Rescuing Verubecestat: An Integrative Molecular Modeling and Simulation Approach for Designing Next-Generation BACE1 Inhibitors
Abstract
1. Introduction
2. Results
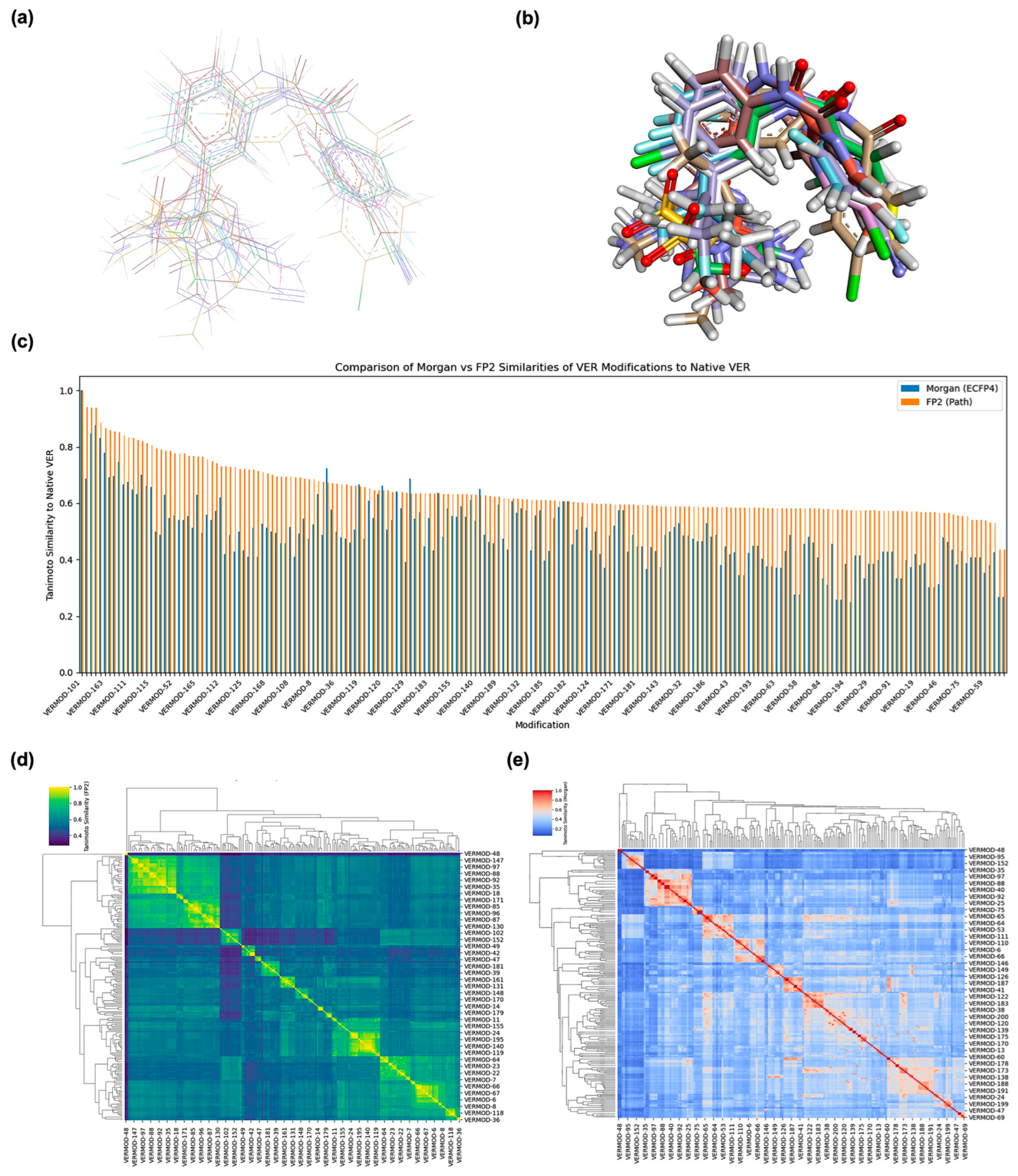
2.1. Structure Alignment and Similarity Analysis
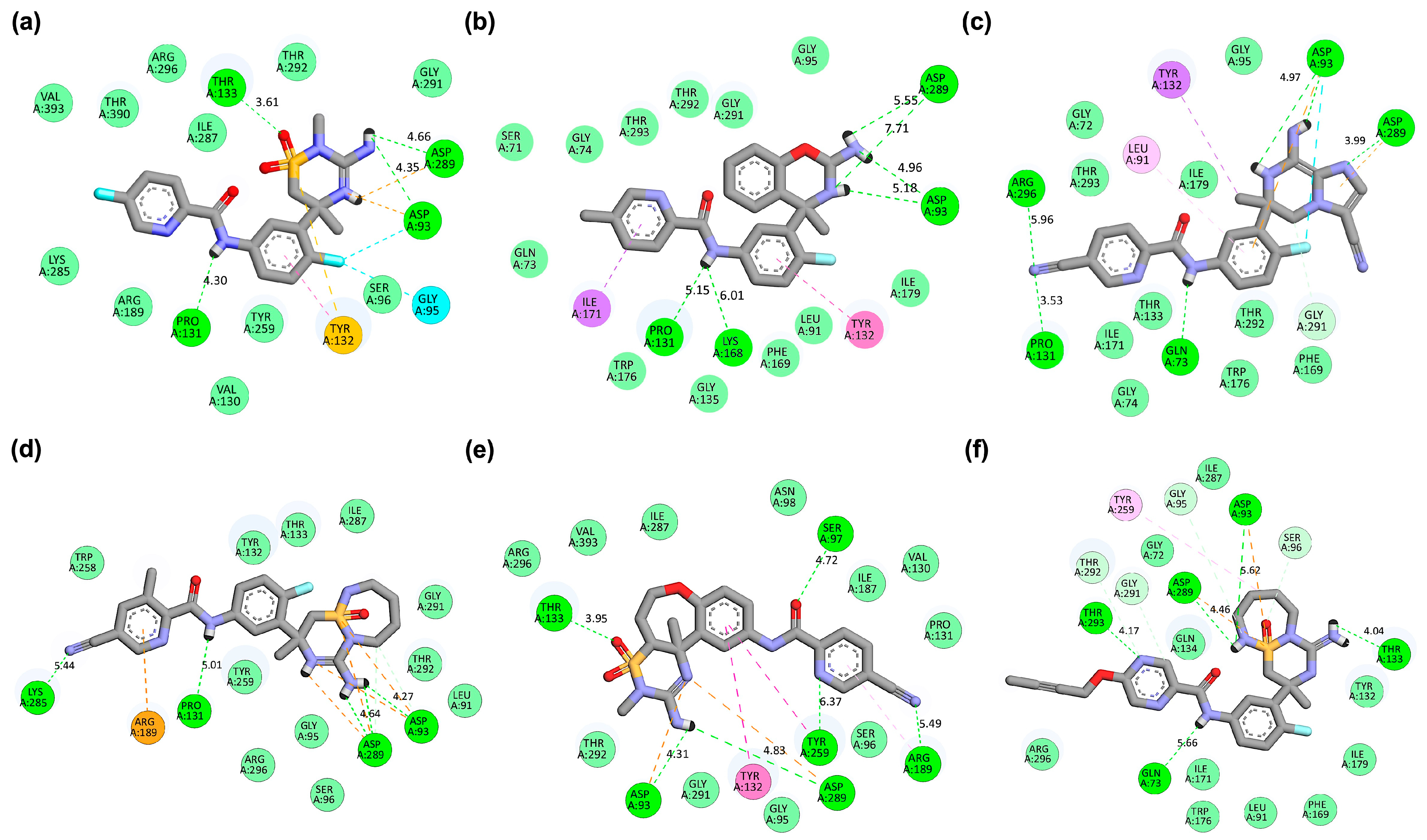
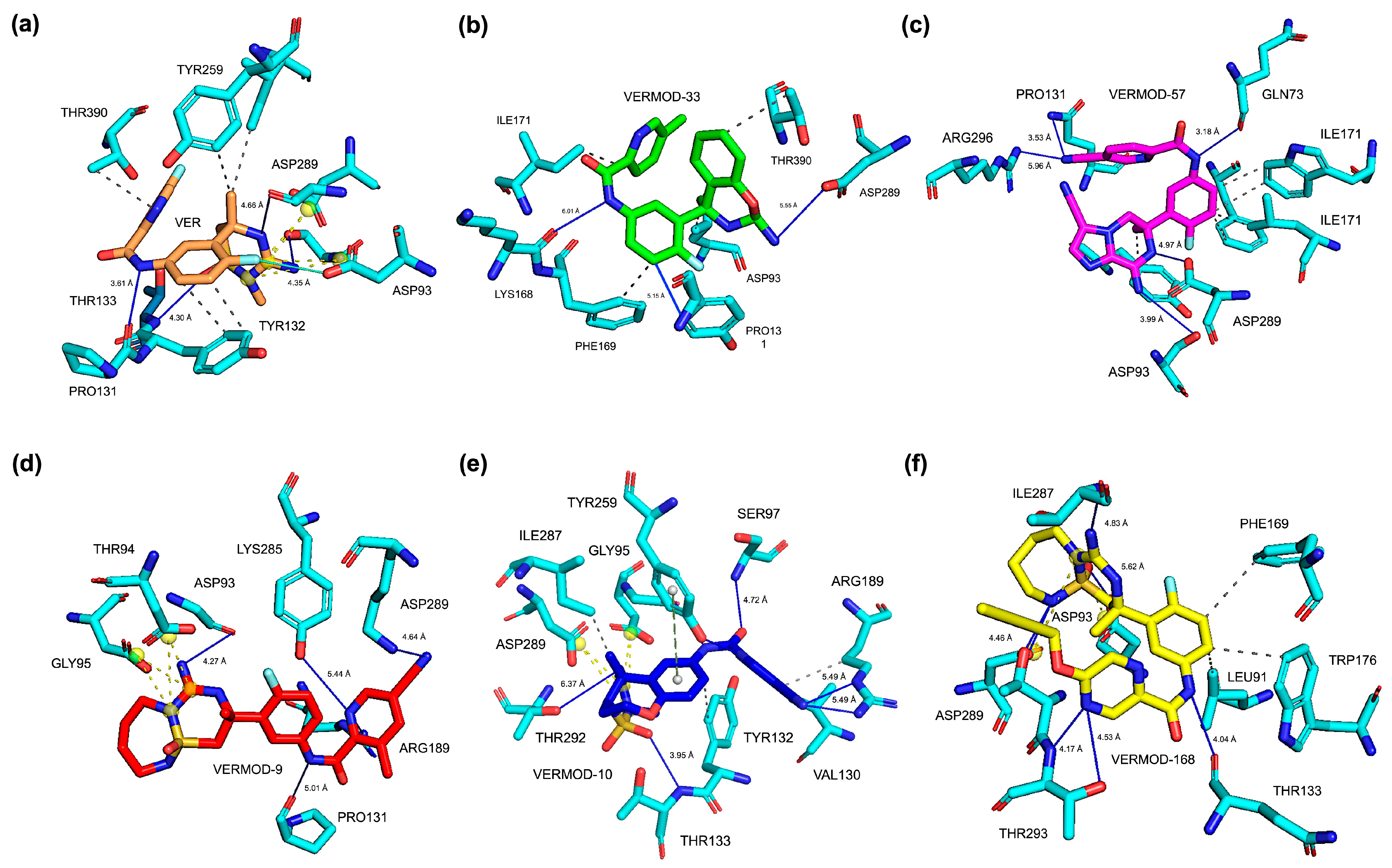
2.2. Molecular Docking Results, Binding Pose, and Binding Affinity Analysis
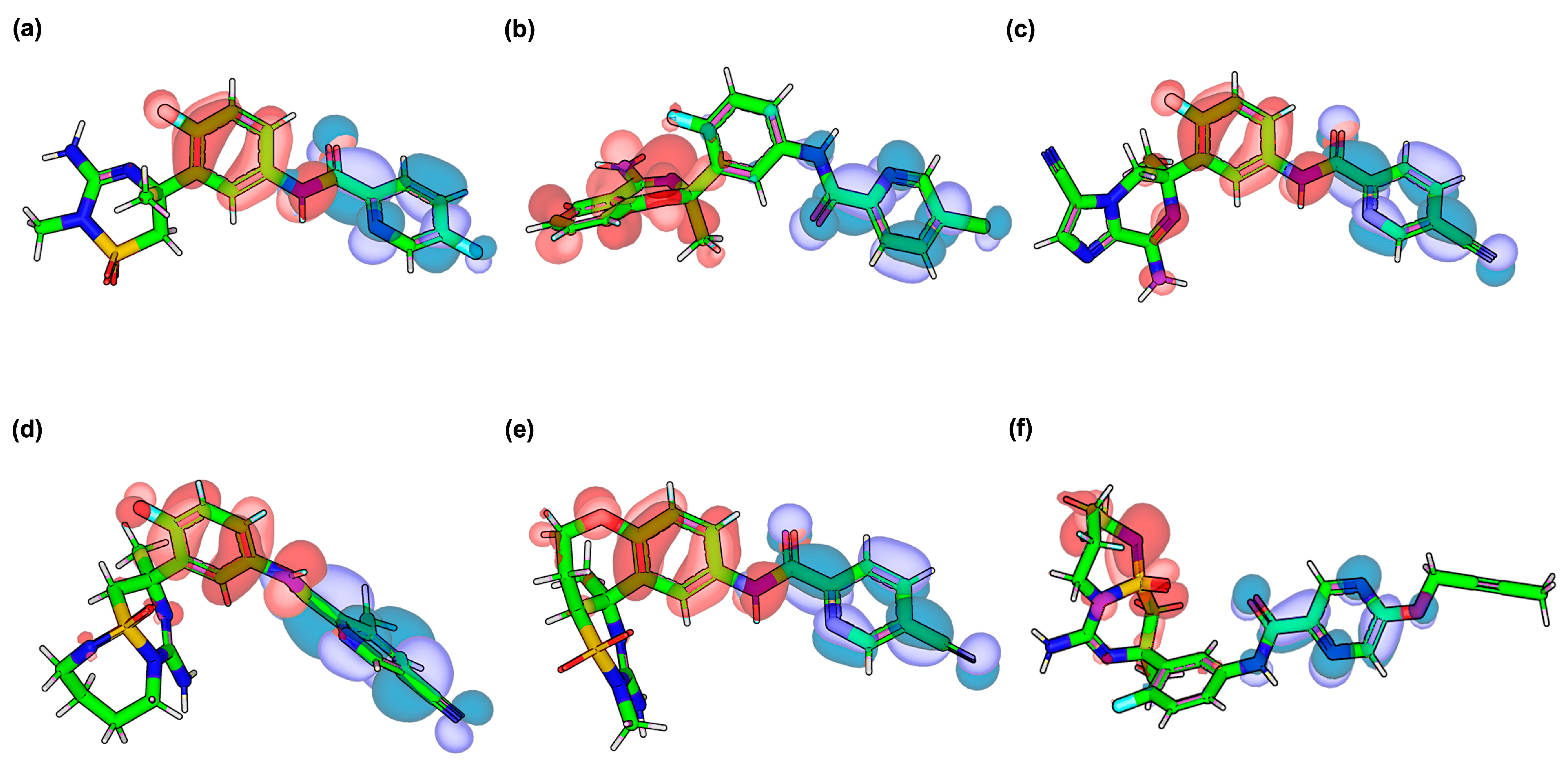
2.3. Frontier Molecular Orbital (HOMO–LUMO) Results of Verubecestat and Its Derivatives
2.4. Pharmacophore Modeling Results
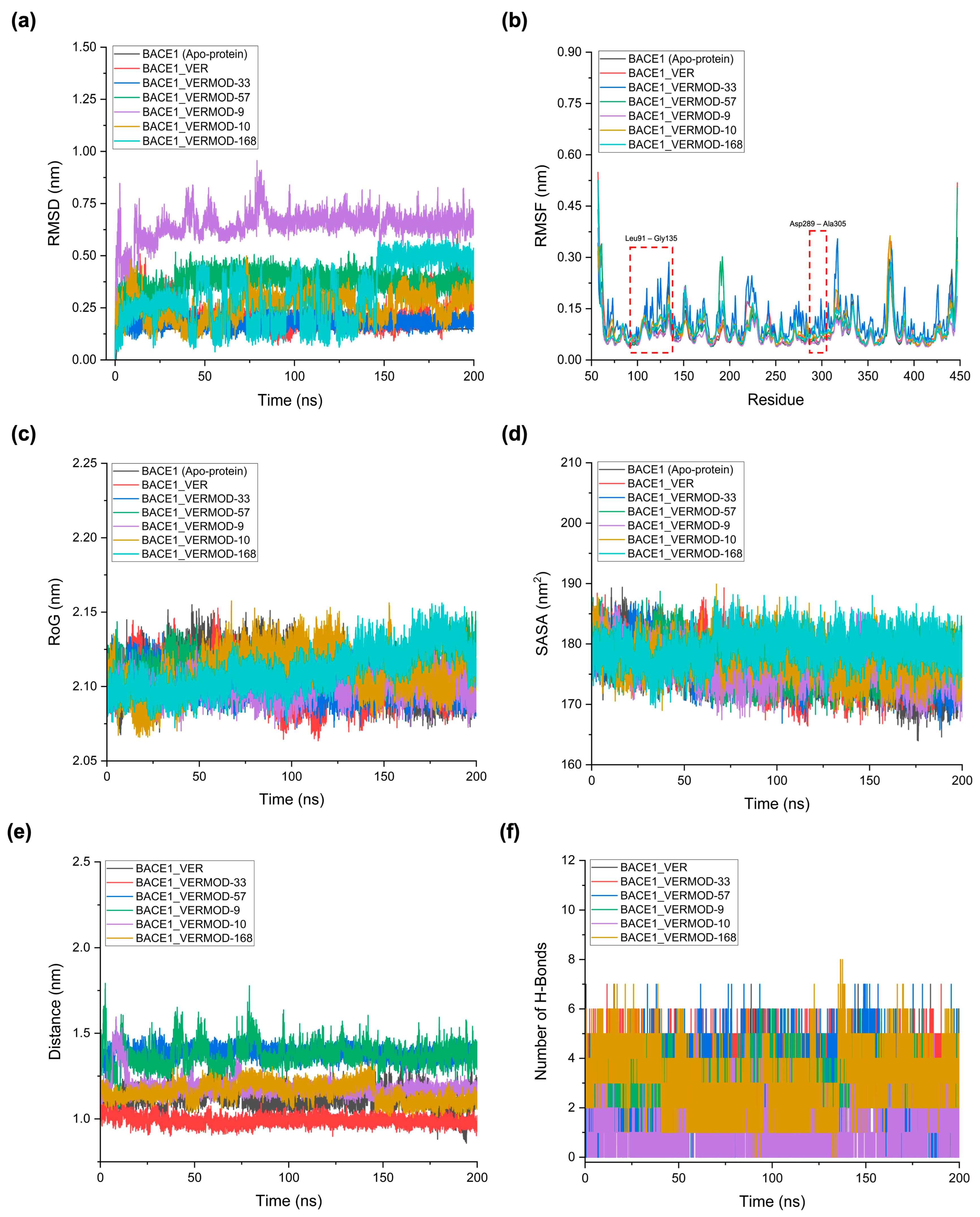
2.5. MD Simulations Reveal Structural Stability and Interaction Profiles of Verubecestat and Its Derivatives
2.6. MM/PBSA Free Energy Analysis and Per-Residue Decomposition
2.7. In Silico Pharmacokinetics and ADMET Profiling of Verubecestat and Its Derivatives
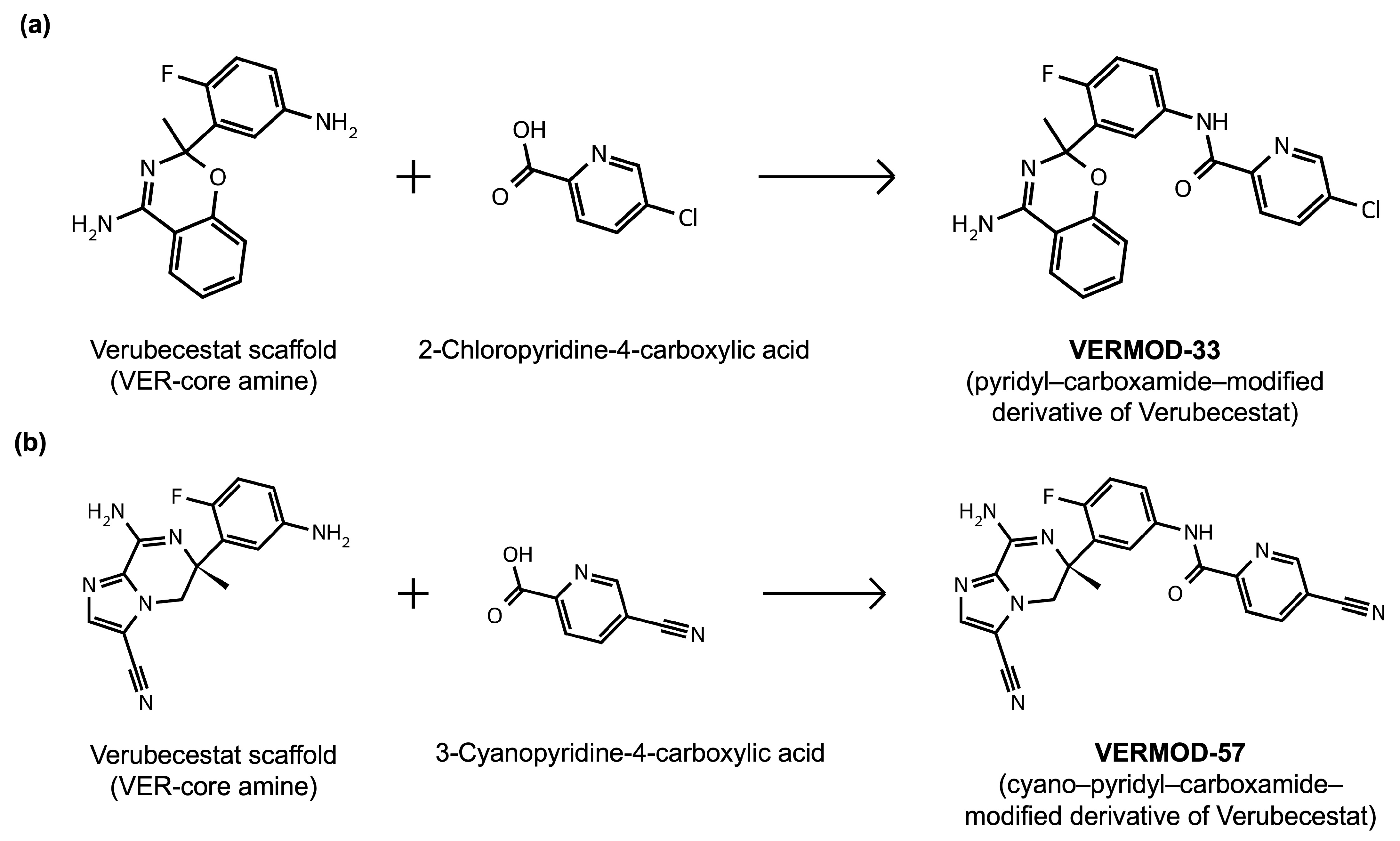
2.8. Results of Retrosynthetic Design for Verubecestat Derivatives
3. Discussion
4. Materials and Methods
4.1. Three-Dimensional Structure Construction, Rational Modifications, and MM2 Energy Minimization
4.2. Three-Dimensional Structure Alignment and Similarity Analysis
4.3. Molecular Docking Simulations and Binding Affinity Analysis
4.4. HOMO–LUMO Analysis of Verubecestat Derivatives
4.5. Three-Dimensional Pharmacophore Modeling
4.6. Molecular Dynamics (MD) Simulation for Structural Stability and Interaction Analysis
4.7. Molecular Mechanics/Poisson–Boltzmann Surface Area (MM/PBSA) Calculations
4.8. In Silico Pharmacokinetics and ADMET Evaluation
4.9. Retrosynthetic Design of Verubecestat Derivatives
5. Limitations and Future Works
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AD | Alzheimer’s disease |
| ADMET | Absorption, distribution, metabolism, excretion, and toxicity |
| AIRs | Ambiguous interaction restraints |
| BACE1 | β-site APP-cleaving enzyme 1 |
| BBB | Blood–brain barrier |
| CNS | Central nervous system |
| DFT | Density functional theory |
| FEP | Free-energy perturbation |
| FMO | Frontier molecular orbital |
| GAFF2 | General amber force field |
| HADDOCK | High ambiguity driven protein-protein docking |
| HBA | Hydrogen bond acceptor |
| HBD | Hydrogen bond donor |
| HOMO | Highest occupied molecular orbital |
| ICT | Intramolecular charge transfer |
| LBD | Ligand-binding domain |
| LUMO | Lowest unoccupied molecular orbital |
| MD | Molecular dynamics |
