14570–14575. [Google Scholar] [CrossRef]
- Yamasaki, S.; Sanada, Y.; Imase, R.; Matsuura, H.; Ueno, D.; Demura, T.; Kato, K. Arabidopsis thaliana cold-regulated 47 gene 5’-untranslated region enables stable high-level expression of transgenes. J. Biosci. Bioeng. 2018, 125, 124–130. [Google Scholar] [CrossRef]
- Bremer, A.; Kent, B.; Hauß, T.; Thalhammer, A.; Yepuri, N.R.; Darwish, T.A.; Garvey, C.J.; Bryant, G.; Hincha, D.K. Intrinsically Disordered Stress Protein COR15A Resides at the Membrane Surface during Dehydration. Biophys. J. 2017, 113, 572–579. [Google Scholar] [CrossRef]
- Yamasaki, Y.; Randall, S.K. Functionality of soybean CBF/DREB1 transcription factors. Plant Sci. Int. J. Exp. Plant Biol. 2016, 246, 80–90. [Google Scholar] [CrossRef]
- Kidokoro, S.; Watanabe, K.; Ohori, T.; Moriwaki, T.; Maruyama, K.; Mizoi, J.; Myint Phyu Sin Htwe, N.; Fujita, Y.; Sekita, S.; Shinozaki, K.; et al. Soybean DREB1/CBF-type transcription factors function in heat and drought as well as cold stress-responsive gene expression. Plant J. Cell Mol. Biol. 2015, 81, 505–518. [Google Scholar] [CrossRef] [PubMed]
- Kao, P.H.; Baiya, S.; Lai, Z.Y.; Huang, C.M.; Jhan, L.H.; Lin, C.J.; Lai, Y.S.; Kao, C.F. An advanced systems biology framework of feature engineering for cold tolerance genes discovery from integrated omics and non-omics data in soybean. Front. Plant Sci. 2022, 13, 1019709. [Google Scholar] [CrossRef]
- Funck, D.; Baumgarten, L.; Stift, M.; von Wirén, N.; Schönemann, L. Differential Contribution of P5CS Isoforms to Stress Tolerance in Arabidopsis. Front. Plant Sci. 2020, 11, 565134. [Google Scholar] [CrossRef]
- Baskaran, A.; Kaari, M.; Venugopal, G.; Manikkam, R.; Joseph, J.; Bhaskar, P.V. Anti freeze proteins (Afp): Properties, sources and applications—A review. Int. J. Biol. Macromol. 2021, 189, 292–305. [Google Scholar] [CrossRef]
- Zhu, F.; Li, M.; Sun, M.; Jiang, X.; Qiao, F. Plant hormone signals regulate trehalose accumulation against osmotic stress in watermelon cells. Protoplasma 2022, 259, 1351–1369. [Google Scholar] [CrossRef]
- Kryukov, A.A.; Gorbunova, A.O.; Kudriashova, T.R.; Yakhin, O.I.; Lubyanov, A.A.; Malikov, U.M.; Shishova, M.F.; Kozhemyakov, A.P.; Yurkov, A.P. Sugar transporters of the SWEET family and their role in arbuscular mycorrhiza. Vavilovskii Zhurnal Genet. I Sel. 2021, 25, 754–760. [Google Scholar] [CrossRef]
- Gautam, T.; Dutta, M.; Jaiswal, V.; Zinta, G.; Gahlaut, V.; Kumar, S. Emerging Roles of SWEET Sugar Transporters in Plant Development and Abiotic Stress Responses. Cells 2022, 11, 1303. [Google Scholar] [CrossRef]
- Blagojevic, D.P.; Grubor-Lajsic, G.N.; Spasic, M.B. Cold defence responses: The role of oxidative stress. Front. Biosci. 2011, 3, 416–427. [Google Scholar] [CrossRef]
- Pandey, S.; Fartyal, D.; Agarwal, A.; Shukla, T.; James, D.; Kaul, T.; Negi, Y.K.; Arora, S.; Reddy, M.K. Abiotic Stress Tolerance in Plants: Myriad Roles of Ascorbate Peroxidase. Front. Plant Sci. 2017, 8, 581. [Google Scholar] [CrossRef]
