Boolean Networks with Classic and New Updating Modes Applied to Genetic Regulation in Some Familial Diseases
Abstract
1. Introduction
2. Results
- -
- The enlargement or, on the contrary, the reduction in the size of their attractor basins (trajectorial robustness);
- -
- The change in the nature of an attractor, for example, the transition from a stationary state to a limit cycle after the passage of a parameter value above a bifurcation threshold (asymptotic robustness);
- -
- The birth of a new attractor, for example, following a modification of the updating rule (structural robustness).
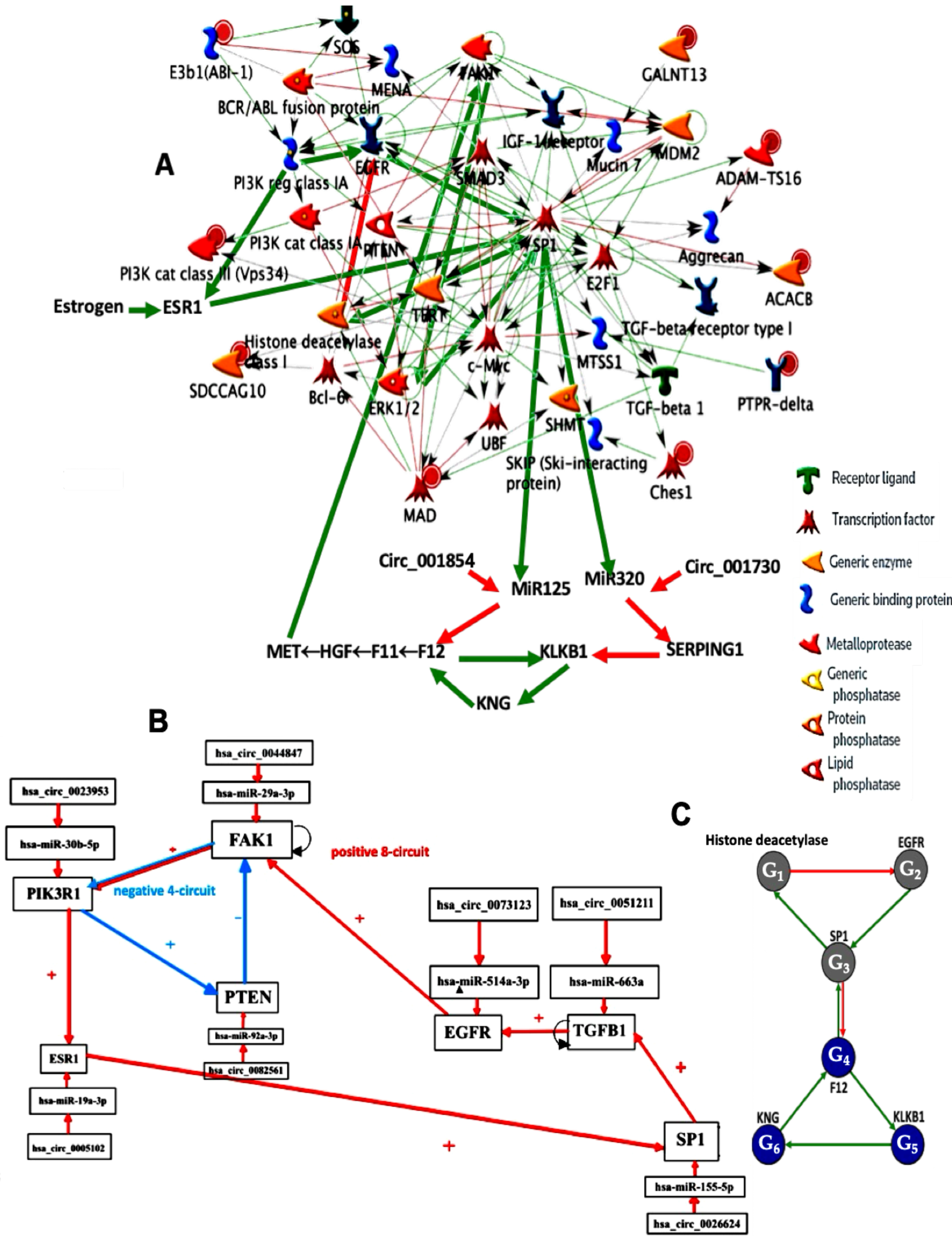
2.1. Familial Angioedema
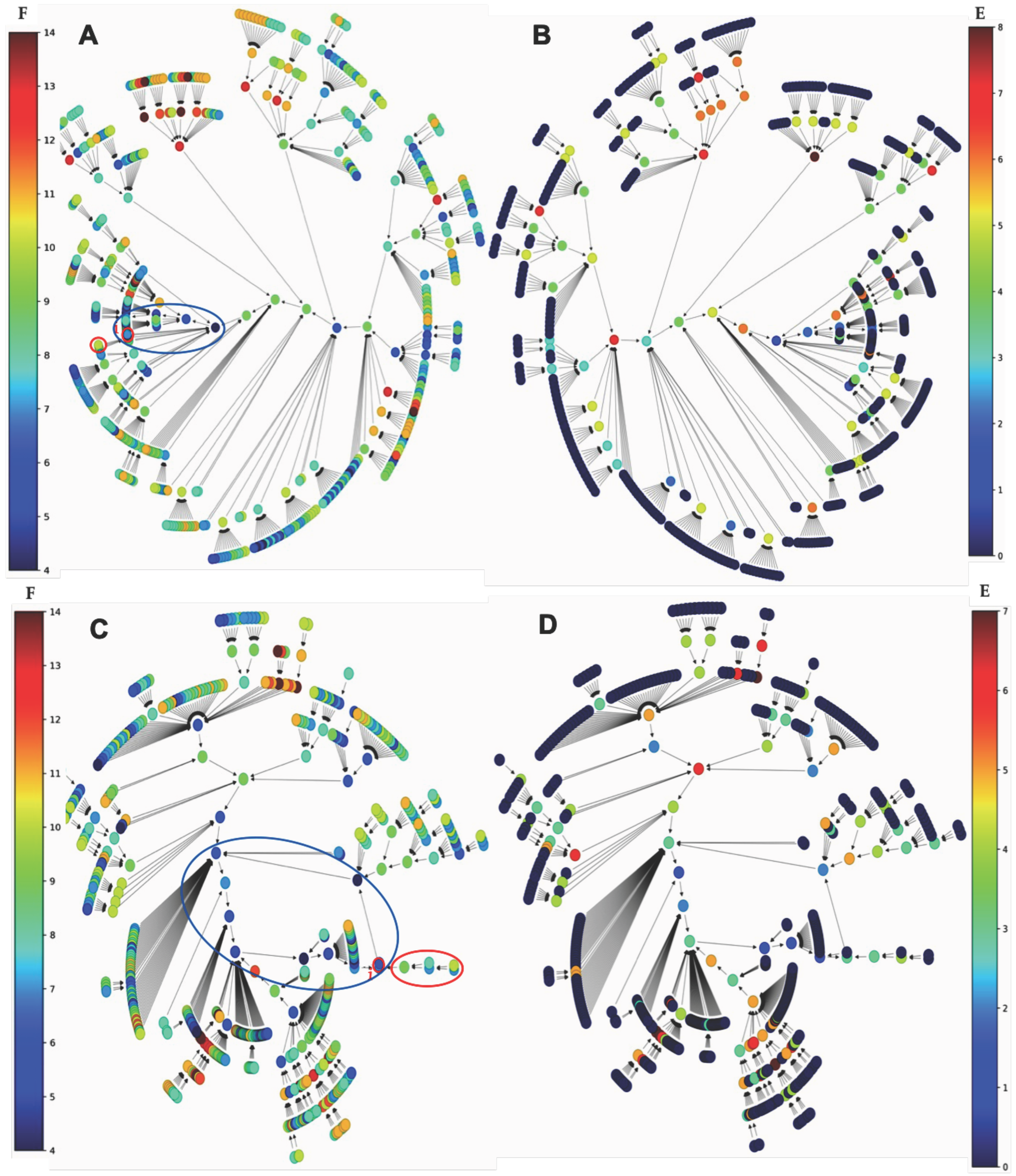
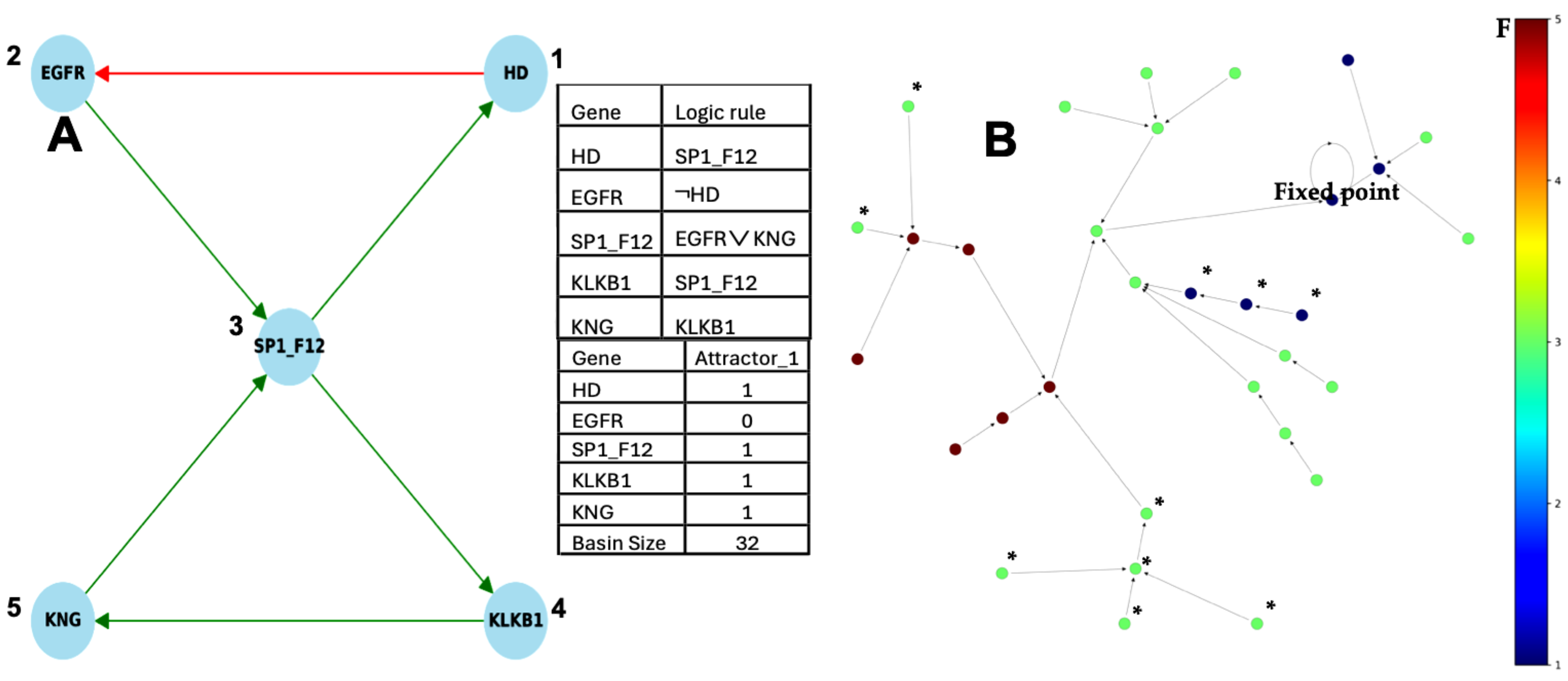
2.1.1. Interaction Graph of the Familial Angioedema Genetic Network
2.1.2. Familial Angioedema Network Dynamics
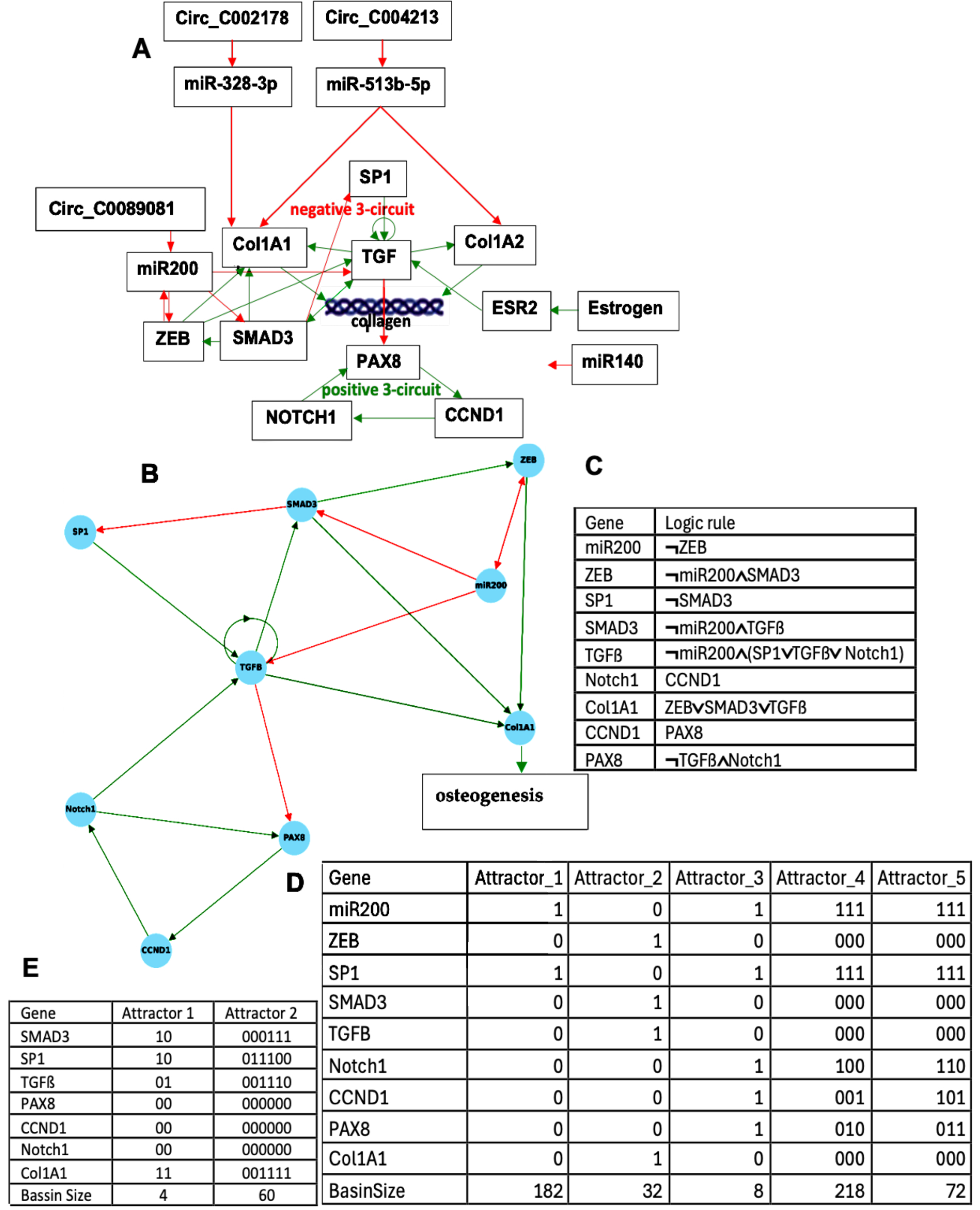
2.2. Osteogenesis Imperfecta
2.2.1. Interaction Graph of the Osteogenesis Imperfecta Genetic Network
2.2.2. Dynamics of the Osteogenesis Imperfecta Network
2.3. Biliary Atresia
2.3.1. Interaction Graph of the Biliary Atresia Genetic Network
2.3.2. Dynamics of the Biliary Atresia Network
3. Discussion
3.1. Intricate Updating Mode
3.2. State-Dependent Updating Mode
- (i)
- A simulation is performed with a classical parallel updating mode and shown the presence of a single attractor, a limit cycle of length 5. Figure 11A (respectively, Figure 11B) shows the frustration (resp. energy) function on the trajectories. The basin of attraction of the configuration i of the limit cycle in Figure 11A, called the isochronal basin of i, is limited to five initial configurations rotating in phase with i on the limit cycle after one iteration.
- (ii)
- the second simulation was performed with the state-dependent updating mode, i.e., in parallel mode if histone genes were expressed (state 1) and sequential otherwise (in the order of the genes in Gene column of Figure 10B).

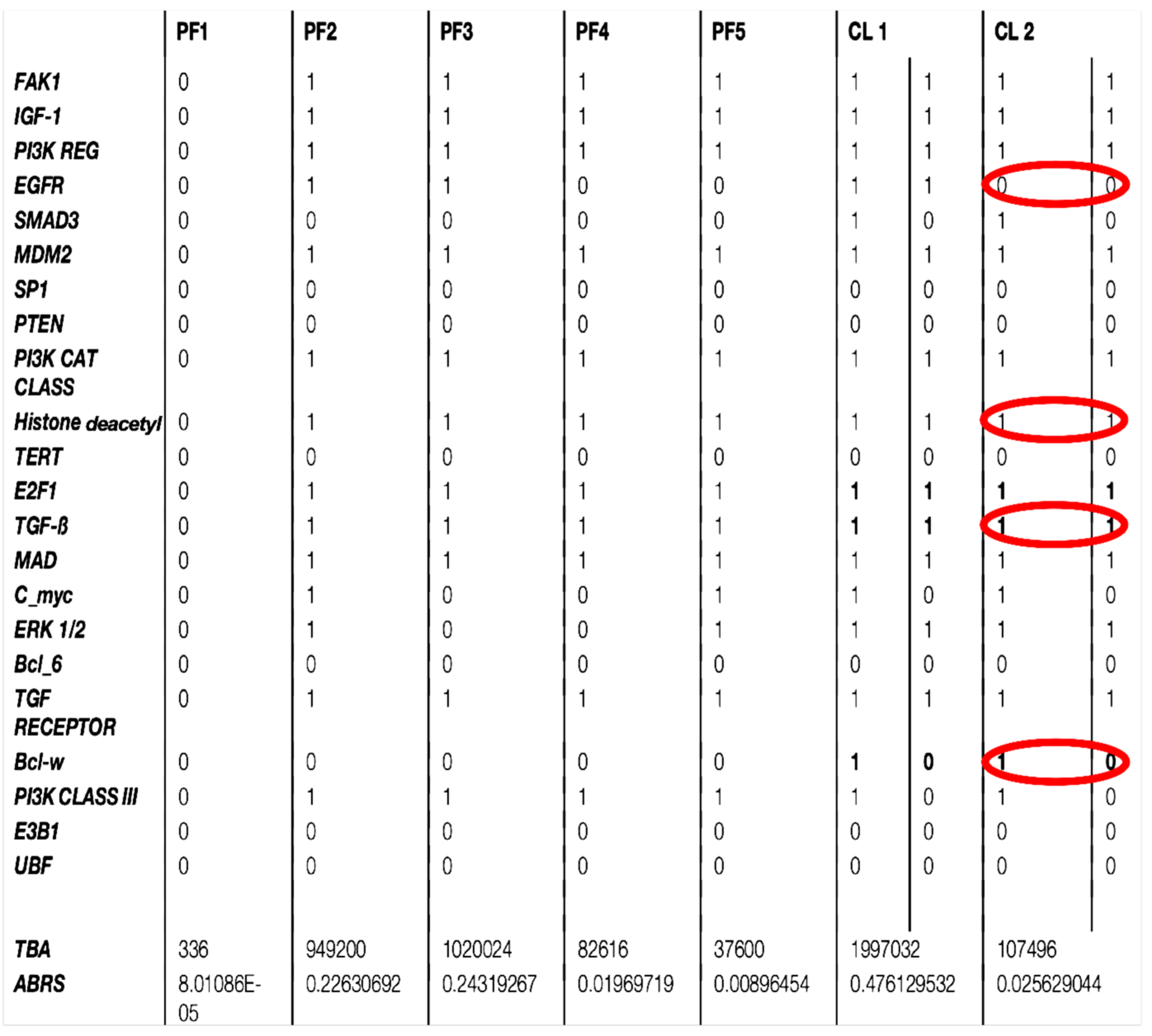
3.3. Comparison Between Updating Modes of the Subnetwork SP1
4. Material and Methods
4.1. Boolean Networks
4.2. Interaction Graph
4.3. Attractors
- (i)
- A is a fixed set for the composed set operator LoB: A = L(B(A));
- (ii)
- there is no B ≠ A, B⊃A and verifying (i), where A is A with shadow trajectories [53];
- (iii)
- there is no C ≠ A, C⊂A and verifying (i) and (ii).
4.4. Updating Modes
- -
- the block-sequential mode, which consists of choosing a partition of N in m disjoint subsets of nodes S1, …, Sm, with ∪k=1,m Sk = N, which are updated sequentially, the nodes of each subset being updated parallelly. If each subset is a singleton, the updating mode is called sequential, the choice of the order being possibly random.
- -
- the block-parallel mode, which consists of choosing a partition of N in m disjoint subsets, which are updated parallelly, the nodes of each subset being updated sequentially. If the partition has only a set, the updating mode is called parallel.
- -
- the block-intricate sequential (respectively, parallel) mode, if the subsets of the partition of N are not obligatory disjoint, i.e., if there exists two indices i and j in {1, …, m}, such as Si ∩ Sj = ∅. The subsets Sk, k∈{1, …, m}, are updated sequentially (respectively, parallelly), nodes of each subset being updated parallelly (respectively, sequentially).