| MM/PBSA | Molecular mechanics/Poisson-Boltzmann surface area |
| NPT | Number of particles, pressure, and temperature |
| NVT | Number of particles, volume, and temperature |
| PME | Particle mesh Ewald |
| PRODIGY | Protein binding energy prediction |
| RMSD | Root mean square deviation |
| RMSF | Root mean square fluctuation |
| RoG | Radius of gyration |
| SASA | Solvent-accessible surface area |
| SCF | Self-consistent field |
| SPCE | Single point charge extended |
| SPR | Surface plasmon resonance |
| STP | Single-trajectory protocol |
References
- Breijyeh, Z.; Karaman, R. Comprehensive Review on Alzheimer’s Disease: Causes and Treatment. Molecules 2020, 25, 5789. [Google Scholar] [CrossRef]
- Guo, T.; Zhang, D.; Zeng, Y.; Huang, T.Y.; Xu, H.; Zhao, Y. Molecular and cellular mechanisms underlying the pathogenesis of Alzheimer’s disease. Mol. Neurodegener. 2020, 15, 40. [Google Scholar] [CrossRef]
- Kamatham, P.T.; Shukla, R.; Khatri, D.K.; Vora, L.K. Pathogenesis, diagnostics, and therapeutics for Alzheimer’s disease: Breaking the memory barrier. Ageing Res. Rev. 2024, 101, 102481. [Google Scholar] [CrossRef] [PubMed]
- Hampel, H.; Hardy, J.; Blennow, K.; Chen, C.; Perry, G.; Kim, S.H.; Villemagne, V.L.; Aisen, P.; Vendruscolo, M.; Iwatsubo, T.; et al. The Amyloid-β Pathway in Alzheimer’s Disease. Mol. Psychiatry 2021, 26, 5481–5503. [Google Scholar] [CrossRef]
- Moloney, C.M.; Lowe, V.J.; Murray, M.E. Visualization of neurofibrillary tangle maturity in Alzheimer’s disease: A clinicopathologic perspective for biomarker research. Alzheimer’s Dement. 2021, 17, 1554–1574. [Google Scholar] [CrossRef]
- Andrew, R.J.; Kellett, K.A.; Thinakaran, G.; Hooper, N.M. A Greek Tragedy: The Growing Complexity of Alzheimer Amyloid Precursor Protein Proteolysis. J. Biol. Chem. 2016, 291, 19235–19244. [Google Scholar] [CrossRef] [PubMed]
- Hampel, H.; Vassar, R.; De Strooper, B.; Hardy, J.; Willem, M.; Singh, N.; Zhou, J.; Yan, R.; Vanmechelen, E.; De Vos, A.; et al. The β-Secretase BACE1 in Alzheimer’s Disease. Biol. Psychiatry 2021, 89, 745–756. [Google Scholar] [CrossRef]
- Vassar, R. BACE1 inhibition as a therapeutic strategy for Alzheimer’s disease. J. Sport. Health Sci. 2016, 5, 388–390. [Google Scholar] [CrossRef]
- Bazzari, F.H.; Bazzari, A.H. BACE1 Inhibitors for Alzheimer’s Disease: The Past, Present and Any Future? Molecules 2022, 27, 8823. [Google Scholar] [CrossRef] [PubMed]