- Rehman, S.; Rashid, A.; Manzoor, M.A.; Li, L.; Sun, W.; Riaz, M.W.; Li, D.; Zhuge, Q. Genome-Wide Evolution and Comparative Analysis of Superoxide Dismutase Gene Family in Cucurbitaceae and Expression Analysis of Lagenaria siceraria Under Multiple Abiotic Stresses. Front. Genet. 2021, 12, 784878. [Google Scholar] [CrossRef]
- Wang, W.B.; Kim, Y.H.; Lee, H.S.; Deng, X.P.; Kwak, S.S.J.P.B.R. Differential antioxidation activities in two alfalfa cultivars under chilling stress. Plant Biotechnol. Rep. 2009, 3, 301–307. [Google Scholar] [CrossRef]
- Ma, Y.; Dai, X.; Xu, Y.; Luo, W.; Zheng, X.; Zeng, D.; Pan, Y.; Lin, X.; Liu, H.; Zhang, D.; et al. COLD1 Confers Chilling Tolerance in Rice. Cell 2015, 160, 1209–1221. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Ohta, M.; Kanrar, S.; Lee, B.H.; Hong, X.; Agarwal, M.; Zhu, J.K. ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003, 17, 1043–1054. [Google Scholar] [CrossRef]
- Jiang, D.; Gu, X.; He, Y. Establishment of the winter-annual growth habit via FRIGIDA-mediated histone methylation at FLOWERING LOCUS C in Arabidopsis. Plant Cell 2009, 21, 1733–1746. [Google Scholar] [CrossRef]
- Johanson, U.; West, J.; Lister, C.; Michaels, S.; Amasino, R.; Dean, C. Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 2000, 290, 344–347. [Google Scholar] [CrossRef]
- Michaels, S.D.; He, Y.; Scortecci, K.C.; Amasino, R.M. Attenuation of FLOWERING LOCUS C activity as a mechanism for the evolution of summer-annual flowering behavior in Arabidopsis. Proc. Natl. Acad. Sci. USA 2003, 100, 10102–10107. [Google Scholar] [CrossRef]
- Li, Z.; Jiang, D.; He, Y. FRIGIDA establishes a local chromosomal environment for FLOWERING LOCUS C mRNA production. Nat. Plants 2018, 4, 836–846. [Google Scholar] [CrossRef]
- Irwin, J.A.; Lister, C.; Soumpourou, E.; Zhang, Y.; Howell, E.C.; Teakle, G.; Dean, C. Functional alleles of the flowering time regulator FRIGIDA in the Brassica oleracea genome. BMC Plant Biol. 2012, 12, 21. [Google Scholar] [CrossRef]
- Choi, K.; Kim, J.; Hwang, H.J.; Kim, S.; Park, C.; Kim, S.Y.; Lee, I. The FRIGIDA complex activates transcription of FLC, a strong flowering repressor in Arabidopsis, by recruiting chromatin modification factors. Plant Cell 2011, 23, 289–303. [Google Scholar] [CrossRef]
- Csorba, T.; Questa, J.I.; Sun, Q.; Dean, C. Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proc. Natl. Acad. Sci. USA 2014, 111, 16160–16165. [Google Scholar] [CrossRef]
- Swiezewski, S.; Liu, F.; Magusin, A.; Dean, C. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 2009, 462, 799–802. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Angel, A.; Howard, M.; Dean, C. Vernalization—A cold-induced epigenetic switch. J. Cell Sci. 2012, 125, 3723–3731. [Google Scholar] [CrossRef] [PubMed]
- Yun, J.Y.; Tamada, Y.; Kang, Y.E.; Amasino, R.M. Arabidopsis trithorax-related3/SET domain GROUP2 is required for the winter-annual habit of Arabidopsis thaliana. Plant Cell Physiol. 2012, 53, 834–846. [Google Scholar] [CrossRef]