- -
- the state-dependent mode, in which the mode of iteration can be chosen at each iteration of the network N depending on the state of nodes of a subset C of N, considered as a clock, e.g., if the nodes of C are in state 0, then the updating mode is sequential inside the subsets Sk, and if they are in state 1, the updating mode is parallel.
4.5. Different Notions of Robustness
- -
- Trajectorial robustness, which corresponds to the existence of a distance threshold respected between original and perturbed trajectories;
- -
- Asymptotic robustness, which corresponds to the conservation of the attractor of any trajectory after a perturbation, even if the transient part of this trajectory is modified;
- -
- Structural robustness, which corresponds to the conservation of the number and nature of the attractors in response to a structural perturbation (change in interaction graph, transition function or updating clock).
4.6. Frustration, Energy and Entropy
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, G.; Shi, H.; Wang, L.; Zhou, M.; Wang, Z.; Liu, X.; Cheng, L.; Li, W.; Li, X. MicroRNA and transcription factor mediated regulatory network analysis reveals critical regulators and regulatory modules in myocardial infarction. PLoS ONE 2015, 10, e0135339. [Google Scholar] [CrossRef]
- De Maat, S.; Björkqvist, J.; Suffritti, C.; Wiesenekker, C.P.; Nagtegaal, W.; Koekman, A.; van Dooremalen, S.; Pasterkamp, G.; de Groot, P.G.; Cicardi, M.; et al. Plasmin is a natural trigger for bradykinin production in patients with hereditary angioedema with factor XII mutations. J. Allergy Clin. Immunol. 2016, 138, 1414–1423. [Google Scholar] [CrossRef]
- Caccia, S.; Suffritti, C.; Carzaniga, T.; Berardelli, R.; Berra, S.; Martorana, V.; Fra, A.; Drouet, C.; Cicardi, M. Intermittent C1-inhibitor deficiency associated with recessive inheritance: Functional and structural insight. Sci. Rep. 2018, 8, 977. [Google Scholar] [CrossRef]
- Bork, K.; Wulff, K.; Steinmüller-Magin, L.; Braenne, I.; Staubach-Renz, P.; Witzke, G.; Hardt, J. Hereditary angioedema with a mutation in the plasminogen gene. Allergy 2018, 73, 442–450. [Google Scholar] [CrossRef]
- Dewald, G. A missense mutation in the plasminogen gene, within the plasminogen kringle 3 domain, in hereditary angioedema with normal C1 inhibitor. Biochem. Biophys. Res. Commun. 2018, 498, 193–198. [Google Scholar] [CrossRef]
- Datta, P.K.; Blake, M.C.; Moses, H.L. Regulation of plasminogen activator inhibitor-1 expression by transforming growth factor-beta -induced physical and functional interactions between smads and Sp1. J. Biol. Chem. 2000, 275, 40014–40019. [Google Scholar] [CrossRef]
- Bafunno, V.; Firinu, D.; D’Apolito, M.; Cordisco, G.; Loffredo, S.; Leccese, A.; Bova, M.; Barca, M.P.; Santacroce, R.; Cicardi, M.; et al. Mutation of the angiopoietin-1 gene (ANGPT1) associates with a new type of hereditary angioedema. J. Allergy Clin. Immunol. 2018, 141, 1009–1017. [Google Scholar] [CrossRef]
