Proteomic Analysis of Midgut of Silkworm Reared on Artificial Diet and Mulberry Leaves and Functional Study of Three UGT Genes
Abstract
1. Introduction
2. Results
2.1. Proteome Differential Expression Analysis
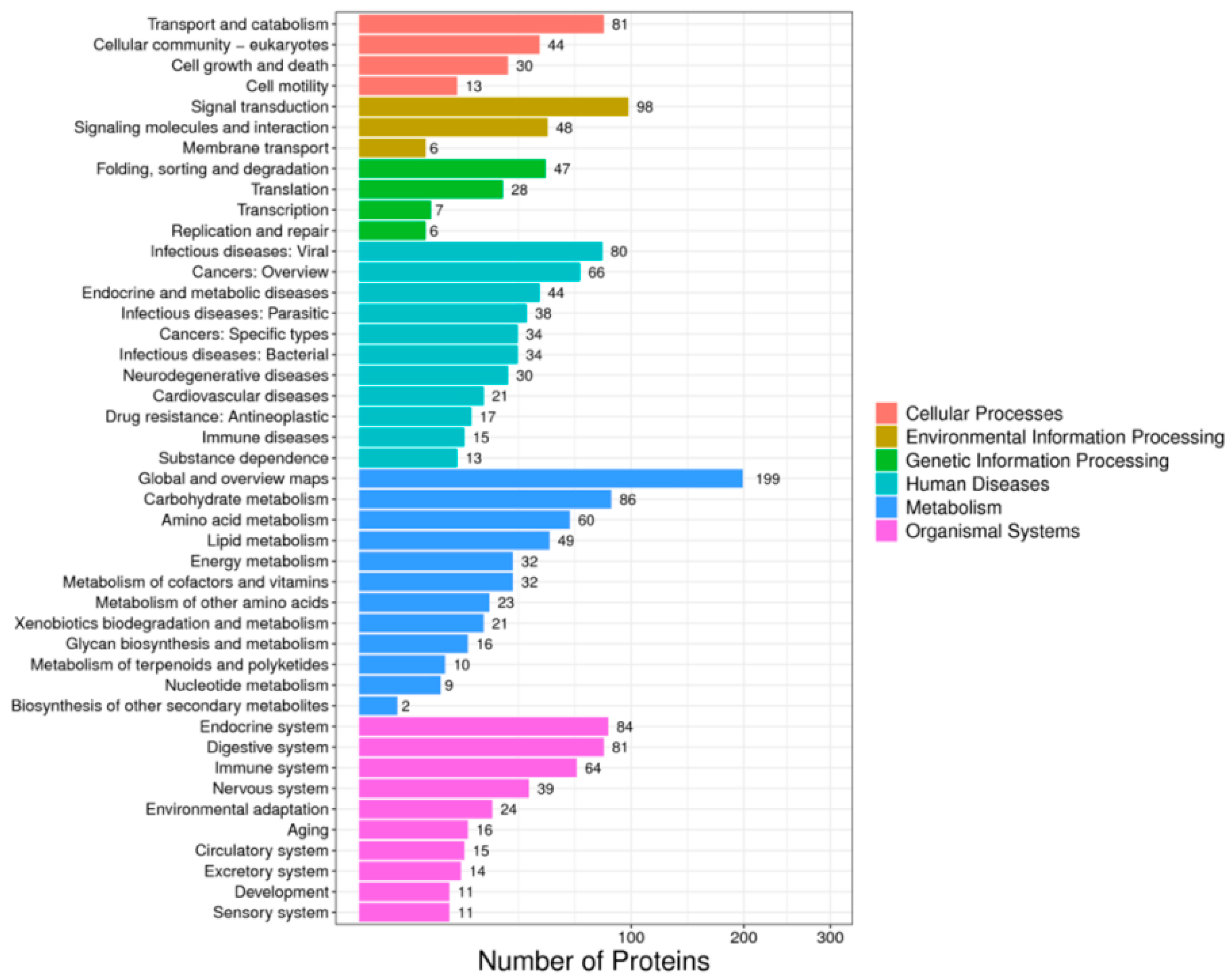
2.2. Statistical Analysis of Differential Protein KEGG Classification
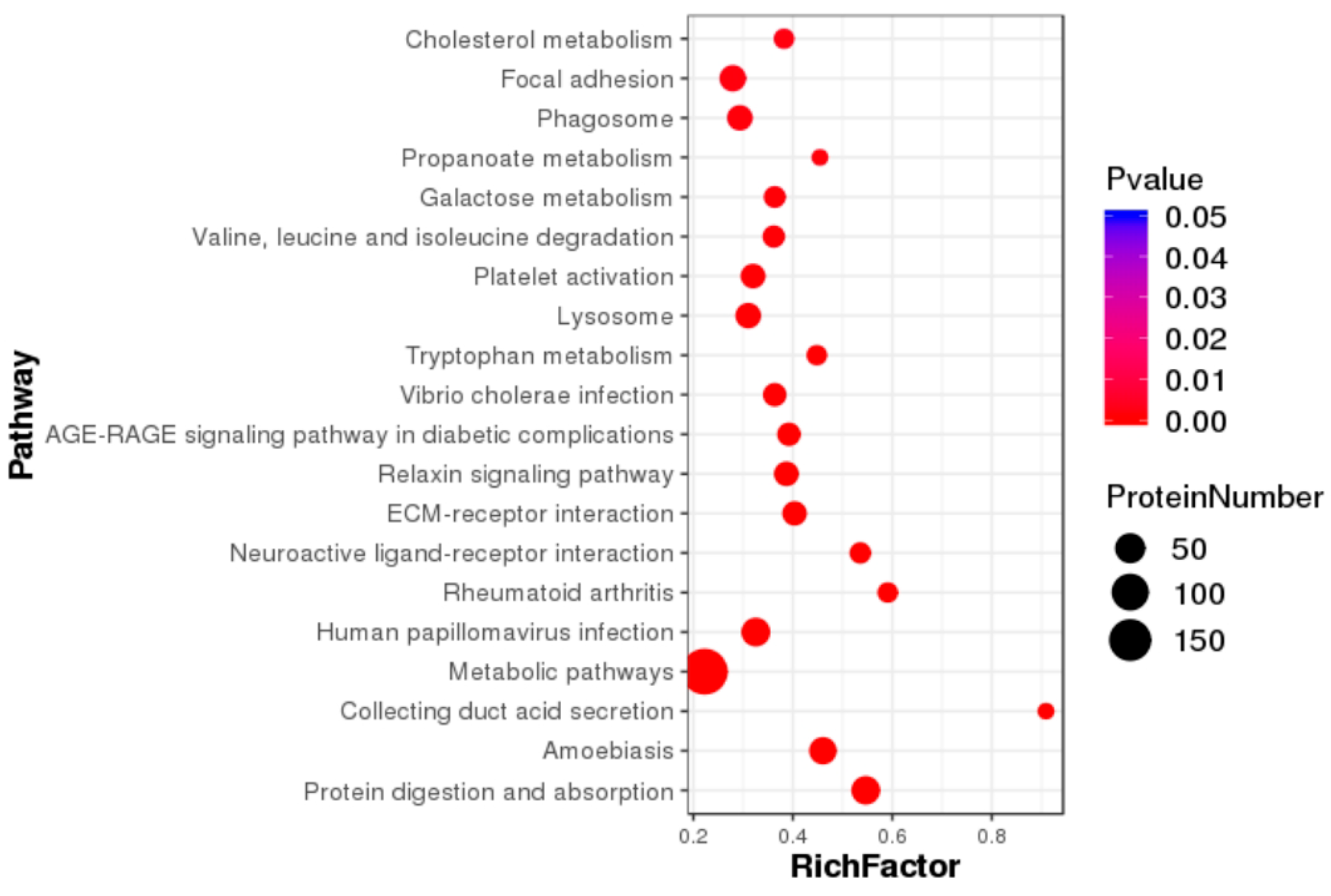
2.3. KEGG Enrichment Analysis of Differential Proteins
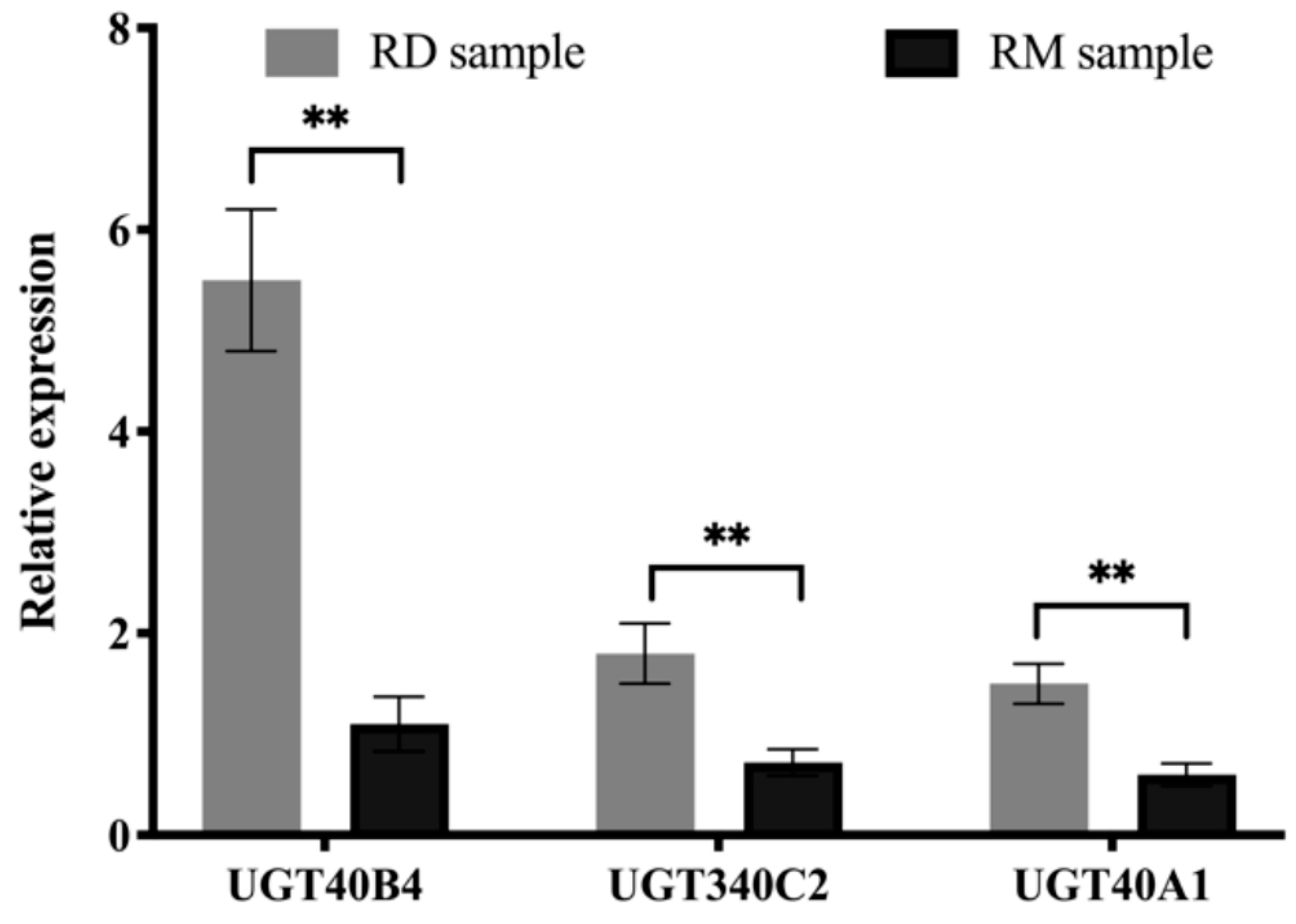
2.4. RT-qPCR Validation Analysis
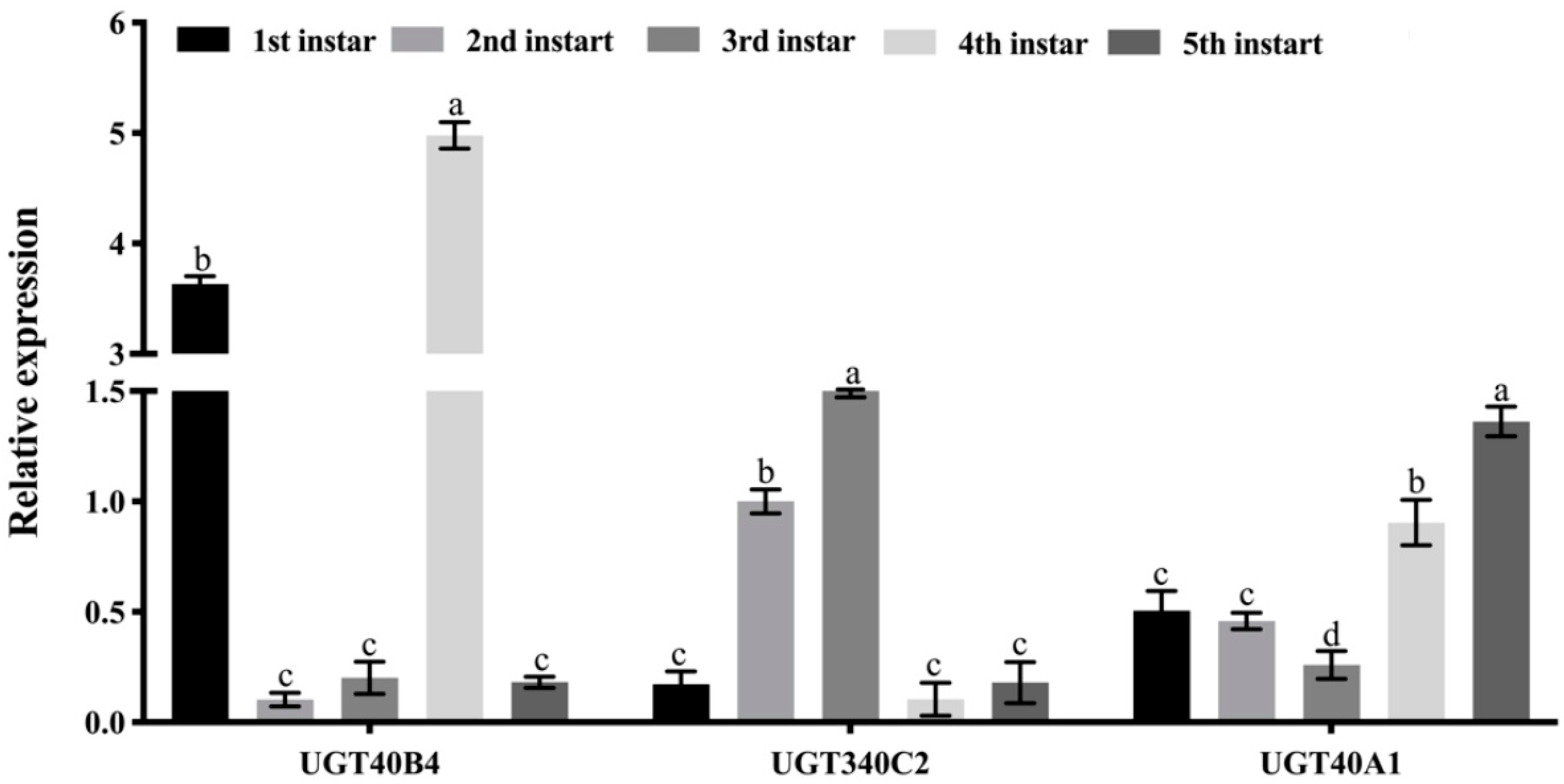
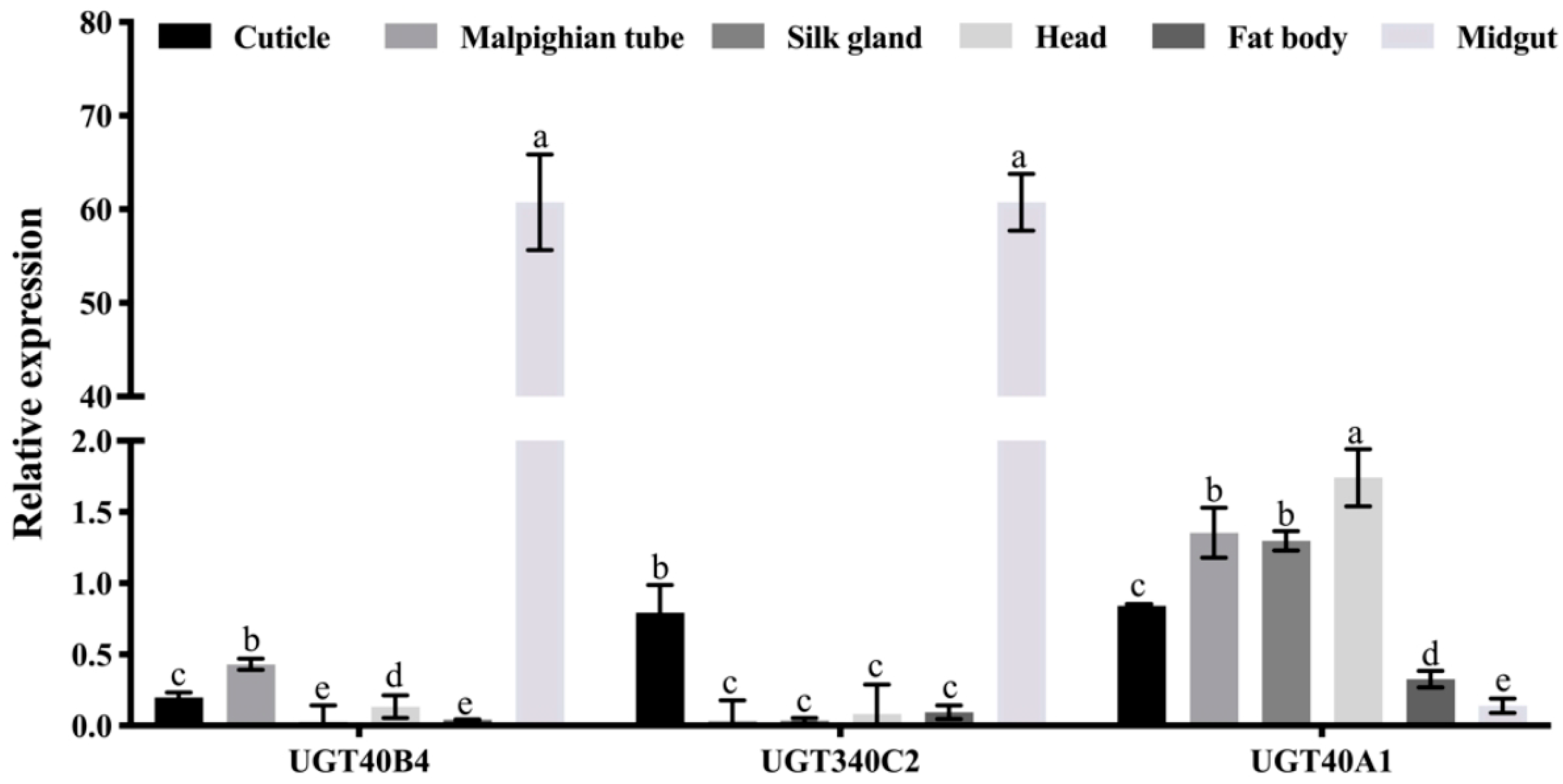
2.5. UGT Gene Expression Profile Analysis
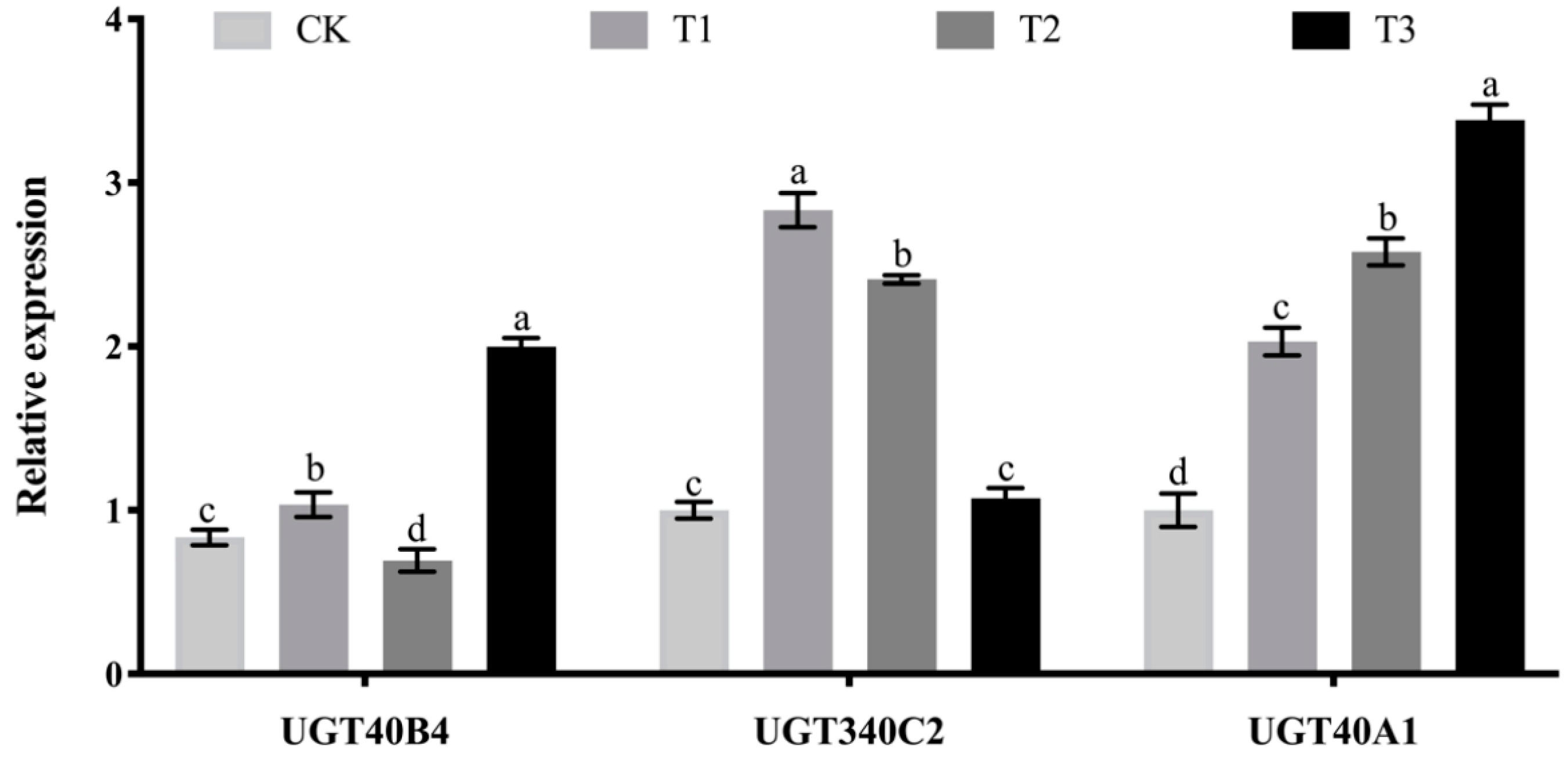
2.6. Analysis of the Effect of Adding Soybean Isoflavones Test
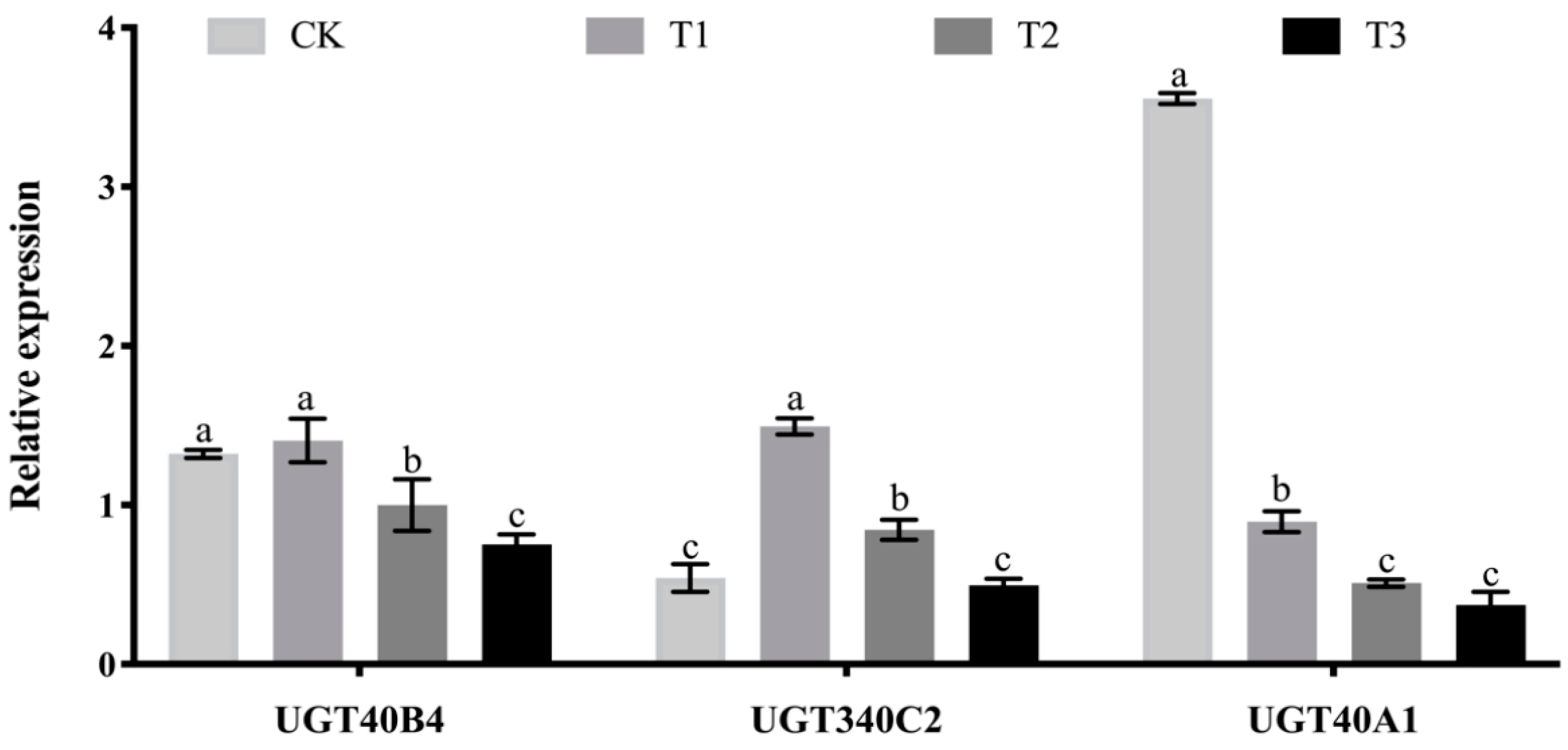
2.7. Analysis of the Effect of Adding Tannic Acid Test
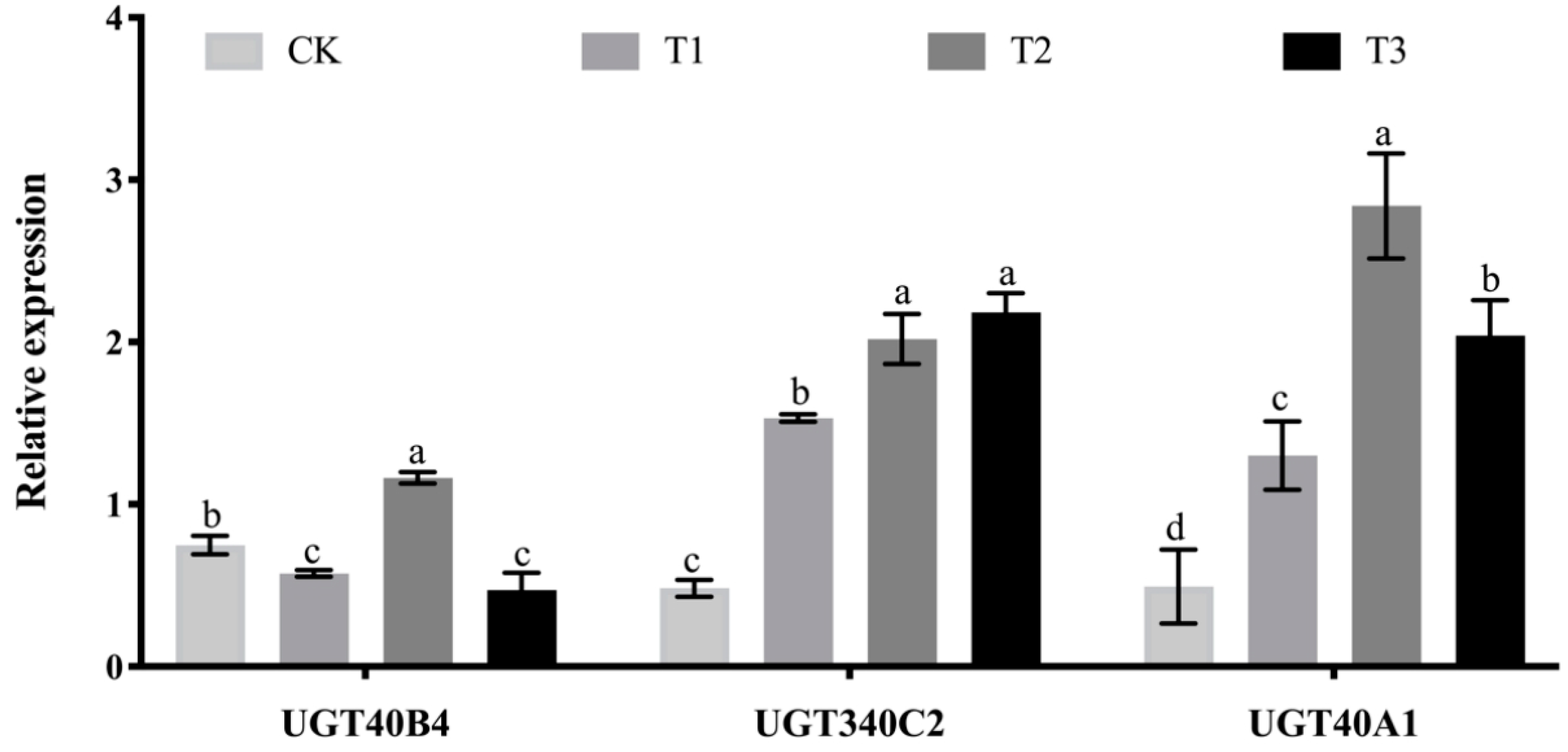
2.8. Analysis of the Effect of Adding Arabinoxylan Test
3. Discussion
4. Materials and Methods
4.1. Test Silkworm Varieties and Feed
4.2. Sample Processing Materials
4.3. Fluorescence Quantitative PCR
4.4. Semi-Synthetic Artificial Feed Feeding Test Materials
4.5. Experiment on Adding Soybean Isoflavones to Semi-Synthetic Feed
4.6. Test of Adding Tannic Acid in Semi-Synthetic Feed
4.7. Experiment on Adding Arabinoxylan to Semi-Synthetic Feed
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Habeanu, M.; Gheorghe, A.; Mihalcea, T. Nutritional Value of Silkworm Pupae (Bombyx mori) with Emphases on Fatty Acids Profile and Their Potential Applications for Humans and Animals. Insects 2023, 14, 254. [Google Scholar] [CrossRef] [PubMed]
