Multi-Omics Analysis Reveals Immune Infiltration and Clinical Significance of Phosphorylation Modification Enzymes in Lung Adenocarcinoma
Abstract
1. Introduction
2. Results
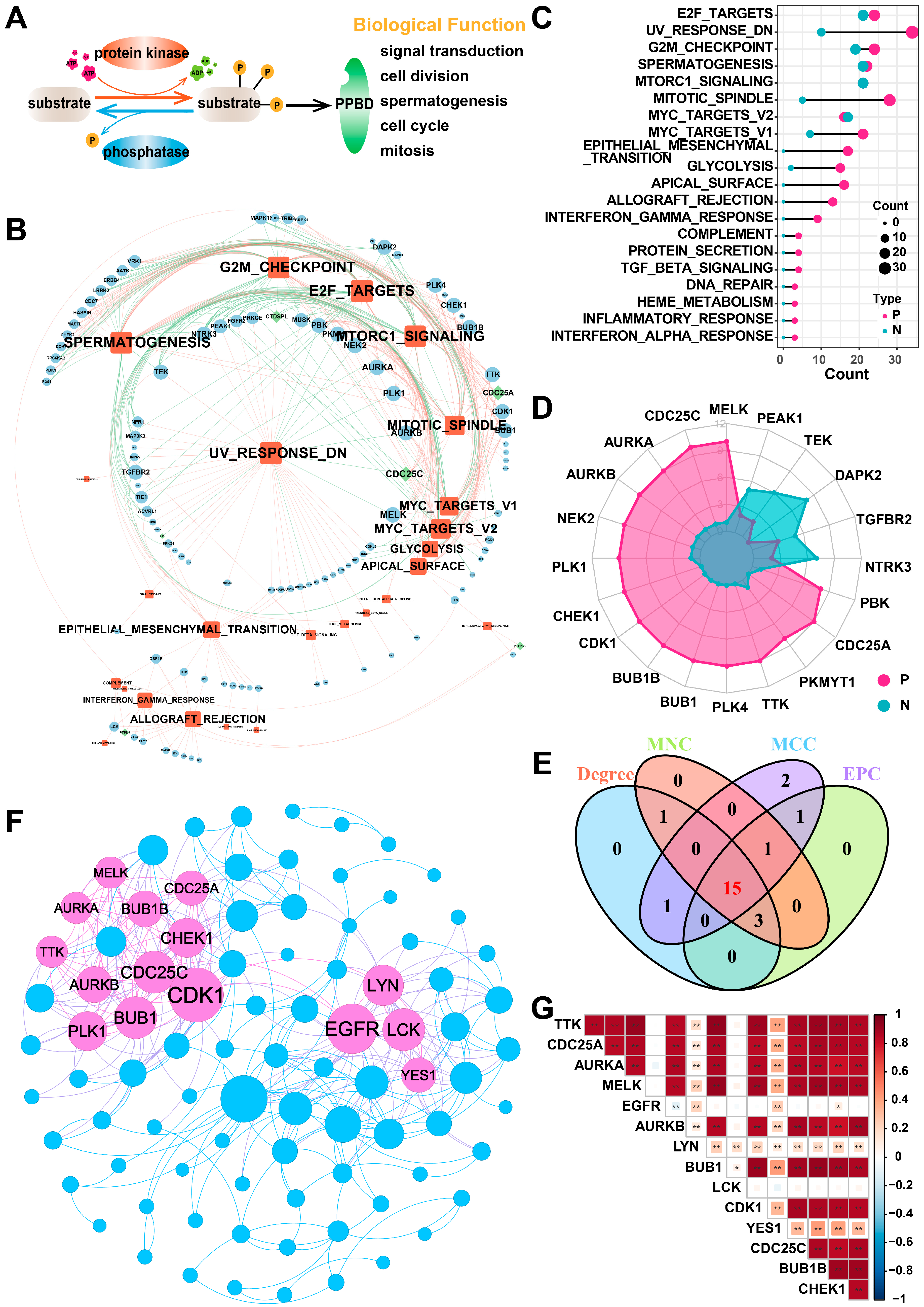
2.1. Identification of Hub PPMEs in Lung Adenocarcinoma
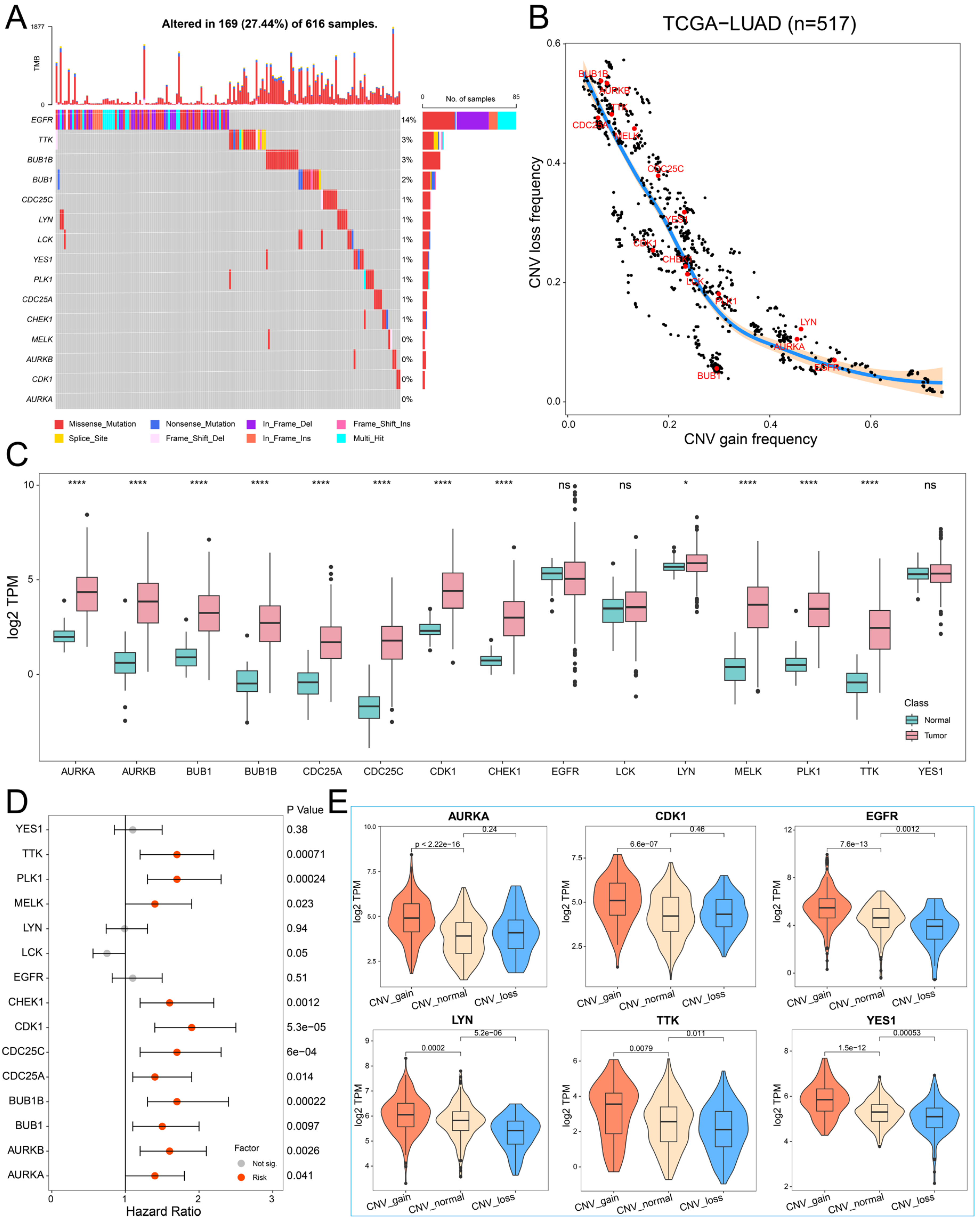
2.2. The Genetic Variation Landscape of Hub PPMEs in Lung Adenocarcinoma
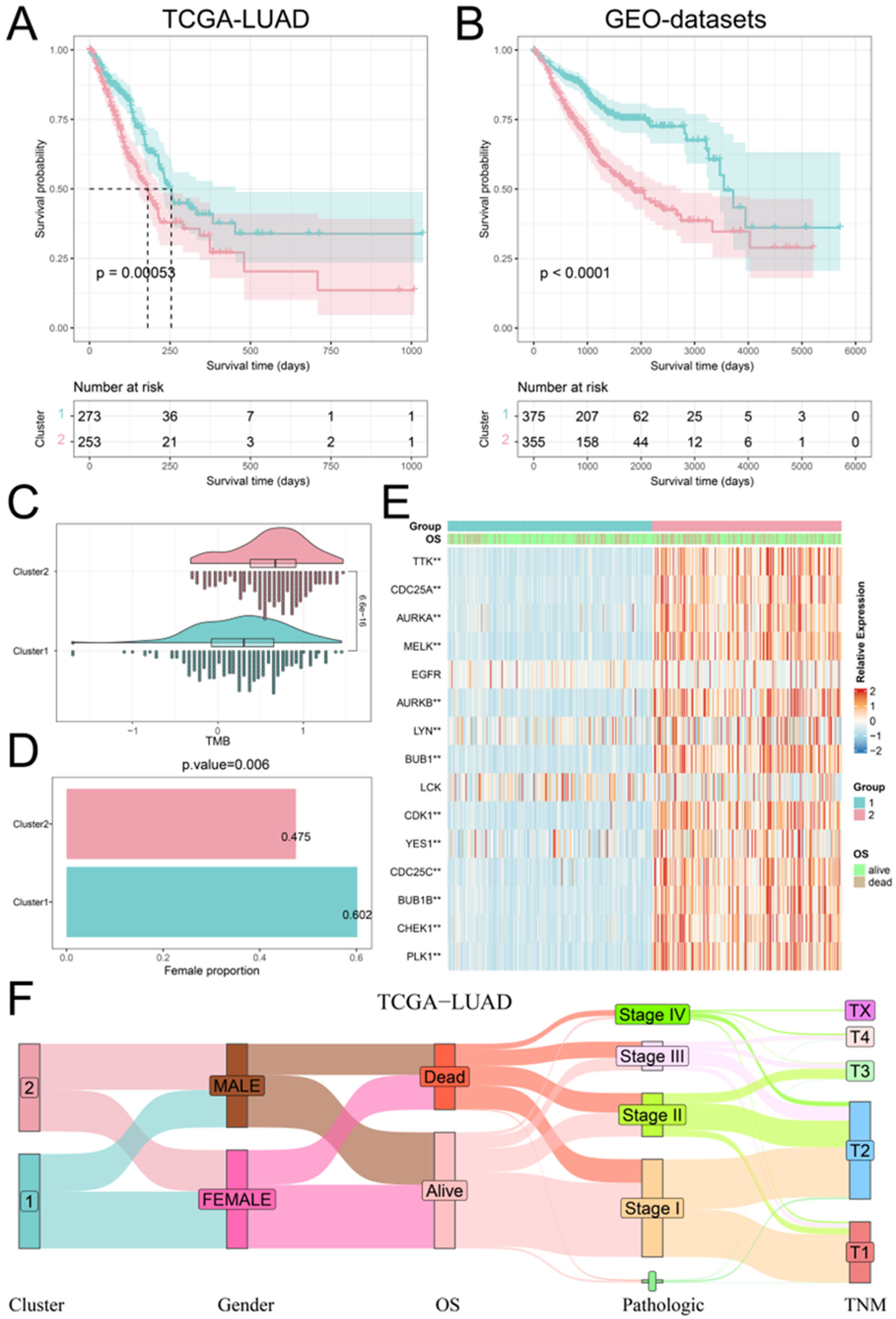
2.3. Recognition of Phosphorylation Modification Patterns Mediated by Hub PPMEs
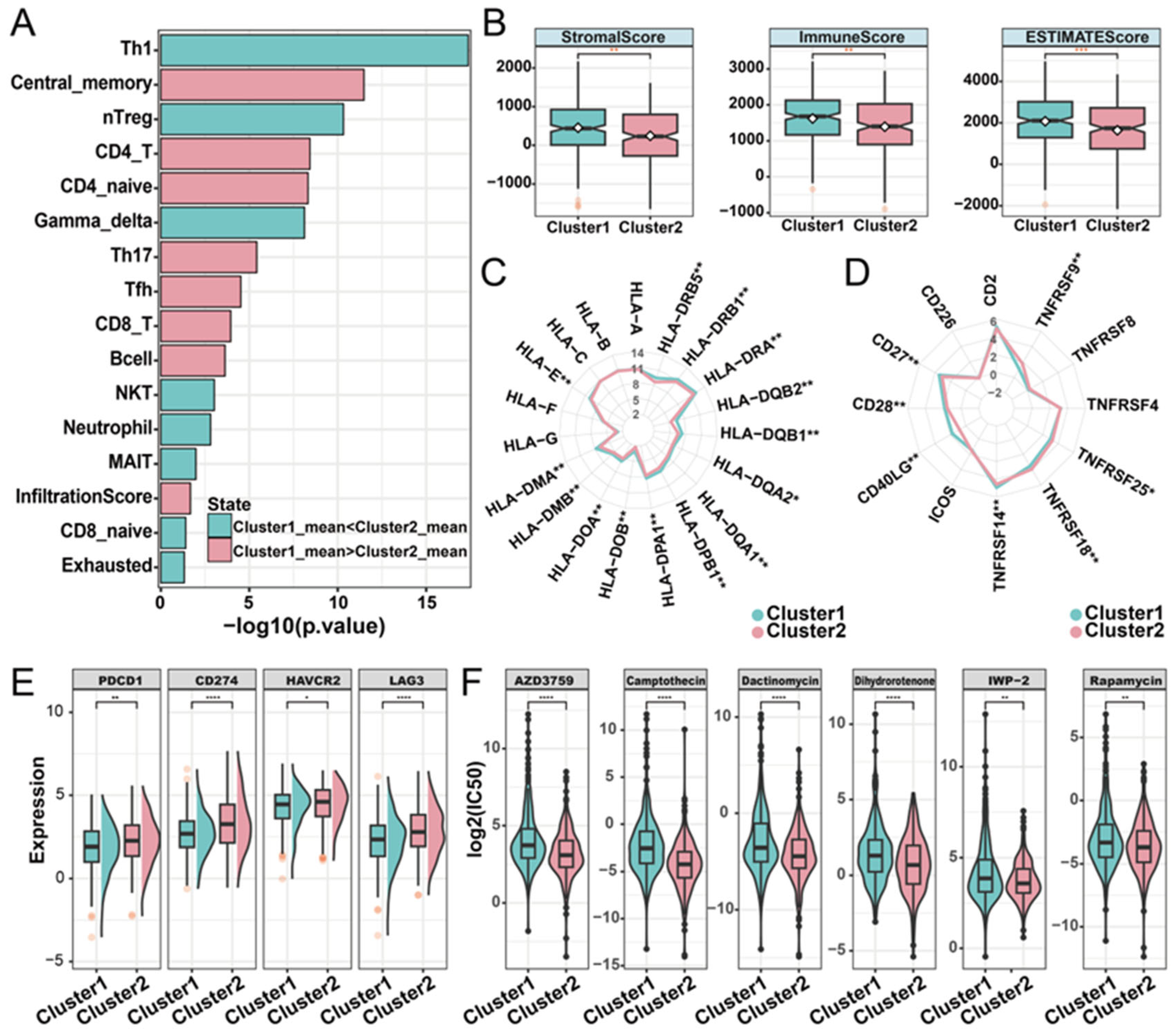
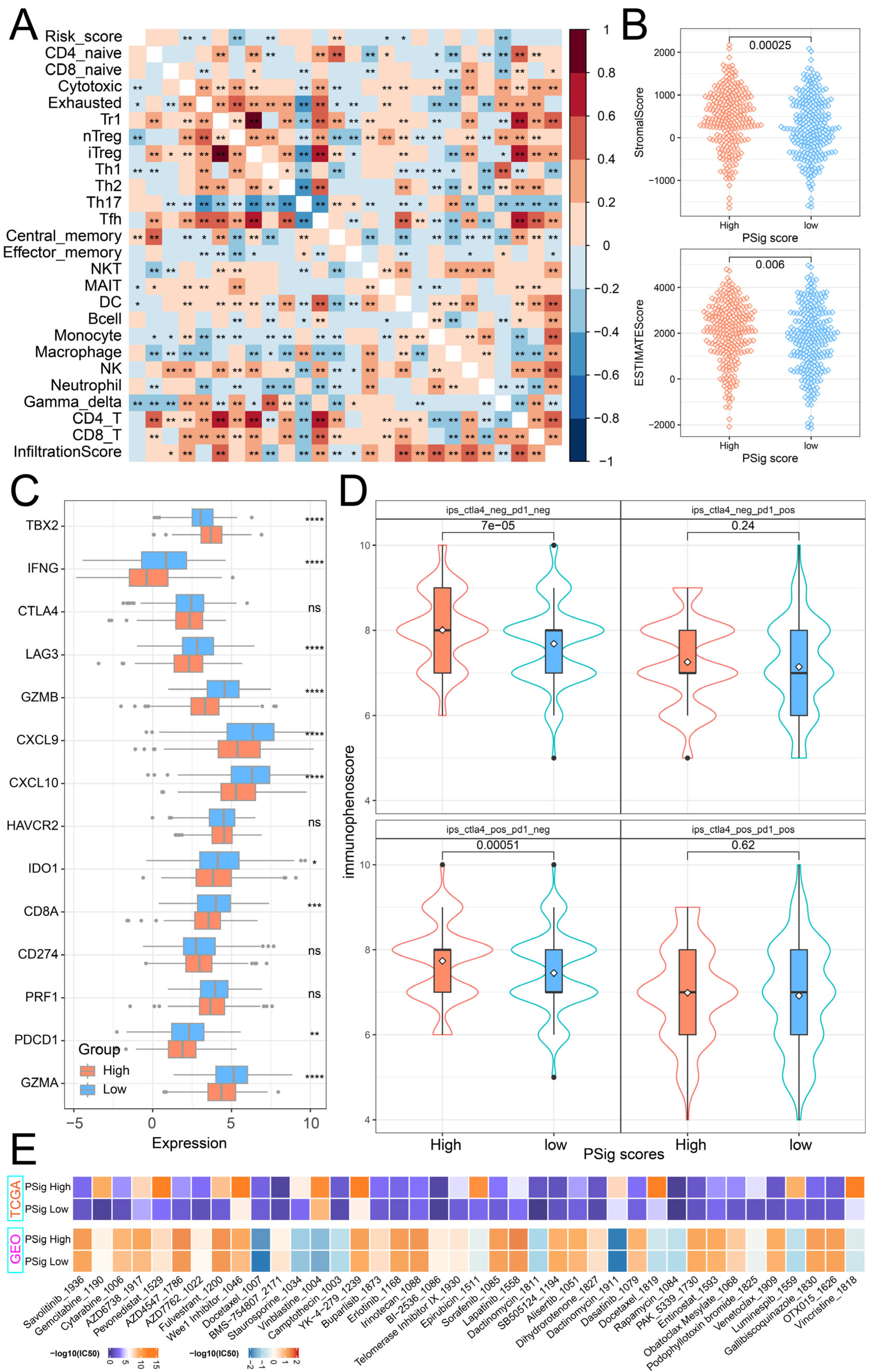
2.4. Distinct Immune Landscapes of Phosphorylation Modification Subtypes
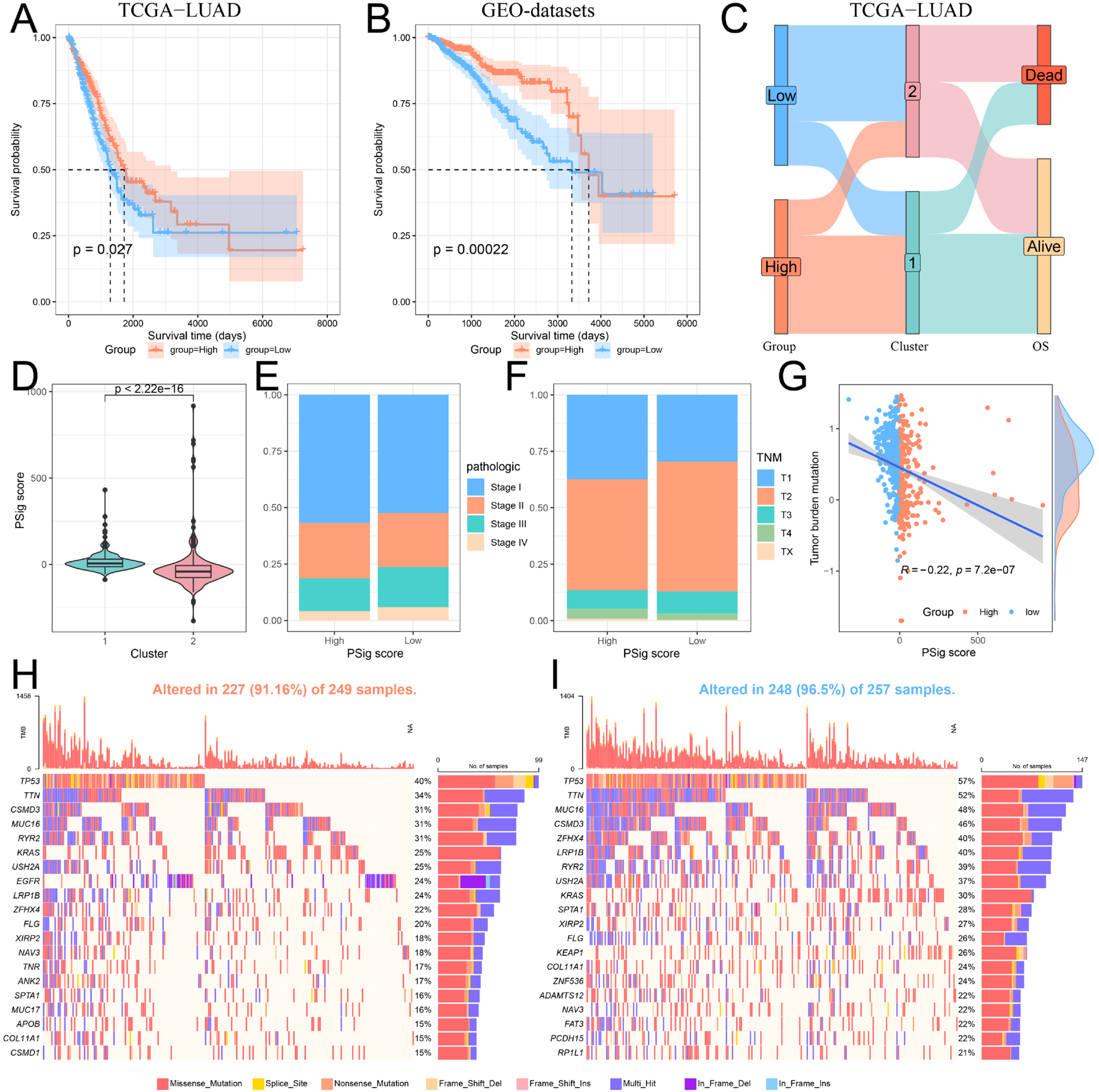
2.5. Construction and Functional Characterization of Hub PPME Signature Scores
2.6. The Role of PSig Scores in Predicting Immunotherapy and Chemotherapeutic Efficacy
3. Discussion
4. Materials and Methods
4.1. Sources and Preprocessing of Publicly Obtainable Datasets
4.2. Identification of Hub PPMEs
4.3. Genetic Variation Analysis of Hub PPMEs
4.4. Unsupervised Clustering and Survival Analysis
4.5. Immune Landscape of Phosphorylation Modification Subtypes
4.6. Construction of PSig Score
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Abbreviation | Full Name |
| BH | Benjamini and Hochberg |
| CNV | Copy number variation |
| CDF | Cumulative distribution function |
| CTLA-4 | Cytotoxic T lymphocyte antigen-4 |
| EPC | Edge percolated component |
| GSVA | Gene set variation analysis |
| GDC | Genomic data commons |
| GTEx | Genotype–Tissue Expression |
| GSK3β | Glycogen synthase kinase 3β |
| HPM | Human proteome map |
| ImmuCellAI | Immune cell abundance identifier |
| ICIs | Immune checkpoint inhibitors |
| IPS | Immunophenoscore |
| LUAD | Lung adenocarcinoma |
| MCC | Maximal clique centrality |
| MNC | Maximum neighborhood component |
| MSigDB | Molecular signatures database |
| NKT | Natural killer T |
| nTreg | Natural regulatory T cell |
| PCC | Pearson correlation coefficient |
| PPs | Phosphatases |
| PPBDs | Phosphoprotein binding domains |
| PCA | Principal component analysis |
| PD-L1 | Programmed death ligand 1 |
| PD-1 | Programmed death protein-1 |
| PKs | Protein kinases |
| PPME | Protein phosphorylation modification enzyme |
| PPI | Protein–protein interaction |
| RMA | Robust multichip average |
| Th17 | T helper 17 |
| TCIA | The Cancer Immunome Atlas |
| TME | Tumor microenvironment |
| TMB | Tumor mutation burden |
| US FDA | United States Food and Drug Administration |
References
- Niu, Z.Y.; Jin, R.S.; Zhang, Y.; Li, H.C. Signaling pathways and targeted therapies in lung squamous cell carcinoma: Mechanisms and clinical trials. Signal Transduct. Target. Ther. 2022, 7, 353. [Google Scholar] [CrossRef]