- Grivceva-Panovska, V.; Košnik, M.; Korošec, P.; Andrejevic, S.; Karadža-Lapic, L.; Rijavec, M. Hereditary angioedema due to C1-inhibitor deficiency in Macedonia: Clinical characteristics, novel SERPING1 mutations and genetic factors modifying the clinical phenotype. Ann. Med. 2018, 50, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Castellano, G.; Divella, C.; Sallustio, F.; Montinaro, V.; Curci, C.; Zanichelli, A.; Bonanni, E.; Suffritti, C.; Caccia, S.; Bossi, F.; et al. A transcriptomics study of hereditary angioedema attacks. J. Allergy Clin. Immunol. 2018, 142, 883–891. [Google Scholar] [CrossRef] [PubMed]
- Ponard, D.; Gaboriaud, C.; Charignon, D.; Ghannam, A.; Wagenaar-Bos, I.G.A.; Roem, D.; López-Lera, A.; López-Trascasa, M.; Tosi, M.; Drouet, C. SERPING1 mutation update: Mutation spectrum and C1 Inhibitor phenotypes. Hum. Mutat. 2020, 41, 38–57. [Google Scholar] [CrossRef] [PubMed]
- Banday, A.Z.; Kaur, A.; Jindal, A.K.; Rawat, A.; Singh, S. An update on the genetics and pathogenesis of hereditary angioedema. Genes Dis. 2020, 7, 75–83. [Google Scholar] [CrossRef]
- Khan, S.; Longhurst, H. Epigenetic alterations on C1-inhibitor expression may influence hereditary angioedema attack frequency and C4 levels. Clin. Exp. Immunol. 2020, 202, 144–145. [Google Scholar] [CrossRef]
- Vatsiou, S.; Zamanakou, M.; Loules, G.; Psarros, F.; Parsopoulou, F.; Csuka, D.; Valerieva, A.; Staevska, M.; Porebski, G.; Obtulowicz, K.; et al. A novel deep intronic SERPING1 variantas a cause of hereditary angioedema due to C1-inhibitor deficiency. Allergol. Int. 2020, 69, 443–449. [Google Scholar] [CrossRef] [PubMed]
- Hujová, P.; Soucek, P.; Grodecká, L.; Grombiríková, H.; Ravcuková, B.; Kuklínek, P.; Hakl, R.; Litzman, J.; Freiberger, T. Deep intronic mutation in SERPING1 caused hereditary angioedema through pseudoexon activation. J. Clin. Immunoogyl. 2020, 40, 435–446. [Google Scholar] [CrossRef] [PubMed]
- Kajdácsi, E.; Jandrasics, Z.; Veszeli, N.; Makó, V.; Koncz, A.; Gulyás, D.; Köhalmi, K.V.; Temesszentandrási, G.; Cervenak, L.; Gál, P.; et al. Patterns of C1-Inhibitor/Plasma Serine Protease Complexes in Healthy Humans and in Hereditary Angioedema Patients. Front. Immunol. 2020, 11, 794. [Google Scholar] [CrossRef]
- Corvillo, F.; de la Morena Barrio, M.E.; Marcos Bravo, C.; López-Trascasa, M.; Vicente, V.; Emsley, J.; Caballero, T.; Corral, J.; Lopez Lera, A. The FXII c.-4T>C Polymorphism as a Disease Modifier in Patients with Hereditary Angioedema Due to the FXIIp.Thr328Lys Variant. Front. Genet. 2020, 11, 1033. [Google Scholar] [CrossRef] [PubMed]
- Santacroce, R.; D’Andrea, G.; Maffione, A.B.; Margaglione, M.; d’Apolito, M. The Genetics of Hereditary Angioedema: A Review. J. Clin. Med. 2021, 10, 2023. [Google Scholar] [CrossRef]
- Yong, P.F.K.; Coulter, T.; El-Shanawany, T.; Garcez, T.; Hackett, S.; Jain, R.; Kiani-Alikhan, S.; Manson, A.; Noorani, S.; Stroud, C.; et al. A National Survey of Hereditary Angioedema and Acquired C1 Inhibitor Deficiency in the United Kingdom. J. Allergy Clin. Immunol. Pract. 2023, 11, 2476–2483. [Google Scholar] [CrossRef]