- Lee, I.; Bleecker, A.; Amasino, R. Analysis of naturally occurring late flowering in Arabidopsis thaliana. Mol. Gen. Genet. MGG 1993, 237, 171–176. [Google Scholar] [CrossRef]
- Clarke, J.H.; Dean, C. Mapping FRI, a locus controlling flowering time and vernalization response in Arabidopsis thaliana. Mol. Gen. Genet. MGG 1994, 242, 81–89. [Google Scholar] [CrossRef]
- Michaels, S.D.; Amasino, R.M. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 1999, 11, 949–956. [Google Scholar] [CrossRef]
- Michaels, S.D.; Himelblau, E.; Kim, S.Y.; Schomburg, F.M.; Amasino, R.M. Integration of flowering signals in winter-annual Arabidopsis. Plant Physiol. 2005, 137, 149–156. [Google Scholar] [CrossRef]
- Searle, I.; He, Y.; Turck, F.; Vincent, C.; Fornara, F.; Kröber, S.; Amasino, R.A.; Coupland, G. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 2006, 20, 898–912. [Google Scholar] [CrossRef]
- Deng, W.; Ying, H.; Helliwell, C.A.; Taylor, J.M.; Peacock, W.J.; Dennis, E.S. FLOWERING LOCUS C (FLC) regulates development pathways throughout the life cycle of Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 6680–6685. [Google Scholar] [CrossRef]
- Zografos, B.R.; Sung, S. Vernalization-mediated chromatin changes. J. Exp. Bot. 2012, 63, 4343–4348. [Google Scholar] [CrossRef]
- Sun, Q.; Csorba, T.; Skourti-Stathaki, K.; Proudfoot, N.J.; Dean, C. R-loop stabilization represses antisense transcription at the Arabidopsis FLC locus. Science 2013, 340, 619–621. [Google Scholar] [CrossRef]
- Rosa, S.; De Lucia, F.; Mylne, J.S.; Zhu, D.; Ohmido, N.; Pendle, A.; Kato, N.; Shaw, P.; Dean, C. Physical clustering of FLC alleles during Polycomb-mediated epigenetic silencing in vernalization. Genes Dev. 2013, 27, 1845–1850. [Google Scholar] [CrossRef]
- Sprent, J.I. 60Ma of legume nodulation. What’s new? What’s changing? J. Exp. Bot. 2008, 59, 1081–1084. [Google Scholar] [CrossRef]
- Guzzo, F.; Portaluppi, P.; Grisi, R.; Barone, S.; Zampieri, S.; Franssen, H.; Levi, M. Reduction of cell size induced by enod40 in Arabidopsis thaliana. J. Exp. Bot. 2005, 56, 507–513. [Google Scholar] [CrossRef]
- Chen, J.; Wang, Z.; Wang, L.; Hu, Y.; Yan, Q.; Lu, J.; Ren, Z.; Hong, Y.; Ji, H.; Wang, H.; et al. The B-type response regulator GmRR11d mediates systemic inhibition of symbiotic nodulation. Nat. Commun. 2022, 13, 7661. [Google Scholar] [CrossRef]
- Peters, N.K.; Verma, D.P. Phenolic compounds as regulators of gene expression in plant-microbe relations. Mol. Plant-Microbe Interact. MPMI 1990, 3, 4–8. [Google Scholar] [CrossRef]
- Ghantasala, S.; Roy Choudhury, S. Nod factor perception: An integrative view of molecular communication during legume symbiosis. Plant Mol. Biol. 2022, 110, 485–509. [Google Scholar] [CrossRef]
- D’Haeze, W.; Holsters, M. Nod factor structures, responses, and perception during initiation of nodule development. Glycobiology 2002, 12, 79r–105r. [Google Scholar] [CrossRef]