- Drosten, M.; Barbacid, M. Targeting KRAS mutant lung cancer: Light at the end of the tunnel. Mol. Oncol. 2022, 16, 1057–1071. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, F.R.; Scagliotti, G.V.; Mulshine, J.L.; Kwon, R.; Curran, W.J.; Wu, Y.-L.; Paz-Ares, L. Lung cancer: Current therapies and new targeted treatments. Lancet 2017, 389, 299–311. [Google Scholar] [CrossRef] [PubMed]
- Denisenko, T.V.; Budkevich, I.N.; Zhivotovsky, B. Cell death-based treatment of lung adenocarcinoma. Cell Death Dis. 2018, 9, 117. [Google Scholar] [CrossRef]
- Behrend, S.J.; Giotopoulou, G.A.; Spella, M.; Stathopoulos, G.T. A role for club cells in smoking-associated lung adenocarcinoma. Eur. Respir. Rev. 2021, 30, 210122. [Google Scholar] [CrossRef] [PubMed]
- Houston, K.A.; Henley, S.J.; Li, J.; White, M.C.; Richards, T.B. Patterns in lung cancer incidence rates and trends by histologic type in the United States, 2004–2009. Lung Cancer 2014, 86, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Kenfield, S.A.; Wei, E.K.; Stampfer, M.J.; Rosner, B.A.; Colditz, G.A. Comparison of aspects of smoking among the four histological types of lung cancer. Tob. Control 2008, 17, 198–204. [Google Scholar] [CrossRef]
- Luo, W.; Tian, P.; Wang, Y.; Xu, H.; Chen, L.; Tang, C.; Shu, Y.; Zhang, S.; Wang, Z.; Zhang, J.; et al. Characteristics of genomic alterations of lung adenocarcinoma in young never-smokers. Int. J. Cancer 2018, 143, 1696–1705. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Cui, Y.; Mo, Q.; Zhang, J.; Zhao, T.; Xu, Y.; Wu, Z.; Sun, D.; Zhang, X.; Li, Y.; et al. Characterization of m6A RNA Methylation Regulators Predicts Survival and Immunotherapy in Lung Adenocarcinoma. Front. Immunol. 2021, 12, 782551. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Fillmore, C.M.; Hammerman, P.S.; Kim, C.F.; Wong, K.-K. Non-small-cell lung cancers: A heterogeneous set of diseases. Nat. Rev. Cancer 2014, 14, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Hunter, T. A journey from phosphotyrosine to phosphohistidine and beyond. Mol. Cell 2022, 82, 2190–2200. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, S.J.; James, D.E.; Mann, M. Protein Phosphorylation: A Major Switch Mechanism for Metabolic Regulation. Trends Endocrinol. Metab. 2015, 26, 676–687. [Google Scholar] [CrossRef]
- Jin, J.; Pawson, T. Modular evolution of phosphorylation-based signalling systems. Philos. Trans. R. Soc. B Biol. Sci. 2012, 367, 2540–2555. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Peng, D.; Zhou, J.; Lin, S.; Wang, C.; Ning, W.; Xu, H.; Deng, W.; Xue, Y. iEKPD 2.0: An update with rich annotations for eukaryotic protein kinases, protein phosphatases and proteins containing phosphoprotein-binding domains. Nucleic Acids Res. 2019, 47, D344–D350. [Google Scholar] [CrossRef]
- Lim, W.A.; Pawson, T. Phosphotyrosine Signaling: Evolving a New Cellular Communication System. Cell 2010, 142, 661–667. [Google Scholar] [CrossRef]