- Vincent, D.; Parsopoulou, F.; Martin, L.; Gaboriaud, C.; Demongeot, J.; Loules, G.; Fischer, S.; Cichon, S.; Germenis, A.E.; Ghannam, A.; et al. Hereditary angioedema with normal C1 inhibitor associated with carboxypeptidase N deficiency. J. Allergy Clin. Immunol. Glob. 2024, 3, 100223. [Google Scholar] [CrossRef]
- Craig, T.; Richwine, K.; Ishmael, F.T. Plasma microRNAs as biomarkers in hereditary angioedema. Ann. Allergy Asthma Immunol. 2024, 132, 723–729. [Google Scholar] [CrossRef]
- Fails, J.M.; Gierer, S.A. Phenotypic Heterogeny of Hereditary Angioedema Within a Single Family. Kans. J. Med. 2025, 18, 49–50. [Google Scholar] [CrossRef]
- Jayasena, C.S.; Ohyama, T.; Segil, N.; Groves, A.K. Notch signaling augments the canonical Wnt pathway to specify the size of the otic placode. Development 2008, 135, 2251–2261. [Google Scholar] [CrossRef]
- Tan, T.; Lu, B.; Zhang, J.; Niu, Y.; Si, W.; Wei, Q.; Ji, W. Notch1 signaling antagonizes transforming growth factor-β pathway and induces apoptosis in rabbit trophoblast stem cells. Stem Cells Dev. 2014, 23, 813–822. [Google Scholar] [CrossRef]
- Ghazi, A.; Grant, J.A. Hereditary angioedema: Epidemiology, management, and role of icatibant. Biol. Targets Ther. 2013, 7, 103–113. [Google Scholar] [CrossRef]
- Charignon, D.; Ponard, D.; de Gennes, C.; Drouet, C.; Ghannam, A. SERPING1 and F12 combined variants in a hereditary angioedema family. Ann. Allergy Asthma Immunol. 2018, 121, 500–502. [Google Scholar] [CrossRef] [PubMed]
- Grafe, I.; Yang, T.; Alexander, S.; Homan, E.P.; Lietman, C.; Jiang, M.M.; Bertin, T.; Munivez, E.; Chen, Y.; Dawson, B.; et al. Excessive transforming growth factor-β signaling is a common mechanism in osteogenesis imperfecta. Nat. Med. 2014, 20, 670–675. [Google Scholar] [CrossRef] [PubMed]
- Palomo, T.; Vilaça, T.; Lazaretti-Castro, M. Osteogenesis imperfecta: Diagnosis and treatment. Curr. Opin. Endocrinol. Diabetes Obes. 2017, 24, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Garajová, I.; Giovannetti, E.; Biasco, G.; Peters, G.J. c-Met as a Target for Personalized Therapy. Transl. Oncogen. 2015, 23, 13–31. [Google Scholar]
- Sagar, R.; Gotherstrom, C.; David, A.L.; Westgren, M. Fetal stem cell transplantation and gene therapy. Best Pract. Res. Clin. Obstet. Gynaecol. 2019, 58, 142–153. [Google Scholar] [CrossRef]
- Rossi, V.; Lee, B.; Marom, R. Osteogenesis imperfecta: Advancements in genetics and treatment. Curr. Opin. Pediatr. 2019, 31, 708–715. [Google Scholar] [CrossRef]
- Surowiec, R.K.; Battle, L.F.; Schlecht, S.H.; Wojtys, E.M.; Caird, M.S.; Kozloff, K.M. Gene Expression Profile and Acute Gene Expression Response to Sclerostin Inhibition in Osteogenesis Imperfecta Bone. JBMR Plus 2020, 4, e10377. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.G.; Thuillier, D.; Chin, E.N.; Alliston, T. Chondrocyte-intrinsic Smad3 represses Runx2-inducible matrix metalloproteinase 13 expression to maintain articular cartilage and prevent osteoarthritis. Arthritis Rheum. 2012, 64, 3278–3289. [Google Scholar] [CrossRef]
- Li, H.; Wang, D.; Yuan, Y.; Min, J. New insights on the MMP-13 regulatory network in the pathogenesis of early osteoarthritis. Arthritis Res. Ther. 2017, 19, 248. [Google Scholar] [CrossRef]