- Suzaki, T.; Nishida, H. Autoregulation of Legume Nodulation by Sophisticated Transcriptional Regulatory Networks. Mol. Plant 2019, 12, 1179–1181. [Google Scholar] [CrossRef]
- Journet, E.P.; El-Gachtouli, N.; Vernoud, V.; de Billy, F.; Pichon, M.; Dedieu, A.; Arnould, C.; Morandi, D.; Barker, D.G.; Gianinazzi-Pearson, V. Medicago truncatula ENOD11: A novel RPRP-encoding early nodulin gene expressed during mycorrhization in arbuscule-containing cells. Mol. Plant-Microbe Interact. MPMI 2001, 14, 737–748. [Google Scholar] [CrossRef]
- Journet, E.P.; Pichon, M.; Dedieu, A.; de Billy, F.; Truchet, G.; Barker, D.G. Rhizobium meliloti Nod factors elicit cell-specific transcription of the ENOD12 gene in transgenic alfalfa. Plant J. Cell Mol. Biol. 1994, 6, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Scheres, B.; Van De Wiel, C.; Zalensky, A.; Horvath, B.; Spaink, H.; Van Eck, H.; Zwartkruis, F.; Wolters, A.M.; Gloudemans, T.; Van Kammen, A.; et al. The ENOD12 gene product is involved in the infection process during the pea-Rhizobium interaction. Cell 1990, 60, 281–294. [Google Scholar] [CrossRef] [PubMed]
- Catoira, R.; Galera, C.; de Billy, F.; Penmetsa, R.V.; Journet, E.P.; Maillet, F.; Rosenberg, C.; Cook, D.; Gough, C.; Dénarié, J. Four genes of Medicago truncatula controlling components of a nod factor transduction pathway. Plant Cell 2000, 12, 1647–1666. [Google Scholar] [CrossRef]
- Oldroyd, G.E.; Long, S.R. Identification and characterization of nodulation-signaling pathway 2, a gene of Medicago truncatula involved in Nod actor signaling. Plant Physiol. 2003, 131, 1027–1032. [Google Scholar] [CrossRef]
- Ehrhardt, D.W.; Wais, R.; Long, S.R. Calcium spiking in plant root hairs responding to Rhizobium nodulation signals. Cell 1996, 85, 673–681. [Google Scholar] [CrossRef]
- Wais, R.J.; Galera, C.; Oldroyd, G.; Catoira, R.; Penmetsa, R.V.; Cook, D.; Gough, C.; Denarié, J.; Long, S.R. Genetic analysis of calcium spiking responses in nodulation mutants of Medicago truncatula. Proc. Natl. Acad. Sci. USA 2000, 97, 13407–13412. [Google Scholar] [CrossRef]
- Walker, S.A.; Viprey, V.; Downie, J.A. Dissection of nodulation signaling using pea mutants defective for calcium spiking induced by nod factors and chitin oligomers. Proc. Natl. Acad. Sci. USA 2000, 97, 13413–13418. [Google Scholar] [CrossRef]
- Miwa, H.; Sun, J.; Oldroyd, G.E.; Downie, J.A. Analysis of calcium spiking using a cameleon calcium sensor reveals that nodulation gene expression is regulated by calcium spike number and the developmental status of the cell. Plant J. Cell Mol. Biol. 2006, 48, 883–894. [Google Scholar] [CrossRef]
- Gleason, C.; Chaudhuri, S.; Yang, T.; Muñoz, A.; Poovaiah, B.W.; Oldroyd, G.E. Nodulation independent of rhizobia induced by a calcium-activated kinase lacking autoinhibition. Nature 2006, 441, 1149–1152. [Google Scholar] [CrossRef]
- Borisov, A.Y.; Madsen, L.H.; Tsyganov, V.E.; Umehara, Y.; Voroshilova, V.A.; Batagov, A.O.; Sandal, N.; Mortensen, A.; Schauser, L.; Ellis, N.; et al. The Sym35 gene required for root nodule development in pea is an ortholog of Nin from Lotus japonicus. Plant Physiol. 2003, 131, 1009–1017. [Google Scholar] [CrossRef]