- Wu, X.; Xu, M.; Geng, M.; Chen, S.; Little, P.J.; Xu, S.; Weng, J. Targeting protein modifications in metabolic diseases: Molecular mechanisms and targeted therapies. Signal Transduct. Target. Ther. 2023, 8, 220. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.-X.; Chen, Q.; Li, F.; Fu, S.; Huang, B.; Zhao, Y. Protein phosphorylation database and prediction tools. Brief. Bioinform. 2023, 24, bbad090. [Google Scholar] [CrossRef] [PubMed]
- Roskoski, R. Properties of FDA-approved small molecule protein kinase inhibitors: A 2023 update. Pharmacol. Res. 2023, 187, 106552. [Google Scholar] [CrossRef] [PubMed]
- Klaeger, S.; Heinzlmeir, S.; Wilhelm, M.; Polzer, H.; Vick, B.; Koenig, P.-A.; Reinecke, M.; Ruprecht, B.; Petzoldt, S.; Meng, C.; et al. The target landscape of clinical kinase drugs. Science 2017, 358, eaan4368. [Google Scholar] [CrossRef] [PubMed]
- Hoekstra, E.; Peppelenbosch, M.P.; Fuhler, G.M. Meeting Report Europhosphatase 2015: Phosphatases as Drug Targets in Cancer. Cancer Res. 2016, 76, 193–196. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Stanford, S.M.; Bottini, N. Targeting protein phosphatases in cancer immunotherapy and autoimmune disorders. Nat. Rev. Drug Discov. 2023, 22, 273–294. [Google Scholar] [CrossRef]
- Quail, D.F.; Joyce, J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013, 19, 1423–1437. [Google Scholar] [CrossRef]
- Marin-Acevedo, J.A.; Dholaria, B.; Soyano, A.E.; Knutson, K.L.; Chumsri, S.; Lou, Y. Next generation of immune checkpoint therapy in cancer: New developments and challenges. J. Hematol. Oncol. 2018, 11, 39. [Google Scholar] [CrossRef]
- Pardoll, D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 2012, 12, 252–264. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Allison, J.P. The future of immune checkpoint therapy. Science 2015, 348, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Goswami, S.; Raychaudhuri, D.; Siddiqui, B.A.; Singh, P.; Nagarajan, A.; Liu, J.; Subudhi, S.K.; Poon, C.; Gant, K.L.; et al. Immune checkpoint therapy—Current perspectives and future directions. Cell 2023, 186, 1652–1669. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Hassan, D.; Aldawsari, H.M.; Molugulu, N.; Shukla, R.; Kesharwani, P. Immune checkpoint inhibitors: A promising anticancer therapy. Drug Discov. Today 2020, 25, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Hayase, E.; Jenq, R.R. Role of the intestinal microbiome and microbial-derived metabolites in immune checkpoint blockade immunotherapy of cancer. Genome Med. 2021, 13, 107. [Google Scholar] [CrossRef] [PubMed]
- Hodi, F.S.; O’Day, S.J.; McDermott, D.F.; Weber, R.W.; Sosman, J.A.; Haanen, J.B.; Gonzalez, R.; Robert, C.; Schadendorf, D.; Hassel, J.C.; et al. Improved Survival with Ipilimumab in Patients with Metastatic Melanoma. N. Engl. J. Med. 2010, 363, 711–723. [Google Scholar] [CrossRef]