- Panthee, S.; Paudel, A.; Hamamoto, H.; Sekimizu, K. Advantages of the Silkworm as an Animal Model for Developing Novel Antimicrobial Agents. Front. Microbiol. 2017, 8, 373. [Google Scholar] [CrossRef] [PubMed]
- Nwibo, D.D.; Hamamoto, H.; Matsumoto, Y.; Kaito, C.; Sekimizu, K. Current use of silkworm larvae (Bombyx mori) as an animal model in pharmaco-medical research. Drug Discov. Ther. 2015, 9, 133–135. [Google Scholar] [CrossRef] [PubMed]
- Xia, Q.; Guo, Y.; Zhang, Z.; Li, D.; Xuan, Z.; Li, Z.; Dai, F.; Li, Y.; Cheng, D.; Li, R.; et al. Complete resequencing of 40 genomes reveals domestication events and genes in silkworm (Bombyx). Science 2009, 326, 433–436. [Google Scholar] [CrossRef]
- Hamamura, Y. Food selection by silkworm larvae. Nature 1959, 183, 1746–1747. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Li, H.; Hao, F.; Li, N.; Liu, X.; Wang, G.; Wang, Y.; Tang, H. Developmental Changes for the Hemolymph Metabolome of Silkworm (Bombyx mori L.). J. Proteome Res. 2015, 14, 2331–2347. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.-h.; Yang, H.-j.; Chen, M.; Lou, C.-f.; Zhang, Y.-z.; Chen, K.-p.; Wang, Y.; Yu, M.-l.; Yu, F.; Li, J.-y.; et al. Comparative Proteomic Analysis between the Domesticated Silkworm (Bombyx mori) Reared on Fresh Mulberry Leaves and on Artificial Diet. J. Proteome Res. 2008, 7, 5103–5111. [Google Scholar] [CrossRef] [PubMed]
- Hayashiya, K.; Naito, K.i.; Nishida, J.; Hamamura, Y. Silkworm Rearing on Artificial Diet. J. Agric. Chem. Soc. Jpn. 1963, 37, 735–737. [Google Scholar] [CrossRef]