- Marsh, J.F.; Rakocevic, A.; Mitra, R.M.; Brocard, L.; Sun, J.; Eschstruth, A.; Long, S.R.; Schultze, M.; Ratet, P.; Oldroyd, G.E. Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol. 2007, 144, 324–335. [Google Scholar] [CrossRef]
- Schauser, L.; Roussis, A.; Stiller, J.; Stougaard, J. A plant regulator controlling development of symbiotic root nodules. Nature 1999, 402, 191–195. [Google Scholar] [CrossRef]
- Miwa, H.; Sun, J.; Oldroyd, G.E.; Downie, J.A. Analysis of Nod-factor-induced calcium signaling in root hairs of symbiotically defective mutants of Lotus japonicus. Mol. Plant-Microbe Interact. MPMI 2006, 19, 914–923. [Google Scholar] [CrossRef]
- Zhenzhen, Q.; Lise, P.; Mehrnoush, N.R.; Marc, L.J.F.i.P.S. Comprehensive Comparative Genomic and Transcriptomic Analyses of the Legume Genes Controlling the Nodulation Process. Front. Plant Sci. 2016, 7, 34. [Google Scholar]
- Walsh, K.B.; Layzell, D.B. Carbon and nitrogen assimilation and partitioning in soybeans exposed to low root temperatures. Plant Physiol. 1986, 80, 249–255. [Google Scholar] [CrossRef]
- van Heerden, P.D.; Kiddle, G.; Pellny, T.K.; Mokwala, P.W.; Jordaan, A.; Strauss, A.J.; de Beer, M.; Schlüter, U.; Kunert, K.J.; Foyer, C.H. Regulation of respiration and the oxygen diffusion barrier in soybean protect symbiotic nitrogen fixation from chilling-induced inhibition and shoots from premature senescence. Plant Physiol. 2008, 148, 316–327. [Google Scholar] [CrossRef]
- Allen, D.J.; Ort, D.R. Impacts of chilling temperatures on photosynthesis in warm-climate plants. Trends Plant Sci. 2001, 6, 36–42. [Google Scholar] [CrossRef]
- Kingston-Smith, A.H.; Harbinson, J.; Williams, J.; Foyer, C.H. Effect of Chilling on Carbon Assimilation, Enzyme Activation, and Photosynthetic Electron Transport in the Absence of Photoinhibition in Maize Leaves. Plant Physiol. 1997, 114, 1039–1046. [Google Scholar] [CrossRef]
- Matamoros, M.A.; Baird, L.M.; Escuredo, P.R.; Dalton, D.A.; Minchin, F.R.; Iturbe-Ormaetxe, I.; Rubio, M.C.; Moran, J.F.; Gordon, A.J.; Becana, M. Stress-induced legume root nodule senescence. Physiological, biochemical, and structural alterations. Plant Physiol. 1999, 121, 97–112. [Google Scholar] [CrossRef]
- Gibson, A.H. Factors in the physical and biological environment affecting nodulation and nitrogen fixation by legumes. Plant Soil 1971, 35, 139–152. [Google Scholar] [CrossRef]
- Funatsuki, H.; Ohnishi, S.J.J.A.R.Q. Recent Advances in Physiological and Genetic Studies on Chilling Tolerance in Soybean. Jpn. Agric. Res. Q. 2012, 43, 95–101. [Google Scholar] [CrossRef]
- Kurosaki, H.; Yumoto, S.J.P.P.S. Effects of Low Temperature and Shading during Flowering on the Yield Components in Soybeans. Plant Prod. Sci. 2003, 6, 17–23. [Google Scholar] [CrossRef]