- Wolchok, J.D.; Kluger, H.; Callahan, M.K.; Postow, M.A.; Rizvi, N.A.; Lesokhin, A.M.; Segal, N.H.; Ariyan, C.E.; Gordon, R.-A.; Reed, K.; et al. Nivolumab plus Ipilimumab in Advanced Melanoma. N. Engl. J. Med. 2013, 369, 122–133. [Google Scholar] [CrossRef]
- Krummel, M.F.; Allison, J.P. Cd28 and Ctla-4 Have Opposing Effects on the Response of T-Cells to Stimulation. J. Exp. Med. 1995, 182, 459–465. [Google Scholar] [CrossRef] [PubMed]
- Freeman, G.J.; Long, A.J.; Iwai, Y.; Bourque, K.; Chernova, T.; Nishimura, H.; Fitz, L.J.; Malenkovich, N.; Okazaki, T.; Byrne, M.C.; et al. Engagement of the Pd-1 Immunoinhibitory Receptor by a Novel B7 Family Member Leads to Negative Regulation of Lymphocyte Activation. J. Exp. Med. 2000, 192, 1027–1034. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Gao, S. Comprehensive Analysis of Clinical Significance, Immune Infiltration and Biological Role of m6A Regulators in Early-Stage Lung Adenocarcinoma. Front. Immunol. 2021, 12, 698236. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Lim, S.-O.; Yamaguchi, H. Oncogenic signaling pathways associated with immune evasion and resistance to immune checkpoint inhibitors in cancer. Semin. Cancer Biol. 2020, 65, 51–64. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Bu, L.; Luo, J.; Guo, J. Targeting protein kinases benefits cancer immunotherapy. Biochim. Et Biophys. Acta (BBA)—Rev. Cancer 2022, 1877, 188738. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Vega, F.; Mina, M.; Armenia, J.; Chatila, W.K.; Luna, A.; La, K.C.; Dimitriadoy, S.; Liu, D.L.; Kantheti, H.S.; Saghafinia, S.; et al. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell 2018, 173, 321–337.e310. [Google Scholar] [CrossRef] [PubMed]
- Pan, S.; Chen, R. Pathological implication of protein post-translational modifications in cancer. Mol. Asp. Med. 2022, 86, 101097. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Qian, W.; Sun, X.; Jiang, S. Small-molecule inhibitors, immune checkpoint inhibitors, and more: FDA-approved novel therapeutic drugs for solid tumors from 1991 to 2021. J. Hematol. Oncol. 2022, 15, 143. [Google Scholar] [CrossRef]
- Yao, Y.; Wang, X.; Guan, J.; Xie, C.; Zhang, H.; Yang, J.; Luo, Y.; Chen, L.; Zhao, M.; Huo, B.; et al. Metabolomic differentiation of benign vs malignant pulmonary nodules with high specificity via high-resolution mass spectrometry analysis of patient sera. Nat. Commun. 2023, 14, 2339. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.; Li, W.; Zheng, X.; Ren, L.; Liu, J.; Li, S.; Wang, J.; Du, G. MELK is an oncogenic kinase essential for metastasis, mitotic progression, and programmed death in lung carcinoma. Signal Transduct. Target. Ther. 2020, 5, 279. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Humeau, J.; Buqué, A.; Zitvogel, L.; Kroemer, G. Immunostimulation with chemotherapy in the era of immune checkpoint inhibitors. Nat. Rev. Clin. Oncol. 2020, 17, 725–741. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Yang, M.; Wang, Q.; Song, F.; Li, X.; Chen, K. The new identified biomarkers determine sensitivity to immune check-point blockade therapies in melanoma. OncoImmunology 2019, 8, 1608132. [Google Scholar] [CrossRef]
- Chen, H.; Chong, W.; Wu, Q.; Yao, Y.; Mao, M.; Wang, X. Association of LRP1B Mutation With Tumor Mutation Burden and Outcomes in Melanoma and Non-small Cell Lung Cancer Patients Treated With Immune Check-Point Blockades. Front. Immunol. 2019, 10, 1113. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, R.; Hei, H. Advances in post-translational modifications of proteins and cancer immunotherapy. Front. Immunol. 2023, 14, 1229397. [Google Scholar] [CrossRef] [PubMed]
- Qian, G.; Guo, J.; Vallega, K.A.; Hu, C.; Chen, Z.; Deng, Y.; Wang, Q.; Fan, S.; Ramalingam, S.S.; Owonikoko, T.K.; et al. Membrane-Associated RING-CH 8 Functions as a Novel PD-L1 E3 Ligase to Mediate PD-L1 Degradation Induced by EGFR Inhibitors. Mol. Cancer Res. 2021, 19, 1622–1634. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Jain, A.D.; Truica, M.I.; Izquierdo-Ferrer, J.; Anker, J.F.; Lysy, B.; Sagar, V.; Luan, Y.; Chalmers, Z.R.; Unno, K.; et al. Small-Molecule MYC Inhibitors Suppress Tumor Growth and Enhance Immunotherapy. Cancer Cell 2019, 36, 483–497.e415. [Google Scholar] [CrossRef] [PubMed]
- Ni, W.; Mo, H.; Liu, Y.; Xu, Y.; Qin, C.; Zhou, Y.; Li, Y.; Li, Y.; Zhou, A.; Yao, S.; et al. Targeting cholesterol biosynthesis promotes anti-tumor immunity by inhibiting long noncoding RNA SNHG29-mediated YAP activation. Mol. Ther. 2021, 29, 2995–3010. [Google Scholar] [CrossRef] [PubMed]
- Tu, J.; Xu, H.; Ma, L.; Li, C.; Qin, W.; Chen, X.; Yi, M.; Sun, L.; Liu, B.; Yuan, X. Nintedanib enhances the efficacy of PD-L1 blockade by upregulating MHC-I and PD-L1 expression in tumor cells. Theranostics 2022, 12, 747–766. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Gao, Y.; Wei, W. Post-translational regulations of PD-L1 and PD-1: Mechanisms and opportunities for combined immunotherapy. Semin. Cancer Biol. 2022, 85, 246–252. [Google Scholar] [CrossRef]
- Tumbrink, H.L.; Heimsoeth, A.; Sos, M.L. The next tier of EGFR resistance mutations in lung cancer. Oncogene 2020, 40, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Foggetti, G.; Li, C.; Cai, H.; Hellyer, J.A.; Lin, W.-Y.; Ayeni, D.; Hastings, K.; Choi, J.; Wurtz, A.; Andrejka, L.; et al. Genetic Determinants of EGFR-Driven Lung Cancer Growth and Therapeutic Response In Vivo. Cancer Discov. 2021, 11, 1736–1753. [Google Scholar] [CrossRef]