- Neumann, U.; Machauer, R.; Shimshek, D.R. The β-secretase (BACE) inhibitor NB-360 in preclinical models: From amyloid-β reduction to downstream disease-relevant effects. Br. J. Pharmacol. 2019, 176, 3435–3446. [Google Scholar] [CrossRef]
- Kennedy, M.E.; Stamford, A.W.; Chen, X.; Cox, K.; Cumming, J.N.; Dockendorf, M.F.; Egan, M.; Ereshefsky, L.; Hodgson, R.A.; Hyde, L.A.; et al. The BACE1 inhibitor verubecestat (MK-8931) reduces CNS β-amyloid in animal models and in Alzheimer’s disease patients. Sci. Transl. Med. 2016, 8, 363ra150. [Google Scholar] [CrossRef]
- Novak, G.; Streffer, J.R.; Timmers, M.; Henley, D.; Brashear, H.R.; Bogert, J.; Russu, A.; Janssens, L.; Tesseur, I.; Tritsmans, L.; et al. Long-term safety and tolerability of atabecestat (JNJ-54861911), an oral BACE1 inhibitor, in early Alzheimer’s disease spectrum patients: A randomized, double-blind, placebo-controlled study and a two-period extension study. Alzheimers Res. Ther. 2020, 12, 58. [Google Scholar] [CrossRef]
- Yan, R. Stepping closer to treating Alzheimer’s disease patients with BACE1 inhibitor drugs. Transl. Neurodegener. 2016, 5, 13. [Google Scholar] [CrossRef]
- Evin, G. Future Therapeutics in Alzheimer’s Disease: Development Status of BACE Inhibitors. BioDrugs 2016, 30, 173–194. [Google Scholar] [CrossRef] [PubMed]
- Egan, M.F.; Kost, J.; Voss, T.; Mukai, Y.; Aisen, P.S.; Cummings, J.L.; Tariot, P.N.; Vellas, B.; van Dyck, C.H.; Boada, M.; et al. Randomized Trial of Verubecestat for Prodromal Alzheimer’s Disease. N. Engl. J. Med. 2019, 380, 1408–1420. [Google Scholar] [CrossRef] [PubMed]
- Yiannopoulou, K.G.; Papageorgiou, S.G. Current and Future Treatments in Alzheimer Disease: An Update. J. Cent. Nerv. Syst. Dis. 2020, 12, 1179573520907397. [Google Scholar] [CrossRef] [PubMed]
- Alotaiq, N.; Dermawan, D. Computational Investigation of Montelukast and Its Structural Derivatives for Binding Affinity to Dopaminergic and Serotonergic Receptors: Insights from a Comprehensive Molecular Simulation. Pharmaceuticals 2025, 18, 559. [Google Scholar] [CrossRef]
- Dermawan, D.; Elbouamri, L.; Chtita, S.; Alotaiq, N. Molecular Insights into Bromocriptine Binding to GPCRs Within Histamine-Linked Signaling Networks: Network Pharmacology, Pharmacophore Modeling, and Molecular Dynamics Simulation. Int. J. Mol. Sci. 2025, 26, 8717. [Google Scholar] [CrossRef]