- Cyranoski, D. Silkworm genome gets solid coverage. Nature 2004. [Google Scholar] [CrossRef]
- Goldsmith, M.R.; Shimada, T.; Abe, H. The genetics and genomics of the silkworm, Bombyx mori. Annu. Rev. Entomol. 2005, 50, 71–100. [Google Scholar] [CrossRef] [PubMed]
- Sehadova, H.; Zavodska, R.; Rouhova, L.; Zurovec, M.; Sauman, I. The Role of Filippi’s Glands in the Silk Moths Cocoon Construction. Int. J. Mol. Sci. 2021, 22, 13523. [Google Scholar] [CrossRef] [PubMed]
- Ge, Q.; Cao, W.; Zhu, F.; Yuan, Y.; Chen, L.; Xu, J.; Li, J.; Chen, H.; Ma, S.; Sun, L.; et al. Genomics and proteomics combined analysis revealed the toxicity response of silkworm Bombyx mori to the environmental pathogen Bacillus cereus ZJ-4. Ecotoxicol. Environ. Saf. 2021, 222, 112467. [Google Scholar] [CrossRef] [PubMed]
- Meech, R.; Hu, D.G.; McKinnon, R.A.; Mubarokah, S.N.; Haines, A.Z.; Nair, P.C.; Rowland, A.; Mackenzie, P.I. The UDP-Glycosyltransferase (UGT) Superfamily: New Members, New Functions, and Novel Paradigms. Physiol. Rev. 2019, 99, 1153–1222. [Google Scholar] [CrossRef]
- Wang, H.; Song, J.; Hunt, B.J.; Zuo, K.; Zhou, H.; Hayward, A.; Li, B.; Xiao, Y.; Geng, X.; Bass, C.; et al. UDP-glycosyltransferases act as key determinants of host plant range in generalist and specialist Spodoptera species. Proc. Natl. Acad. Sci. USA 2024, 121, e2402045121. [Google Scholar] [CrossRef]
- Yuan, X.; Li, R.; He, W.; Xu, W.; Xu, W.; Yan, G.; Xu, S.; Chen, L.; Feng, Y.; Li, H. Progress in Identification of UDP-Glycosyltransferases for Ginsenoside Biosynthesis. J. Nat. Prod. 2024, 87, 1246–1267. [Google Scholar] [CrossRef]