- Chaugule, S.; Constantinou, C.K.; John, A.A.; Micha, D.; Eekhoff, M.; Gravallese, E.; Gao, G.; Shim, J.H. Comprehensive Review of Osteogenesis Imperfecta: Current Treatments and Future Innovations. Hum. Gene Ther. 2025, 36, 597–617. [Google Scholar] [CrossRef]
- Dubuisson, L.; Lepreux, S.; Bioulac-Sage, P.; Balabaud, C.; Costa, A.M.; Rosenbaum, J.; Desmoulière, A. Expression and cellular localization of fibrillin-1 in normal and pathological human liver. J. Hepatol. 2001, 34, 514–522. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Huang, J.; Liu, Y. The extracellular matrix glycoprotein fibrillin-1 in health and disease. Front. Cell Dev. Biol. 2024, 11, 1302285. [Google Scholar] [CrossRef]
- Dong, R.; Dong, K.; Wang, X.; Chen, G.; Shen, C.; Zheng, S. Interleukin-33 overexpression is associated with gamma-glutamyl transferase in biliary atresia. Cytokine 2013, 61, 433–437. [Google Scholar] [CrossRef]
- Bessho, K.; Mourya, R.; Shivakumar, P.; Walters, S.; Magee, J.C.; Rao, M.; Jegga, A.G.; Bezerra, J.A. Gene expression signature for biliary atresia and a role for interleukin-8 in pathogenesis of experimental disease. Hepatology 2014, 60, 211–223. [Google Scholar] [CrossRef]
- Asai, A.; Malladi, S.; Misch, J.; Pan, X.; Malladi, P.; Diehl, A.M.; Whitington, P.F. Elaboration of tubules with active hedgehog drives parenchymal fibrogenesis in gestational alloimmune liver disease. Hum. Pathol. 2015, 46, 84–93. [Google Scholar] [CrossRef]
- Moyer, K.; Kaimal, V.; Pacheco, C.; Mourya, R.; Xu, H.; Shivakumar, P.; Chakraborty, R.; Rao, M.; Magee, J.C.; Bove, K.; et al. Staging of biliary atresia at diagnosis by molecular profiling of the liver. Genome Med. 2010, 2, 33. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Yang, L.; Liu, H.; Pang, S.; Chen, Y.; Fu, J.; Chen, Y.; Wen, Z.; Zhang, R.; Zhu, B.; et al. Identification of circulating MicroRNAs in biliary atresia by next-generation sequencing. J. Pediatr. Gastroenterol. Nutr. 2016, 63, 518–523. [Google Scholar] [CrossRef]
- Zhang, X.; Du, G.; Xu, Y.; Li, X.; Fan, W.; Chen, J.; Liu, C.; Zern, M.A.; Mu, Y.; Liu, P. Inhibition of notch signaling pathway prevents cholestatic liver fibrosis by decreasing the differentiation of hepatic progenitor cells into cholangiocytes. Lab Investig. 2016, 96, 350–360. [Google Scholar] [CrossRef]
- Kanta, J. Elastin in the liver. Front. Physiol. 2016, 7, 491. [Google Scholar] [CrossRef]
- Zhang, X.; Xu, Y.; Chen, J.M.; Liu, C.; Du, G.L.; Zhang, H.; Chen, G.; Jiang, S.L.; Liu, C.H.; Mu, Y.P.; et al. Huang Qi decoction prevents bdl-induced liver fibrosis through inhibition of notch signaling activation. Am. J. Chin. Med. 2017, 45, 85–104. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Tang, S.; Yang, L.; Li, K. Inhibition of the notch signaling pathway reduces the differentiation of hepatic progenitor cells into cholangiocytes in biliary atresia. Cell Physiol. Biochem. 2018, 49, 1115–1123. [Google Scholar] [CrossRef]
- Jiang, J.; Wang, J.; Shen, Z.; Lu, X.; Chen, G.; Huang, Y.; Dong, R.; Zheng, S. Serum MMP-7 in the diagnosis of biliary atresia. Pediatrics 2019, 144, e20190902. [Google Scholar] [CrossRef] [PubMed]
- Verhulst, S.; Roskams, T.; Sancho-Bru, P.; van Grunsven, L.A. Meta-analysis of human and mouse biliary epithelial cell gene profiles. Cells 2019, 8, 1117. [Google Scholar] [CrossRef]