- Fan, P.-D.; Narzisi, G.; Jayaprakash, A.D.; Venturini, E.; Robine, N.; Smibert, P.; Germer, S.; Yu, H.A.; Jordan, E.J.; Paik, P.K.; et al. YES1 amplification is a mechanism of acquired resistance to EGFR inhibitors identified by transposon mutagenesis and clinical genomics. Proc. Natl. Acad. Sci. USA 2018, 115, E6030–E6038. [Google Scholar] [CrossRef]
- Zhou, X.; Yuan, Y.; Kuang, H.; Tang, B.; Zhang, H.; Zhang, M. BUB1B (BUB1 Mitotic Checkpoint Serine/Threonine Kinase B) promotes lung adenocarcinoma by interacting with Zinc Finger Protein ZNF143 and regulating glycolysis. Bioengineered 2022, 13, 2471–2485. [Google Scholar] [CrossRef]
- Osmanbeyoglu, H.U.; Huang, K.; Zhang, Y.; Gong, H.; Qiao, Z.; Wang, T.; Zhao, W.; Huang, L.; Zhou, X. Inferring evolutionary trajectories from cross-sectional transcriptomic data to mirror lung adenocarcinoma progression. PLOS Comput. Biol. 2023, 19, e1011122. [Google Scholar] [CrossRef]
- Jang, H.-R.; Shin, S.-B.; Kim, C.-H.; Won, J.-Y.; Xu, R.; Kim, D.-E.; Yim, H. PLK1/vimentin signaling facilitates immune escape by recruiting Smad2/3 to PD-L1 promoter in metastatic lung adenocarcinoma. Cell Death Differ. 2021, 28, 2745–2764. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.-B.; Jang, H.-R.; Xu, R.; Won, J.-Y.; Yim, H. Active PLK1-driven metastasis is amplified by TGF-β signaling that forms a positive feedback loop in non-small cell lung cancer. Oncogene 2019, 39, 767–785. [Google Scholar] [CrossRef] [PubMed]
- Lv, G.; Shi, Q.; Zhang, T.; Li, J.; Kalashova, J.; Long, Y.; Sun, Y.; Li, C.; Choudhry, N.; Li, H.; et al. 2-Phenoxy-3, 4′-bipyridine derivatives inhibit AURKB-dependent mitotic processes by disrupting its localization. Eur. J. Med. Chem. 2023, 245, 114904. [Google Scholar] [CrossRef] [PubMed]
- Ribas, A.; Wolchok, J.D. Cancer immunotherapy using checkpoint blockade. Science 2018, 359, 1350–1355. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Yu, Q.; Song, T.; Wang, Z.; Song, L.; Yang, Y.; Shao, J.; Li, J.; Ni, Y.; Chao, N.; et al. The heterogeneous immune landscape between lung adenocarcinoma and squamous carcinoma revealed by single-cell RNA sequencing. Signal Transduct. Target. Ther. 2022, 7, 289. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Getz, G.; Wheeler, D.A.; Mardis, E.R.; McLellan, M.D.; Cibulskis, K.; Sougnez, C.; Greulich, H.; Muzny, D.M.; Morgan, M.B.; et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 2008, 455, 1069–1075. [Google Scholar] [CrossRef]
- Pleasance, E.D.; Cheetham, R.K.; Stephens, P.J.; McBride, D.J.; Humphray, S.J.; Greenman, C.D.; Varela, I.; Lin, M.-L.; Ordóñez, G.R.; Bignell, G.R.; et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature 2009, 463, 191–196. [Google Scholar] [CrossRef]
- Levantini, E.; Maroni, G.; Del Re, M.; Tenen, D.G. EGFR signaling pathway as therapeutic target in human cancers. Semin. Cancer Biol. 2022, 85, 253–275. [Google Scholar] [CrossRef]