- Wang, S.; Huang, W.; Li, M.; Wang, N.; Liu, X.; Chen, M.; Peng, X. RpUGT344J7 is involved in the reproduction switch of Rhopalosiphum padi with holocyclic life cycle. Insect Sci. 2024, 31, 1073–1089. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.F.; Chai, C.L.; Zhang, Z.; Liu, Z.H.; Dai, F.Y.; Lu, C.; Xiang, Z.H. The UDP-glucosyltransferase multigene family in Bombyx mori. BMC Genom. 2008, 9, 563. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.J.; Vogel, H.; Heckel, D.G. Comparative analysis of the UDP-glycosyltransferase multigene family in insects. Insect Biochem. Mol. Biol. 2012, 42, 133–147. [Google Scholar] [CrossRef]
- Yu, B.; Yang, Q.; Wei, J.; Pan, G.; Li, C.; Zhou, Z. UDP-Glucosyltransferases Induced by Nosema bombycis Provide Resistance to Microsporidia in Silkworm (Bombyx mori). Insects 2021, 12, 799. [Google Scholar] [CrossRef]
- Xu, X.; Wang, M.; Wang, Y.; Sima, Y.; Zhang, D.; Li, J.; Yin, W.; Xu, S. Green cocoons in silkworm Bombyx mori resulting from the quercetin 5-O-glucosyltransferase of UGT86, is an evolved response to dietary toxins. Mol. Biol. Rep. 2013, 40, 3631–3639. [Google Scholar] [CrossRef] [PubMed]
- Xin, S.; Zhang, W. Construction and analysis of the protein-protein interaction network for the olfactory system of the silkworm Bombyx mori. Arch. Insect Biochem. Physiol. 2020, 105, e21737. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Montalvo, A.D.; Collins, J.M.; Wang, D. Quantitative analysis of the UDP-glucuronosyltransferase transcriptome in human tissues. Pharmacol. Res. Perspect. 2023, 11, e01154. [Google Scholar] [CrossRef] [PubMed]