- Zhou, H.; Skolnick, J. Utility of the Morgan Fingerprint in Structure-Based Virtual Ligand Screening. J. Phys. Chem. B 2024, 128, 5363–5370. [Google Scholar] [CrossRef]
- Egan, M.F.; Kost, J.; Tariot, P.N.; Aisen, P.S.; Cummings, J.L.; Vellas, B.; Sur, C.; Mukai, Y.; Voss, T.; Furtek, C.; et al. Randomized Trial of Verubecestat for Mild-to-Moderate Alzheimer’s Disease. N. Engl. J. Med. 2018, 378, 1691–1703. [Google Scholar] [CrossRef]
- Knopman, D.S. Lowering of Amyloid-Beta by β-Secretase Inhibitors—Some Informative Failures. N. Engl. J. Med. 2019, 380, 1476–1478. [Google Scholar] [CrossRef]
- Scott, J.D.; Li, S.W.; Brunskill, A.P.J.; Chen, X.; Cox, K.; Cumming, J.N.; Forman, M.; Gilbert, E.J.; Hodgson, R.A.; Hyde, L.A.; et al. Discovery of the 3-Imino-1,2,4-thiadiazinane 1,1-Dioxide Derivative Verubecestat (MK-8931)–A β-Site Amyloid Precursor Protein Cleaving Enzyme 1 Inhibitor for the Treatment of Alzheimer’s Disease. J. Med. Chem. 2016, 59, 10435–10450. [Google Scholar] [CrossRef]
- Ghosh, A.K.; Osswald, H.L. BACE1 (β-secretase) inhibitors for the treatment of Alzheimer’s disease. Chem. Soc. Rev. 2014, 43, 6765–6813. [Google Scholar] [CrossRef]
- Ghosh, A.K.; Ghosh, K.; Brindisi, M.; Lendy, E.K.; Yen, Y.C.; Kumaragurubaran, N.; Huang, X.; Tang, J.; Mesecar, A.D. Design, synthesis, X-ray studies, and biological evaluation of novel BACE1 inhibitors with bicyclic isoxazoline carboxamides as the P3 ligand. Bioorg. Med. Chem. Lett. 2018, 28, 2605–2610. [Google Scholar] [CrossRef]
- Kumar, A.; Roy, S.; Tripathi, S.; Sharma, A. Molecular docking based virtual screening of natural compounds as potential BACE1 inhibitors: 3D QSAR pharmacophore mapping and molecular dynamics analysis. J. Biomol. Struct. Dyn. 2016, 34, 239–249. [Google Scholar] [CrossRef]
- Ugbaja, S.C.; Lawal, I.A.; Abubakar, B.H.; Mushebenge, A.G.; Lawal, M.M.; Kumalo, H.M. Allostery Inhibition of BACE1 by Psychotic and Meroterpenoid Drugs in Alzheimer’s Disease Therapy. Molecules 2022, 27, 4372. [Google Scholar] [CrossRef]
- Ghosh, A.K.; Kumaragurubaran, N.; Hong, L.; Kulkarni, S.; Xu, X.; Miller, H.B.; Reddy, D.S.; Weerasena, V.; Turner, R.; Chang, W.; et al. Potent memapsin 2 (beta-secretase) inhibitors: Design, synthesis, protein-ligand X-ray structure, and in vivo evaluation. Bioorg. Med. Chem. Lett. 2008, 18, 1031–1036. [Google Scholar] [CrossRef]