- Ohnishi, S.; Miyoshi, T.; Shirai, S.J.E.; Botany, E. Low temperature stress at different flower developmental stages affects pollen development, pollination, and pod set in soybean. Environ. Exp. Bot. 2010, 69, 56–62. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, Y.; Li, K.; Zou, Y.; Chen, L.; Li, X. Identification of Cold-Responsive miRNAs and Their Target Genes in Nitrogen-Fixing Nodules of Soybean. Int. J. Mol. Sci. 2014, 15, 13596–13614. [Google Scholar] [CrossRef]
- Manasa, L.S.; Panigrahy, M.; Panigrahi, K.C.; Rout, G.R. Overview of Cold Stress Regulation in Plants. Bot. Rev. 2022, 88, 359–387. [Google Scholar] [CrossRef]
- Høgslund, N.; Radutoiu, S.; Krusell, L.; Voroshilova, V.; Hannah, M.A.; Goffard, N.; Sanchez, D.H.; Lippold, F.; Ott, T.; Sato, S.; et al. Dissection of symbiosis and organ development by integrated transcriptome analysis of lotus japonicus mutant and wild-type plants. PLoS ONE 2009, 4, e6556. [Google Scholar] [CrossRef]
- De Luis, A.; Markmann, K.; Cognat, V.; Holt, D.B.; Charpentier, M.; Parniske, M.; Stougaard, J.; Voinnet, O. Two microRNAs linked to nodule infection and nitrogen-fixing ability in the legume Lotus japonicus. Plant Physiol. 2012, 160, 2137–2154. [Google Scholar] [CrossRef]
- Chen, W.; Wang, P.; Wang, D.; Shi, M.; Xia, Y.; He, Q.; Dang, J.; Guo, Q.; Jing, D.; Liang, G. EjFRI, FRIGIDA (FRI) Ortholog from Eriobotrya japonica, Delays Flowering in Arabidopsis. Int. J. Mol. Sci. 2020, 21, 1087. [Google Scholar] [CrossRef]
- Kong, X.; Luo, L.; Zhao, J.; Chen, Q.; Chang, G.; Huang, J.; Yang, Y.; Hu, X. Expression of FRIGIDA in root inhibits flowering in Arabidopsis thaliana. J. Exp. Bot. 2019, 70, 5101–5114. [Google Scholar] [CrossRef]
- Michaels, S.D.; Amasino, R.M. Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell 2001, 13, 935–941. [Google Scholar] [CrossRef]
- Sheldon, C.C.; Burn, J.E.; Perez, P.P.; Metzger, J.; Edwards, J.A.; Peacock, W.J.; Dennis, E.S. The FLF MADS box gene: A repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 1999, 11, 445–458. [Google Scholar] [CrossRef]
- Patra, R.K.; Pant, L.M.; Pradhan, K. Response of soybean to inoculation with rhizobial strains: Effect on growth, yield, N uptake and soil N status. World J. Agric. Sci. 2012, 8, 51–54. [Google Scholar]
- Geraldo, N.; Bäurle, I.; Kidou, S.; Hu, X.; Dean, C. FRIGIDA delays flowering in Arabidopsis via a cotranscriptional mechanism involving direct interaction with the nuclear cap-binding complex. Plant Physiol. 2009, 150, 1611–1618. [Google Scholar] [CrossRef]
- Duzan, H.M.; Mabood, F.; Souleimanov, A.; Smith, D.L. Nod Bj-V (C18:1, MeFuc) production by Bradyrhizobium japonicum (USDA110, 532C) at suboptimal growth temperatures. J. Plant Physiol. 2006, 163, 107–111. [Google Scholar] [CrossRef]
- Janczarek, M.; Adamczyk, P.; Gromada, A.; Polakowski, C.; Wengerska, K.; Bieganowski, A. Adaptation of Rhizobium leguminosarum sv. trifolii strains to low temperature stress in both free-living stage and during symbiosis with clover. Sci. Total Environ. 2024, 951, 175554. [Google Scholar] [CrossRef]