- Al-Yazeedi, T.; Muhammad, A.; Irving, H.; Ahn, S.J.; Hearn, J.; Wondji, C.S. Overexpression and nonsynonymous mutations of UDP-glycosyltransferases are potentially associated with pyrethroid resistance in Anopheles funestus. Genomics 2024, 116, 110798. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Liu, X.; Tang, H.; Li, M.; Gao, P.; Peng, X.; Chen, M. UGT2B13 and UGT2C1 are involved in lambda-cyhalothrin resistance in Rhopalosiphum padi. Pestic. Biochem. Physiol. 2023, 194, 105528. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Xiao, T.; Lu, K. Contribution of UDP-glycosyltransferases to chlorpyrifos resistance in Nilaparvata lugens. Pestic. Biochem. Physiol. 2023, 190, 105321. [Google Scholar] [CrossRef]
- Zeng, X.; Pan, Y.; Tian, F.; Li, J.; Xu, H.; Liu, X.; Chen, X.; Gao, X.; Peng, T.; Bi, R.; et al. Functional validation of key cytochrome P450 monooxygenase and UDP-glycosyltransferase genes conferring cyantraniliprole resistance in Aphis gossypii Glover. Pestic. Biochem. Physiol. 2021, 176, 104879. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.W.; Xing, X.R.; Wu, F.A.; Wang, J.; Sheng, S. UDP-glycosyltransferases contribute to the tolerance of parasitoid wasps towards insecticides. Pestic. Biochem. Physiol. 2021, 179, 104967. [Google Scholar] [CrossRef]