- Quoseena, M.; Vuppaladadium, S.; Hussain, S.; Banu, S.; Bharathi, S.; Idris, M.M. Functional role of annexins in zebrafish caudal fin regeneration—A gene knockdown approach in regenerating tissue. Biochimie 2020, 175, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yuan, H.; Zhao, M.; Fang, L. Identification of signature of gene expression in biliary atresia using weighted gene co-expression network analysis. Medicine 2022, 101, e30232. [Google Scholar] [CrossRef]
- Choe, B.H.; Kim, K.M.; Kwon, S.; Lee, K.S.; Koo, J.H.; Lee, H.M.; Kim, M.K.; Kim, J.C. The pattern of differentially expressed genes in biliary atresia. J. Korean Med. Sci. 2003, 18, 392–396. [Google Scholar] [CrossRef]
- Nizery, L.; Chardot, C.; Sissaoui, S.; Capito, C.; Henrion-Caude, A.; Debray, D.; Girard, M. Biliary atresia: Clinical advances and perspectives. Clin. Res. Hepatol. Gastroenterol. 2016, 40, 281–287. [Google Scholar] [CrossRef]
- Bolia, R. Machine Learning in Biliary Atresia: Taking a Cautious Step into the Future. Indian J. Pediatr. 2025, 92, 114–115. [Google Scholar] [CrossRef] [PubMed]
- Bowen, R. Limit sets for axiom A diffeomorphisms. J. Differ. Equ. 1975, 18, 333–339. [Google Scholar] [CrossRef]
- Poincaré, H. Sur Les courbes définies par une équation différentielle. J. Math. Pures Appliquées 1886, 2, 151–217. [Google Scholar]
- Hilbert, D. Sur les Problèmes Futurs des Mathématiques: Les 23 Problèmes; J. Gabay: Paris, France, 1990. [Google Scholar]
- Delbrück, M. Unités biologiques douées de continuité génétique. Colloques Int. CNRS 1949, 8, 33–35. [Google Scholar]
- BoolNet. Available online: https://sysbio.uni-ulm.de (accessed on 15 February 2025).
- Network-Design. Available online: https://github.com/houssembk30/Network-Design (accessed on 5 May 2025).
- Espinoza, A. Available online: https://github.com/aer-neo/Frustration-and-Energy-Tool-Boolean-Networks/tree/5c9e7fe (accessed on 15 July 2025).
- Metacore. Available online: https://clarivate.com/life-sciences-healthcare (accessed on 15 January 2025).
- Hyun, K.; Jeon, J.; Park, K.; Kim, J. Writing, erasing and reading histone lysine methylations. Exp. Mol. Med. 2017, 49, e324. [Google Scholar] [CrossRef]
- Szczepanek, J.; Tretyn, A. MicroRNA-Mediated Regulation of Histone-Modifying Enzymes in Cancer: Mechanisms and Therapeutic Implications. Biomolecules 2023, 13, 1590. [Google Scholar] [CrossRef]
- Medscape. Available online: https://emedicine.medscape.com/article/135604-overview?form=fpf (accessed on 22 February 2025).
- NCBI. Available online: https://www.ncbi.nlm.nih.gov/nucleotide?cmd=search (accessed on 23 February 2025).
- Signor. Available online: https://signor.uniroma2.it/relation_result.php?id=SIGNOR-ST11&organism=human (accessed on 22 February 2025).
- Thom, R. Stabilité Structurelle et Morphogenèse; Benjamin: New York, NY, USA, 1972. [Google Scholar]
- Cosnard, M.; Demongeot, J. On the definitions of attractors. Lect. Notes Maths 1985, 1163, 23–31. [Google Scholar]
- Kauffman, S. Homeostasis and Differentiation in Random Genetic Control Networks. Nature 1969, 224, 177–178. [Google Scholar] [CrossRef] [PubMed]
- Thomas, R. Boolean formalization of genetic control circuits. J. Theor. Biol. 1973, 42, 563–585. [Google Scholar] [CrossRef]