- Poustini, K.; Mabood, F.; Smith, D.L. Low root zone temperature effects on bean (Phaseolus vulgaris L.) plants inoculated with Rhizobium leguminosarum bv. phaseoli pre-incubated with methyl jasmonate and/or genistein. Acta Agric. Scand. Sect. B Soil Plant Sci. 2005, 55, 293–298. [Google Scholar] [CrossRef]
- Wang, Y.; Li, P.; Cao, X.; Wang, X.; Zhang, A.; Li, X. Identification and expression analysis of miRNAs from nitrogen-fixing soybean nodules. Biochem. Biophys. Res. Commun. 2009, 378, 799–803. [Google Scholar] [CrossRef]
- Wang, X.; Chang, X.; Jing, Y.; Zhao, J.; Fang, Q.; Sun, M.; Zhang, Y.; Li, W.; Li, Y. Identification and functional prediction of soybean CircRNAs involved in low-temperature responses. J. Plant Physiol. 2020, 250, 153188. [Google Scholar] [CrossRef]
- Hussain, M.A.; Li, S.; Gao, H.; Feng, C.; Sun, P.; Sui, X.; Jing, Y.; Xu, K.; Zhou, Y.; Zhang, W.; et al. Comparative analysis of physiological variations and genetic architecture for cold stress response in soybean germplasm. Front. Plant Sci. 2022, 13, 1095335. [Google Scholar] [CrossRef]
- Dong, Z.; Wang, H.; Li, X.; Ji, H. Enhancement of plant cold tolerance by soybean RCC1 family gene GmTCF1a. BMC Plant Biol. 2021, 21, 369. [Google Scholar] [CrossRef]
- Hammad, M.O. Simplified protocol modification of TRIzol method for extraction of high-quality RNA yield from RNase-rich rat pancreas. Process Biochem. 2023, 130, 464–471. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Jian, B.; Liu, B.; Bi, Y.; Hou, W.; Wu, C.; Han, T. Validation of internal control for gene expression study in soybean by quantitative real-time PCR. BMC Mol. Biol. 2008, 9, 59. [Google Scholar] [CrossRef]
- Gao, J. Guide to Experiments in Plant Physiology; Institution of Higher Education Press: Beijing, China, 2006. [Google Scholar]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.; He, L.; Li, H.; Tao, N.; Chang, T.; Wang, D.; Lu, Y.; Li, Z.; Mai, C.; Zhao, X.; et al. Role of GmFRI-1 in Regulating Soybean Nodule Formation Under Cold Stress. Int. J. Mol. Sci. 2025, 26, 879. https://doi.org/10.3390/ijms26030879
Zhang H, He L, Li H, Tao N, Chang T, Wang D, Lu Y, Li Z, Mai C, Zhao X, et al. Role of GmFRI-1 in Regulating Soybean Nodule Formation Under Cold Stress. International Journal of Molecular Sciences. 2025; 26(3):879. https://doi.org/10.3390/ijms26030879
Chicago/Turabian StyleZhang, Hongcai, Lin He, Huiyun Li, Nengfu Tao, Tianda Chang, Dongmei Wang, Yichu Lu, Zhenying Li, Chunhai Mai, Xiaorui Zhao, and et al. 2025. "Role of GmFRI-1 in Regulating Soybean Nodule Formation Under Cold Stress" International Journal of Molecular Sciences 26, no. 3: 879. https://doi.org/10.3390/ijms26030879
APA StyleZhang, H., He, L., Li, H., Tao, N., Chang, T., Wang, D., Lu, Y., Li, Z., Mai, C., Zhao, X., Niu, B., Ma, J., & Wang, L. (2025). Role of GmFRI-1 in Regulating Soybean Nodule Formation Under Cold Stress. International Journal of Molecular Sciences, 26(3), 879. https://doi.org/10.3390/ijms26030879