- Chen, X.; Xia, J.; Shang, Q.; Song, D.; Gao, X. UDP-glucosyltransferases potentially contribute to imidacloprid resistance in Aphis gossypii glover based on transcriptomic and proteomic analyses. Pestic. Biochem. Physiol. 2019, 159, 98–106. [Google Scholar] [CrossRef]
- Chen, N.; Miao, P.P.; Guo, C.E.; Chen, H.Y.; Ma, P.K.; Li, H.P.; Zhu, H.Y.; Gao, X.; Zhang, Y.J. In vitro effects of Genkwa Flos chloroform extract on activity of human liver microsomes UGTs and UGT1A1. Zhongguo Zhong Yao Za Zhi 2016, 41, 3296–3302. [Google Scholar] [CrossRef]
- Bozzolan, F.; Siaussat, D.; Maria, A.; Durand, N.; Pottier, M.A.; Chertemps, T.; Maibeche-Coisne, M. Antennal uridine diphosphate (UDP)-glycosyltransferases in a pest insect: Diversity and putative function in odorant and xenobiotics clearance. Insect Mol. Biol. 2014, 23, 539–549. [Google Scholar] [CrossRef] [PubMed]
- Ma, K.; Tang, Q.; Liang, P.; Li, J.; Gao, X. UDP-Glycosyltransferases from the UGT344 Family Are Involved in Sulfoxaflor Resistance in Aphis gossypii Glover. Insects 2021, 12, 356. [Google Scholar] [CrossRef] [PubMed]
- Rasouli, E.; Jahanian, R. Improved performance and immunological responses as the result of dietary genistein supplementation of broiler chicks. Animal 2015, 9, 1473–1480. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Luo, J.; Zhu, X.; Gao, X.; Hua, H.; Cui, J. Transcriptomic analysis of salivary gland and proteomic analysis of oral secretion in Helicoverpa armigera under cotton plant leaves, gossypol, and tannin stresses. Genomics 2022, 114, 110267. [Google Scholar] [CrossRef]