- Thomas, R. On the relation between the logical structure of systems and their ability to generate multiple steady states or sustained oscillations. In Numerical Methods in the Study of Critical Phenomena, Proceedings of a Colloquium, Carry-le-Rouet, France, 2–4 June 1980; Springer Series in Synergetics; Springer: Berlin/Heidelberg, Germany, 1980; Volume 9, pp. 1–23. [Google Scholar]
- Demongeot, J.; Noual, M.; Sené, S. Combinatorics of Boolean automata circuits dynamics. Discret. Appl. Math. 2012, 160, 398–415. [Google Scholar] [CrossRef]
- Demongeot, J.; Hazgui, H.; Henrion-Caude, A. Genetic regulatory networks: Focus on attractors of their dynamics. In Computational Biology, Bioinformatics and Systems Biology; Tran, Q.N., Arabnia, H.R., Eds.; Elsevier: New York, NY, USA, 2015; pp. 135–165. [Google Scholar]
- Demongeot, J.; Jelassi, M.; Hazgui, H.; Ben Miled, S.; Bellamine Ben Saoud, N.; Taramasco, C. Biological Networks Entropies: Examples in Neural Memory Networks, Genetic Regulation Networks and Social Epidemic Networks. Entropy 2016, 20, 36. [Google Scholar] [CrossRef] [PubMed]
- Demongeot, J.; Aracena, J.; Ben Lamine, S.; Meignen, S.; Tonnelier, A.; Thomas, R. Dynamical systems and biological regulations. In Complex Systems; Goles, E., Martinez, S., Eds.; Kluwer: Amsterdam, The Netherlands, 2001; pp. 105–151. [Google Scholar]
- Aracena, J.; Demongeot, J.; Goles, E. On limit cycles of monotone functions with symmetric connection graphs. Theor. Comp. Sci. 2005, 322, 237–244. [Google Scholar] [CrossRef]
- Demongeot, J.; Elena, A.; Sené, S. Robustness in neural and genetic networks. Acta Biotheoretica 2008, 56, 27–49. [Google Scholar] [CrossRef]
- Demongeot, J.; Ben Amor, H.; Gillois, P.; Noual, M.; Sené, S. Robustness of regulatory networks. A Generic Approach with Applications at Different Levels: Physiologic, Metabolic and Genetic. Int. J. Mol. Sci. 2009, 10, 4437–4473. [Google Scholar] [CrossRef]
- Aracena, J.; Demongeot, J.; Fanchon, E.; Montalva, M. On the number of different dynamics in Boolean networks with deterministic update schedules. Math. Biosci. 2013, 242, 188–194. [Google Scholar] [CrossRef] [PubMed]
- Aracena, J.; Demongeot, J.; Fanchon, E.; Montalva, M. On the number of update digraphs and its relation with the feedback arc sets and tournaments. Discret. Appl. Math. 2013, 161, 1345–1355. [Google Scholar] [CrossRef]
- Demongeot, J.; Sené, S. About block-parallel Boolean networks: A position paper. Nat. Comput. 2020, 19, 5–13. [Google Scholar] [CrossRef]
- Demongeot, J.; Melliti, T.; Noual, M.; Regnault, D.; Sené, S. On Boolean Isolated Cycles and Tangential Double-Cycles Dynamics. In Automata and Complexity; Adamatzky, A., Ed.; Springer Series Emergence, Complexity, Computation; Springer: Berlin, Germany, 2022; Volume 42, pp. 9–19. [Google Scholar]
- Donoso-Leiva, I.; Goles, E.; Ríos-Wilson, M.; Sené, S. Impact of (a) synchronism on ECA: Towards a new classification. Chaos Solitons Fractals 2025, 199, 116601. [Google Scholar] [CrossRef]
- Demongeot, J.; Goles, E.; ben Khalfallah, H.; Montalva-Medel, M.; Sené, S. Robustness of Boolean networks to update modes: An application to hereditary angioedema. arXiv 2025, arXiv:2505.14923. [Google Scholar] [CrossRef]