- GTEx Consortium. Genetic effects on gene expression across human tissues. Nature 2017, 550, 204–213. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.-S.; Pinto, S.M.; Getnet, D.; Nirujogi, R.S.; Manda, S.S.; Chaerkady, R.; Madugundu, A.K.; Kelkar, D.S.; Isserlin, R.; Jain, S.; et al. A draft map of the human proteome. Nature 2014, 509, 575–581. [Google Scholar] [CrossRef] [PubMed]
- Colaprico, A.; Silva, T.C.; Olsen, C.; Garofano, L.; Cava, C.; Garolini, D.; Sabedot, T.S.; Malta, T.M.; Pagnotta, S.M.; Castiglioni, I.; et al. TCGAbiolinks: An R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016, 44, e71. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.; Mounir, M.; Lucchetta, M.; Silva, T.C.; Olsen, C.; Bontempi, G.; Chen, X.; Noushmehr, H.; Colaprico, A.; Papaleo, E. New functionalities in the TCGAbiolinks package for the study and integration of cancer data from GDC and GTEx. PLOS Comput. Biol. 2019, 15, e1006701. [Google Scholar] [CrossRef]
- Leek, J.T.; Johnson, W.E.; Parker, H.S.; Jaffe, A.E.; Storey, J.D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 2012, 28, 882–883. [Google Scholar] [CrossRef] [PubMed]
- Davis, S.; Meltzer, P.S. GEOquery: A bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007, 23, 1846–1847. [Google Scholar] [CrossRef] [PubMed]
- Selamat, S.A.; Chung, B.S.; Girard, L.; Zhang, W.; Zhang, Y.; Campan, M.; Siegmund, K.D.; Koss, M.N.; Hagen, J.A.; Lam, W.L.; et al. Genome-scale analysis of DNA methylation in lung adenocarcinoma and integration with mRNA expression. Genome Res 2012, 22, 1197–1211. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Foreman, O.; Wigle, D.A.; Kosari, F.; Vasmatzis, G.; Salisbury, J.L.; van Deursen, J.; Galardy, P.J. USP44 regulates centrosome positioning to prevent aneuploidy and suppress tumorigenesis. J Clin Invest 2012, 122, 4362–4374. [Google Scholar] [CrossRef]
- Director’s Challenge Consortium for the Molecular Classification of Lung Adenocarcinoma; Shedden, K.; Taylor, J.M.; Enkemann, S.A.; Tsao, M.S.; Yeatman, T.J.; Gerald, W.L.; Eschrich, S.; Jurisica, I.; Giordano, T.J.; et al. Gene expression-based survival prediction in lung adenocarcinoma: A multi-site, blinded validation study. Nat. Med. 2008, 14, 822–827. [Google Scholar] [CrossRef] [PubMed]
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-Seq data. Bmc Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef] [PubMed]
- Chin, C.H.; Chen, S.H.; Wu, H.H.; Ho, C.W.; Ko, M.T.; Lin, C.Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. Bmc Syst Biol 2014, 8, S11. [Google Scholar] [CrossRef] [PubMed]
- Mayakonda, A.; Lin, D.-C.; Assenov, Y.; Plass, C.; Koeffler, H.P. Maftools: Efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018, 28, 1747–1756. [Google Scholar] [CrossRef] [PubMed]