- Yang, Z.; Deng, M.; Wang, W.; Xiao, T.; Peng, H.; Huang, Z.; Lu, K. Characterization and functional analysis of UDP-glycosyltransferases reveal their contribution to phytochemical flavone tolerance in Spodoptera litura. Int. J. Biol. Macromol. 2024, 261, 129745. [Google Scholar] [CrossRef] [PubMed]
- Schupfer, E.; Pak, S.C.; Wang, S.; Micalos, P.S.; Jeffries, T.; Ooi, S.L.; Golombick, T.; Harris, G.; El-Omar, E. The effects and benefits of arabinoxylans on human gut microbiota—A narrative review. Food Biosci. 2021, 43, 101267. [Google Scholar] [CrossRef]
- Li, L.; Yan, S.; Liu, S.; Wang, P.; Li, W.; Yi, Y.; Qin, S. In-depth insight into correlations between gut microbiota and dietary fiber elucidates a dietary causal relationship with host health. Food Res. Int. 2023, 172, 113133. [Google Scholar] [CrossRef] [PubMed]
| Group | Weight (g) | Cocoon Weight (g) | Cocoon Layer Weight (g) | Pupae Weight (g) | Cocoon Layer Rate (%) |
|---|---|---|---|---|---|
| CK | 15.21 a | 1.41 d | 0.28 c | 1.13 d | 19.61 a |
| T1 | 11.35 b | 1.48 c | 0.29 bc | 1.19 c | 19.73 a |
| T2 | 11.98 b | 1.57 b | 0.31 b | 1.26 b | 19.80 a |
| T3 | 15.97 a | 1.64 a | 0.32 a | 1.32 a | 19.62 a |
| Group | Weight (g) | Cocoon Weight (g) | Cocoon Layer Weight (g) | Pupae Weight (g) | Cocoon Layer Rate (%) |
|---|---|---|---|---|---|
| CK | 19.09 a | 1.45 b | 0.25 b | 1.20 b | 17.76 a |
| T1 | 18.54 b | 1.60 a | 0.30 a | 1.30 a | 18.70 a |
| T2 | 14.51 c | 1.36 c | 0.21 c | 1.15 b | 15.50 a |
| T3 | 12.72 d |
| Group | Weight (g) | Cocoon Weight (g) | Cocoon Layer Weight (g) | Pupae Weight (g) | Cocoon Layer Rate (%) |