- Neumann, U.; Ufer, M.; Jacobson, L.H.; Rouzade-Dominguez, M.L.; Huledal, G.; Kolly, C.; Lüönd, R.M.; Machauer, R.; Veenstra, S.J.; Hurth, K.; et al. The BACE-1 inhibitor CNP520 for prevention trials in Alzheimer’s disease. EMBO Mol. Med. 2018, 10, e9316. [Google Scholar] [CrossRef] [PubMed]
- Otani, T.; Hattori, Y.; Akaji, K.; Kobayashi, K. Macrocyclic BACE1 inhibitors with hydrophobic cross-linked structures: Optimization of ring size and ring structure. Bioorg. Med. Chem. 2021, 52, 116517. [Google Scholar] [CrossRef] [PubMed]
- Mullard, A. BACE inhibitor bust in Alzheimer trial. Nat. Rev. Drug Discov. 2017, 16, 155. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.; Bansoad, A.V.; Singh, R.; Khatik, G.L. BACE1: A Key Regulator in Alzheimer’s Disease Progression and Current Development of its Inhibitors. Curr. Neuropharmacol. 2022, 20, 1174–1193. [Google Scholar] [CrossRef]
- Yan, R.; Vassar, R. Targeting the β secretase BACE1 for Alzheimer’s disease therapy. Lancet Neurol. 2014, 13, 319–329. [Google Scholar] [CrossRef]
- Medina, M.; Avila, J. New perspectives on the role of tau in Alzheimer’s disease. Implications for therapy. Biochem. Pharmacol. 2014, 88, 540–547. [Google Scholar] [CrossRef]
- Cummings, J.; Lee, G.; Ritter, A.; Sabbagh, M.; Zhong, K. Alzheimer’s disease drug development pipeline: 2020. Alzheimers Dement 2020, 6, e12050. [Google Scholar] [CrossRef]
- Ly, P.T.; Wu, Y.; Zou, H.; Wang, R.; Zhou, W.; Kinoshita, A.; Zhang, M.; Yang, Y.; Cai, F.; Woodgett, J.; et al. Inhibition of GSK3β-mediated BACE1 expression reduces Alzheimer-associated phenotypes. J. Clin. Investig. 2013, 123, 224–235. [Google Scholar] [CrossRef]
- Dassault Systèmes BIOVIA. BIOVIA Discovery Studio, 2024; Dassault Systèmes: San Diego, CA, USA, 2024. [Google Scholar]
- Venkatraman, V.; Gaiser, J.; Demekas, D.; Roy, A.; Xiong, R.; Wheeler, T.J. Do Molecular Fingerprints Identify Diverse Active Drugs in Large-Scale Virtual Screening? (No). Pharmaceuticals 2024, 17, 992. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Gao, K.; Nguyen, D.D.; Chen, X.; Jiang, Y.; Wei, G.-W.; Pan, F. Algebraic graph-assisted bidirectional transformers for molecular property prediction. Nat. Commun. 2021, 12, 3521. [Google Scholar] [CrossRef]
- Orsi, M.; Reymond, J.-L. One chiral fingerprint to find them all. J. Cheminform. 2024, 16, 53. [Google Scholar] [CrossRef]
- Landrum, G. RDKit: Open-Source Cheminformatics [Computer Software]. 2024. Available online: https://www.rdkit.org (accessed on 14 May 2025).