- Combe, P.; Nencka, H. Frustration and overblocking on graphs. Math. Comp. Model. 1997, 26, 307–309. [Google Scholar] [CrossRef]
- Demongeot, J.; Waku, J. Robustness in biological regulatory networks II: Application to genetic threshold Boolean random regulatory networks. Comptes Rendus Mathématique I 2012, 350, 225–228. [Google Scholar] [CrossRef]
- Cappello, C.; Naserasr, R.; Steffen, E.; Wang, Z. Critically 3-frustrated signed graphs. Discret. Math. 2025, 348, 183–193. [Google Scholar] [CrossRef]
- Ortiz-Tavárez, J.M.; Yang, Z.; Kotov, N.; Mao, X. Statistical Mechanics of Frustrated Assemblies and Incompatible Graphs. Phys. Rev. Lett. 2025, 134, 147401. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Sheng, K.; Chen, C.; Qiu, A. Machine learning-assisted flexible dual modal sensor for multi-sensing detection and target object recognition in the grasping process. Lab Chip 2025, 25, 2247–2255. [Google Scholar] [CrossRef] [PubMed]




| Transcript Identity | Intensity Normal | Intensity BA | Ratio BA/Normal |
|---|---|---|---|
| Bcl-w | 16.7 | 28.8 | |
| Laminin BP (binding protein) | 0.7 | 7.7 | 11 |
| HRS (HGF-regulated tyrosine kinase substrate) | 0.5 | 3.5 | |
| Thymosin ß4 | 0.6 | 4.1 | 6.5 |
| Thymosin ß10 | 0.5 | 2.5 | 5.4 |
| TGF-ß | 0.6 | 2.1 | |
| TIMP-1 (tissue inhibitor of metalloproteinase) | 1.8 | 3.9 | 2.2 |
| SRP4 (signal recognition protein) | 2 | 7.1 | 3.6 |
| SRP9 | 1.7 | 5.6 | 3.3 |
| SNAP 45 (soluble NSF attachment protein) | 1.2 | 5.6 | 4.5 |
| Alu RNA BP | 0.8 | 6.9 | 8.5 |
| Supt5h (human homologue of yeast transcription factor SPT5) | 1.9 | 4.5 | 2.3 |
| Elf-2α kinase | 4.5 | 12.1 | 2.7 |
| HSP 27 (heat shock protein) | 2.1 | 3.7 | 1.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Demongeot, J.; Diallo, A.K.; Hazgui, H.; Jelassi, M.; Kelloufi, F.; ben Khalfallah, H.; Espinoza, A.; Montalva-Medel, M. Boolean Networks with Classic and New Updating Modes Applied to Genetic Regulation in Some Familial Diseases. Int. J. Mol. Sci. 2025, 26, 11976. https://doi.org/10.3390/ijms262411976
Demongeot J, Diallo AK, Hazgui H, Jelassi M, Kelloufi F, ben Khalfallah H, Espinoza A, Montalva-Medel M. Boolean Networks with Classic and New Updating Modes Applied to Genetic Regulation in Some Familial Diseases. International Journal of Molecular Sciences. 2025; 26(24):11976. https://doi.org/10.3390/ijms262411976
Chicago/Turabian StyleDemongeot, Jacques, Abdoul Khadir Diallo, Hana Hazgui, Mariem Jelassi, Fatine Kelloufi, Houssem ben Khalfallah, Alonso Espinoza, and Marco Montalva-Medel. 2025. "Boolean Networks with Classic and New Updating Modes Applied to Genetic Regulation in Some Familial Diseases" International Journal of Molecular Sciences 26, no. 24: 11976. https://doi.org/10.3390/ijms262411976
APA StyleDemongeot, J., Diallo, A. K., Hazgui, H., Jelassi, M., Kelloufi, F., ben Khalfallah, H., Espinoza, A., & Montalva-Medel, M. (2025). Boolean Networks with Classic and New Updating Modes Applied to Genetic Regulation in Some Familial Diseases. International Journal of Molecular Sciences, 26(24), 11976. https://doi.org/10.3390/ijms262411976