|---|---|---|---|---|---|
| CK | 15.21 c | 1.41 b | 0.28 b | 1.13 b | 19.61 a |
| T1 | 16.41 b | 1.53 a | 0.31 a | 1.22 a | 19.85 a |
| T2 | 17.99 a | 1.41 b | 0.27 b | 1.14 b | 19.39 a |
| T3 | 11.35 d | 1.29 c | 0.25 c | 1.04 c | 19.63 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, S.; Zhang, X.; Liang, J.; Huang, S.; Huang, B.; Ren, C.; Gao, H.; Liu, Q. Proteomic Analysis of Midgut of Silkworm Reared on Artificial Diet and Mulberry Leaves and Functional Study of Three UGT Genes. Int. J. Mol. Sci. 2025, 26, 1309. https://doi.org/10.3390/ijms26031309
Zhang S, Zhang X, Liang J, Huang S, Huang B, Ren C, Gao H, Liu Q. Proteomic Analysis of Midgut of Silkworm Reared on Artificial Diet and Mulberry Leaves and Functional Study of Three UGT Genes. International Journal of Molecular Sciences. 2025; 26(3):1309. https://doi.org/10.3390/ijms26031309
Chicago/Turabian StyleZhang, Shengxiang, Xinran Zhang, Jiawen Liang, Shuxian Huang, Bokai Huang, Chunjiu Ren, Huiju Gao, and Qingxin Liu. 2025. "Proteomic Analysis of Midgut of Silkworm Reared on Artificial Diet and Mulberry Leaves and Functional Study of Three UGT Genes" International Journal of Molecular Sciences 26, no. 3: 1309. https://doi.org/10.3390/ijms26031309
APA StyleZhang, S., Zhang, X., Liang, J., Huang, S., Huang, B., Ren, C., Gao, H., & Liu, Q. (2025). Proteomic Analysis of Midgut of Silkworm Reared on Artificial Diet and Mulberry Leaves and Functional Study of Three UGT Genes. International Journal of Molecular Sciences, 26(3), 1309. https://doi.org/10.3390/ijms26031309