- Guex, N.; Peitsch, M.C. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis 1997, 18, 2714–2723. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Jabłońska, J.; Pravda, L.; Vařeková, R.S.; Thornton, J.M. PDBsum: Structural summaries of PDB entries. Protein Sci. 2018, 27, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, C.; Boelens, R.; Bonvin, A.M.J.J. HADDOCK: A Protein–Protein Docking Approach Based on Biochemical or Biophysical Information. J. Am. Chem. Soc. 2003, 125, 1731–1737. [Google Scholar] [CrossRef] [PubMed]
- van Zundert, G.C.P.; Rodrigues, J.; Trellet, M.; Schmitz, C.; Kastritis, P.L.; Karaca, E.; Melquiond, A.S.J.; van Dijk, M.; de Vries, S.J.; Bonvin, A. The HADDOCK2.2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes. J. Mol. Biol. 2016, 428, 720–725. [Google Scholar] [CrossRef]
- Vangone, A.; Bonvin, A. PRODIGY: A Contact-based Predictor of Binding Affinity in Protein-protein Complexes. Bio-protocol 2017, 7, e2124. [Google Scholar] [CrossRef]
- Panda, P.; Enhanced HOMO-LUMO Pipeline. GitHub. 2025. Available online: https://github.com/pritampanda15/Omixium_YouTube_Channel/tree/main/HOMO_LUMO (accessed on 14 May 2025).
- Tirado-Rives, J.; Jorgensen, W.L. Performance of B3LYP Density Functional Methods for a Large Set of Organic Molecules. J. Chem. Theory Comput. 2008, 4, 297–306. [Google Scholar] [CrossRef]
- Bidault, X.; Chaudhuri, S. How Accurate Can Crystal Structure Predictions Be for High-Energy Molecular Crystals? Molecules 2023, 28, 4471. [Google Scholar] [CrossRef]
- Khalid, M.; Zafar, M.; Hussain, S.; Asghar, M.A.; Khera, R.A.; Imran, M.; Abookleesh, F.L.; Akram, M.Y.; Ullah, A. Influence of End-Capped Modifications in the Nonlinear Optical Amplitude of Nonfullerene-Based Chromophores with a D–π–A Architecture: A DFT/TDDFT Study. ACS Omega 2022, 7, 23532–23548. [Google Scholar] [CrossRef] [PubMed]
- Pei, Z.; Ou, Q.; Mao, Y.; Yang, J.; Lande, A.; Plasser, F.; Liang, W.; Shuai, Z.; Shao, Y. Elucidating the Electronic Structure of a Delayed Fluorescence Emitter via Orbital Interactions, Excitation Energy Components, Charge-Transfer Numbers, and Vibrational Reorganization Energies. J. Phys. Chem. Lett. 2021, 12, 2712–2720. [Google Scholar] [CrossRef] [PubMed]
- de Aguiar, A.S.N.; de Carvalho, L.B.R.; Gomes, C.M.; Castro, M.M.; Martins, F.S.; Borges, L.L. Computational Insights into the Antioxidant Activity of Luteolin: Density Functional Theory Analysis and Docking in Cytochrome P450 17A1. Pharmaceuticals 2025, 18, 410. [Google Scholar] [CrossRef]
- Wolber, G.; Langer, T. LigandScout: 3-D pharmacophores derived from protein-bound ligands and their use as virtual screening filters. J. Chem. Inf. Model. 2005, 45, 160–169. [Google Scholar] [CrossRef]
- Pronk, S.; Páll, S.; Schulz, R.; Larsson, P.; Bjelkmar, P.; Apostolov, R.; Shirts, M.R.; Smith, J.C.; Kasson, P.M.; van der Spoel, D.; et al. GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 2013, 29, 845–854. [Google Scholar] [CrossRef] [PubMed]
- Alotaiq, N.; Dermawan, D.; Elwali, N.E. Leveraging Therapeutic Proteins and Peptides from Lumbricus Earthworms: Targeting SOCS2 E3 Ligase for Cardiovascular Therapy through Molecular Dynamics Simulations. Int. J. Mol. Sci. 2024, 25, 10818. [Google Scholar] [CrossRef]
- Alotaiq, N.; Dermawan, D. Evaluation of Structure Prediction and Molecular Docking Tools for Therapeutic Peptides in Clinical Use and Trials Targeting Coronary Artery Disease. Int. J. Mol. Sci. 2025, 26, 462. [Google Scholar] [CrossRef]
- Dermawan, D.; Alotaiq, N. Unveiling Pharmacological Mechanisms of Bombyx mori (Abresham), a Traditional Arabic Unani Medicine for Ischemic Heart Disease: An Integrative Molecular Simulation Study. Pharmaceutics 2025, 17, 295. [Google Scholar] [CrossRef]
- Schrödinger. The PyMOL Molecular Graphics System. 2020. Available online: https://www.pymol.org/ (accessed on 25 May 2025).