- Wilkerson, M.D.; Hayes, D.N. ConsensusClusterPlus: A class discovery tool with confidence assessments and item tracking. Bioinformatics 2010, 26, 1572–1573. [Google Scholar] [CrossRef]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.R.; Zhang, Q.; Lei, Q.; Luo, M.; Xie, G.Y.; Wang, H.; Guo, A.Y. ImmuCellAI: A Unique Method for Comprehensive T-Cell Subsets Abundance Prediction and its Application in Cancer Immunotherapy. Adv. Sci. 2020, 7, 1902880. [Google Scholar] [CrossRef]
- Yoshihara, K.; Shahmoradgoli, M.; Martínez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W.; Treviño, V.; Shen, H.; Laird, P.W.; Levine, D.A.; et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013, 4, 2612. [Google Scholar] [CrossRef]
- Maeser, D.; Gruener, R.F.; Huang, R.S. oncoPredict: An R package for predicting in vivo or cancer patient drug response and biomarkers from cell line screening data. Brief. Bioinform. 2021, 22, bbab260. [Google Scholar] [CrossRef]
- Zhang, B.; Wu, Q.; Li, B.; Wang, D.; Wang, L.; Zhou, Y.L. m6A regulator-mediated methylation modification patterns and tumor microenvironment infiltration characterization in gastric cancer. Mol. Cancer 2020, 19, 53. [Google Scholar] [CrossRef]
- Chong, W.; Shang, L.; Liu, J.; Fang, Z.; Du, F.; Wu, H.; Liu, Y.; Wang, Z.; Chen, Y.; Jia, S.; et al. m6A regulator-based methylation modification patterns characterized by distinct tumor microenvironment immune profiles in colon cancer. Theranostics 2021, 11, 2201–2217. [Google Scholar] [CrossRef] [PubMed]
- Charoentong, P.; Finotello, F.; Angelova, M.; Mayer, C.; Efremova, M.; Rieder, D.; Hackl, H.; Trajanoski, Z. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep. 2017, 18, 248–262. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Long, D.; Ding, Y.; Wang, P.; Wei, L.; Ma, K. Multi-Omics Analysis Reveals Immune Infiltration and Clinical Significance of Phosphorylation Modification Enzymes in Lung Adenocarcinoma. Int. J. Mol. Sci. 2025, 26, 1066. https://doi.org/10.3390/ijms26031066
Long D, Ding Y, Wang P, Wei L, Ma K. Multi-Omics Analysis Reveals Immune Infiltration and Clinical Significance of Phosphorylation Modification Enzymes in Lung Adenocarcinoma. International Journal of Molecular Sciences. 2025; 26(3):1066. https://doi.org/10.3390/ijms26031066
Chicago/Turabian StyleLong, Deyu, Yanheng Ding, Peng Wang, Lili Wei, and Ketao Ma. 2025. "Multi-Omics Analysis Reveals Immune Infiltration and Clinical Significance of Phosphorylation Modification Enzymes in Lung Adenocarcinoma" International Journal of Molecular Sciences 26, no. 3: 1066. https://doi.org/10.3390/ijms26031066
APA StyleLong, D., Ding, Y., Wang, P., Wei, L., & Ma, K. (2025). Multi-Omics Analysis Reveals Immune Infiltration and Clinical Significance of Phosphorylation Modification Enzymes in Lung Adenocarcinoma. International Journal of Molecular Sciences, 26(3), 1066. https://doi.org/10.3390/ijms26031066