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Fogolari, F.; Brigo, A.; Molinari, H. Protocol for MM/PBSA Molecular Dynamics Simulations of Proteins. Biophys. J. 2003, 85, 159–166. [Google Scholar] [CrossRef]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert. Opin. Drug Discov. 2015, 10, 449–461. [Google Scholar] [CrossRef]
- Wang, J.; Hou, T. Develop and test a solvent accessible surface area-based model in conformational entropy calculations. J. Chem. Inf. Model. 2012, 52, 1199–1212. [Google Scholar] [CrossRef] [PubMed]
- Doni Dermawan, F.A.; Elwali, N.E.; Alotaiq, N. Therapeutic potential of earthworm-derived proteins: Targeting NEDD4 for cardiovascular disease intervention. J. Appl. Pharm. Sci. 2024, 15, 216–232. [Google Scholar] [CrossRef]
- Valdés-Tresanco, M.; Valdés-Tresanco, M.; Valiente, P.; Moreno Frias, E. gmx_MMPBSA: A New Tool to Perform End-State Free Energy Calculations with GROMACS. J. Chem. Theory Comput. 2021, 17, 6281–6291. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Sander, T.; Freyss, J.; von Korff, M.; Rufener, C. DataWarrior: An open-source program for chemistry aware data visualization and analysis. J. Chem. Inf. Model. 2015, 55, 460–473. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger. Schrödinger Release 2025-3: QikProp; Schrödinger, LLC: New York, NY, USA, 2025. [Google Scholar]
- Tu, Z.; Choure, S.J.; Fong, M.H.; Roh, J.; Levin, I.; Yu, K.; Joung, J.F.; Morgan, N.; Li, S.C.; Sun, X.; et al. ASKCOS: Open-Source, Data-Driven Synthesis Planning. Acc. Chem. Res. 2025, 58, 1764–1775. [Google Scholar] [CrossRef] [PubMed]
- Schwaller, P.; Hoover, B.; Reymond, J.L.; Strobelt, H.; Laino, T. Extraction of organic chemistry grammar from unsupervised learning of chemical reactions. Sci. Adv. 2021, 7, eabe4166. [Google Scholar] [CrossRef]


| Molecule | Modification Category | Tanimoto Similarity (FP2) | SMILES | 2D Structure |
|---|---|---|---|---|
| VER | N/A | 1.00 | C[C@]1(CS(=O)(=O)N(C(=N1)N)C)C2=C(C=CC(=C2)NC(=O)C3=NC=C(C=C3)F)F |  |
| VERMOD-10 | Alkoxy Substitution | 0.69 | CN1C(N)=N[C@@]2(C)[C@H](CCOC3=C2C=C(NC(=O)C2=NC=C(C=C2)C#N)C=C3)S1(=O)=O | 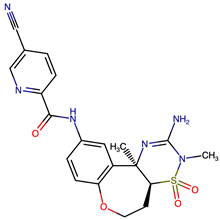 |
| VERMOD-33 | Alkyl Substitution | 0.56 | CC1(OC2=C(C=CC=C2)C(N)=N1)C1=CC(NC(=O)C2=CC=C(Cl)C=N2)=CC=C1F | 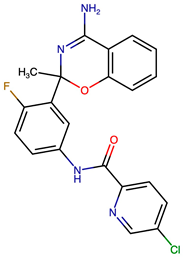 |
| VERMOD-36 | Bioisosteric Replacement | 0.67 | CN=[S@@]1(=O)C[C@](C)(NC(=N)N1C)C1=C(F)C=CC(NC(=O)C2=NC=C(Cl)C=C2)=C1 |  |
| VERMOD-57 | Carbonyl Swap | 0.59 | C[C@]1(CN2C(=CN=C2C(N)=N1)C#N)C1=C(F)C=CC(NC(=O)C2=NC=C(C=C2)C#N)=C1 |  |
| VERMOD-72 | Halogen Substitution | 0.78 | COC1=CN=C(C=N1)C(=S)NC1=CC(F)=C(F)C(=C1)[C@]1(C)CS(=O)(=O)N(C)C(N)=N1 | 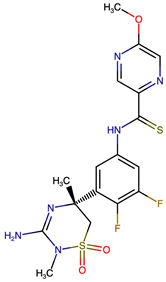 |
| VERMOD-94 | Halogenated Heterocycle | 0.57 | NC1=N[C@@](CF)([C@H]2C[C@H]2O1)C1=CC(NC(=O)C2=NC=C(C=C2Cl)C#N)=CC=C1F | 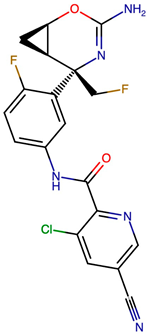 |
| VERMOD-168 | Ring Modification | 0.70 | CC#CCOC1=CN=C(C=N1)C(=O)NC1=CC(=C(F)C=C1)[C@]1(C)C[S@@]2(=O)=NCCCCN2C(N)=N1 |  |
| Complex | HADDOCK Score (a.u.) | Binding Energy (kcal/mol) | Van der Waals Energy | Electrostatic Energy | Desolvation Energy | RMSD | Hydrogen Bonds |
|---|---|---|---|---|---|---|---|
| BACE1_VER | −44.1 ± 0.5 | −8.44 | −29.1 ± 0.6 | −133.4 ± 1.6 | −1.7 ± 0.4 | 0.1 ± 0.1 | Asp93, Pro131, Thr133, Asp289 |
| BACE1_VERMOD-33 | −53.9 ± 0.9 | −10.42 | −30.0 ± 0.8 | −197.8 ± 8.2 | −5.0 ± 0.4 | 0.1 ± 0.0 | Asp93, Pro131, Lys168, Asp289 |
| BACE1_VERMOD-57 | −62.2 ± 0.7 | −10.11 | −36.9 ± 0.3 | −240.0 ± 10.5 | −1.4 ± 0.4 | 0.1 ± 0.0 | Gln73, Asp93, Pro131, Asp289, Arg296 |
| BACE1_VERMOD-9 | −54.9 ± 0.5 | −9.79 | −31.5 ± 0.8 | −216.5 ± 24.7 | −2.6 ± 1.6 | 0.1 ± 0.1 | Asp93, Pro131, Lys285, Asp289 |
| BACE1_VERMOD-10 | −52.7 ± 2.2 | −9.79 | −33.2 ± 1.0 | −202.6 ± 3.5 | −1.0 ± 0.5 | 0.1 ± 0.0 | Asp93, Ser97, Thr133, Arg189, Tyr259, Asp289 |
| BACE1_VERMOD-168 | −51.8 ± 1.2 | −9.05 | −37.8 ± 0.5 | −122.7 ± 10.3 | −3.5 ± 0.3 | 0.1 ± 0.0 | Gln73, Asp93, Thr133, Asp289, Thr293 |
| Complex | CC | CO | CN | CX | OO | OX | NO | NN | NX | XX |
|---|---|---|---|---|---|---|---|---|---|---|
| BACE1_VER | 1770 | 818 | 1028 | 336 | 91 | 117 | 254 | 145 | 102 | 0 |
| BACE1_VERMOD-33 | 2454 | 1000 | 1083 | 132 | 63 | 39 | 190 | 108 | 33 | 0 |
| BACE1_VERMOD-57 | 2332 | 903 | 1484 | 144 | 34 | 41 | 330 | 230 | 35 | 0 |
| BACE1_VERMOD-9 | 1991 | 805 | 1216 | 224 | 57 | 75 | 271 | 178 | 60 | 0 |
| BACE1_VERMOD-10 | 2170 | 1100 | 1216 | 97 | 139 | 39 | 320 | 162 | 29 | 0 |
| BACE1_VERMOD-168 | 2431 | 1149 | 1428 | 258 | 107 | 81 | 332 | 195 | 63 | 0 |
| Molecule | HOMO (eV) | LUMO (eV) | Gap (eV) | Dipole (D) |
|---|---|---|---|---|
| VER | −6.20 | −1.90 | 4.30 | 2.52 |
| VERMOD-33 | −5.85 | −1.98 | 3.87 | 1.91 |
| VERMOD-57 | −6.45 | −2.71 | 3.74 | 2.99 |
| VERMOD-9 | −6.08 | −2.46 | 3.62 | 4.43 |
| VERMOD-10 | −5.79 | −2.50 | 3.29 | 5.13 |
| VERMOD-168 | −5.67 | −1.74 | 3.93 | 1.80 |
| Complex | Average RMSD (nm) | Average RMSF (nm) | Average RoG (nm) | Average SASA (nm2) | Average Distance (nm) | Number of Hydrogen Bonds Between the Ligand-Receptor |
|---|---|---|---|---|---|---|
| BACE1 (Apo-protein) | 0.15 ± 0.01 | 0.08 ± 0.04 | 2.11 ± 0.01 | 176.08 ± 3.40 | N/A | N/A |
| BACE1_VER | 0.21 ± 0.06 | 0.09 ± 0.06 | 2.10 ± 0.01 | 176.59 ± 3.44 | 1.11 ± 0.06 | 3.24 ± 0.78 |
| BACE1_VERMOD-33 | 0.18 ± 0.02 | 0.11 ± 0.05 | 2.10 ± 0.01 | 176.52 ± 3.05 | 0.99 ± 0.03 | 4.27 ± 0.60 |
| BACE1_VERMOD-57 | 0.37 ± 0.04 | 0.08 ± 0.06 | 2.11 ± 0.01 | 176.87 ± 2.94 | 1.38 ± 0.03 | 2.78 ± 1.30 |
| BACE1_VERMOD-9 | 0.64 ± 0.07 | 0.07 ± 0.04 | 2.10 ± 0.01 | 176.37 ± 2.88 | 1.37 ± 0.07 | 2.74 ± 0.79 |
| BACE1_VERMOD-10 | 0.26 ± 0.06 | 0.09 ± 0.04 | 2.11 ± 0.01 | 177.61 ± 2.79 | 1.18 ± 0.05 | 1.04 ± 0.92 |
| BACE1_VERMOD-168 | 0.31 ± 0.14 | 0.09 ± 0.05 | 2.11 ± 0.01 | 179.22 ± 2.45 | 1.15 ± 0.05 | 3.05 ± 1.21 |
| Complex | MM/PBSA Free Binding Energy ΔG_Binding (kcal/mol) |
|---|---|
| BACE1_VER | −35.33 ± 5.21 |
| BACE1_VERMOD-33 | −51.12 ± 4.99 |
| BACE1_VERMOD-57 | −43.85 ± 4.42 |
| BACE1_VERMOD-9 | −21.79 ± 5.31 |
| BACE1_VERMOD-10 | −33.77 ± 4.41 |
| BACE1_VERMOD-168 | −37.55 ± 6.41 |
| Parameter | VER | VERMOD-33 | VERMOD-57 | VERMOD-9 | VERMOD-10 | VERMOD-168 |
|---|---|---|---|---|---|---|
| Molecular Weight (g/mol) | 409.42 | 410.83 | 414.40 | 469.54 | 440.48 | 499.57 |
| Hydrogen Bond Acceptors (HBA) | 8 | 6 | 9 | 9 | 10 | 10 |
| Hydrogen Bond Donors (HBD) | 2 | 2 | 2 | 2 | 2 | 2 |
| cLogP | 0.48 | 3.48 | 1.32 | 1.51 | 0.63 | 1.56 |
| Total Surface Area | 279.25 | 293.18 | 312.90 | 341.37 | 311.02 | 371.98 |
| Polar Surface Area (PSA) | 126.13 | 89.60 | 145.77 | 145.21 | 159.15 | 143.54 |
| Relative PSA | 0.34 | 0.25 | 0.34 | 0.31 | 0.38 | 0.30 |
| Mutagenic | None | None | None | None | None | None |
| Tumorigenic | None | None | None | None | None | None |
| Reproductive Effective | None | None | None | None | None | None |
| Irritant | None | None | None | None | None | None |
| Shape Index | 0.53 | 0.55 | 0.55 | 0.51 | 0.52 | 0.57 |
| Molecular Flexibility | 0.42 | 0.37 | 0.37 | 0.43 | 0.30 | 0.40 |
| Molecular Complexity | 0.85 | 0.87 | 0.89 | 0.92 | 0.94 | 0.92 |
| Solvent Accessible Surface Area (SASA) | 526.22 | 548.33 | 505.21 | 674.14 | 591.97 | 677.86 |
| Hydrophobic Component of SASA (FOSA) | 285.82 | 335.24 | 248.62 | 455.55 | 362.03 | 497.79 |
| Hydrophilic Component of SASA (FISA) | 176.87 | 127.11 | 230.17 | 191.89 | 229.93 | 156.92 |
| Percent Human Oral Absorption | 13.87 | 29.12 | 27.42 | 15.64 | 17.13 | 18.51 |
| QPlogHERG | −5.36 | −5.70 | −7.13 | −7.48 | −6.69 | −7.19 |
| QPPCaco | 3.23 | 9.58 | 1.01 | 2.33 | 1.01 | 4.99 |
| QPlogBB | −0.23 | 0.27 | −0.61 | −0.80 | −1.05 | −0.55 |
| QPPMDCK | 3.03 | 13.03 | 5.41 | 3.35 | 3.39 | 4.92 |
| QPlogKp | −8.23 | −7.51 | −9.02 | −8.41 | −9.02 | −7.58 |
| QPlogKhsa | −0.44 | −0.17 | −0.48 | −0.41 | −0.63 | −0.50 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dermawan, D.; Alotaiq, N. Rescuing Verubecestat: An Integrative Molecular Modeling and Simulation Approach for Designing Next-Generation BACE1 Inhibitors. Int. J. Mol. Sci. 2025, 26, 12143. https://doi.org/10.3390/ijms262412143
Dermawan D, Alotaiq N. Rescuing Verubecestat: An Integrative Molecular Modeling and Simulation Approach for Designing Next-Generation BACE1 Inhibitors. International Journal of Molecular Sciences. 2025; 26(24):12143. https://doi.org/10.3390/ijms262412143
Chicago/Turabian StyleDermawan, Doni, and Nasser Alotaiq. 2025. "Rescuing Verubecestat: An Integrative Molecular Modeling and Simulation Approach for Designing Next-Generation BACE1 Inhibitors" International Journal of Molecular Sciences 26, no. 24: 12143. https://doi.org/10.3390/ijms262412143
APA StyleDermawan, D., & Alotaiq, N. (2025). Rescuing Verubecestat: An Integrative Molecular Modeling and Simulation Approach for Designing Next-Generation BACE1 Inhibitors. International Journal of Molecular Sciences, 26(24), 12143. https://doi.org/10.3390/ijms262412143







