Analysis of qPCR Data: From PCR Efficiency to Absolute Target Quantity
Abstract
1. Introduction
2. The qPCR Basics
2.1. PCR Kinetics
2.2. Fluorescence Monitoring During the PCR
2.3. Common Analysis of qPCR Data
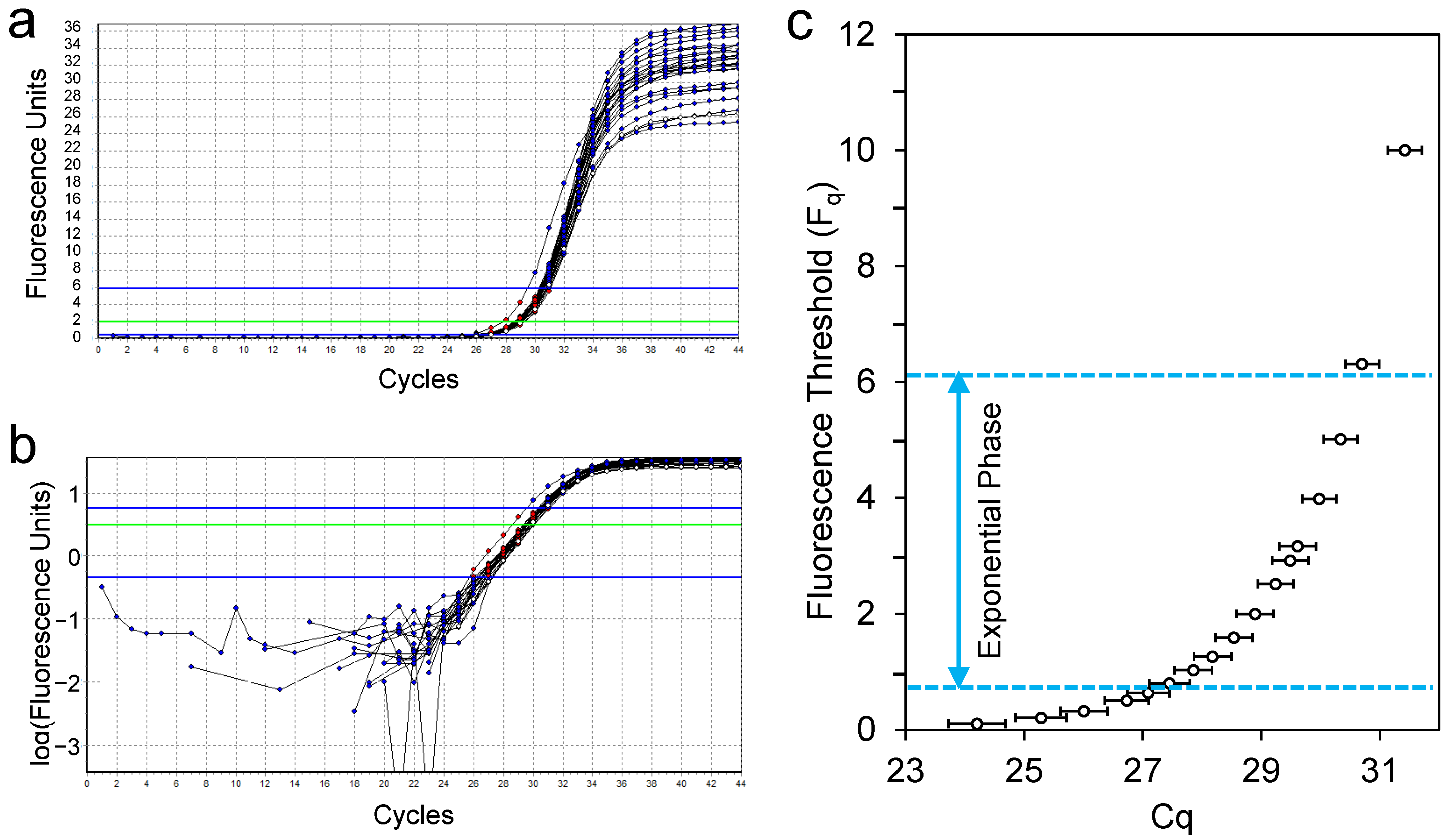
2.3.1. Removing Baseline Fluorescence
2.3.2. Setting the Quantification Threshold
2.3.3. Determining the PCR Efficiency
2.3.4. Comparison of qPCR Data Analysis Methods
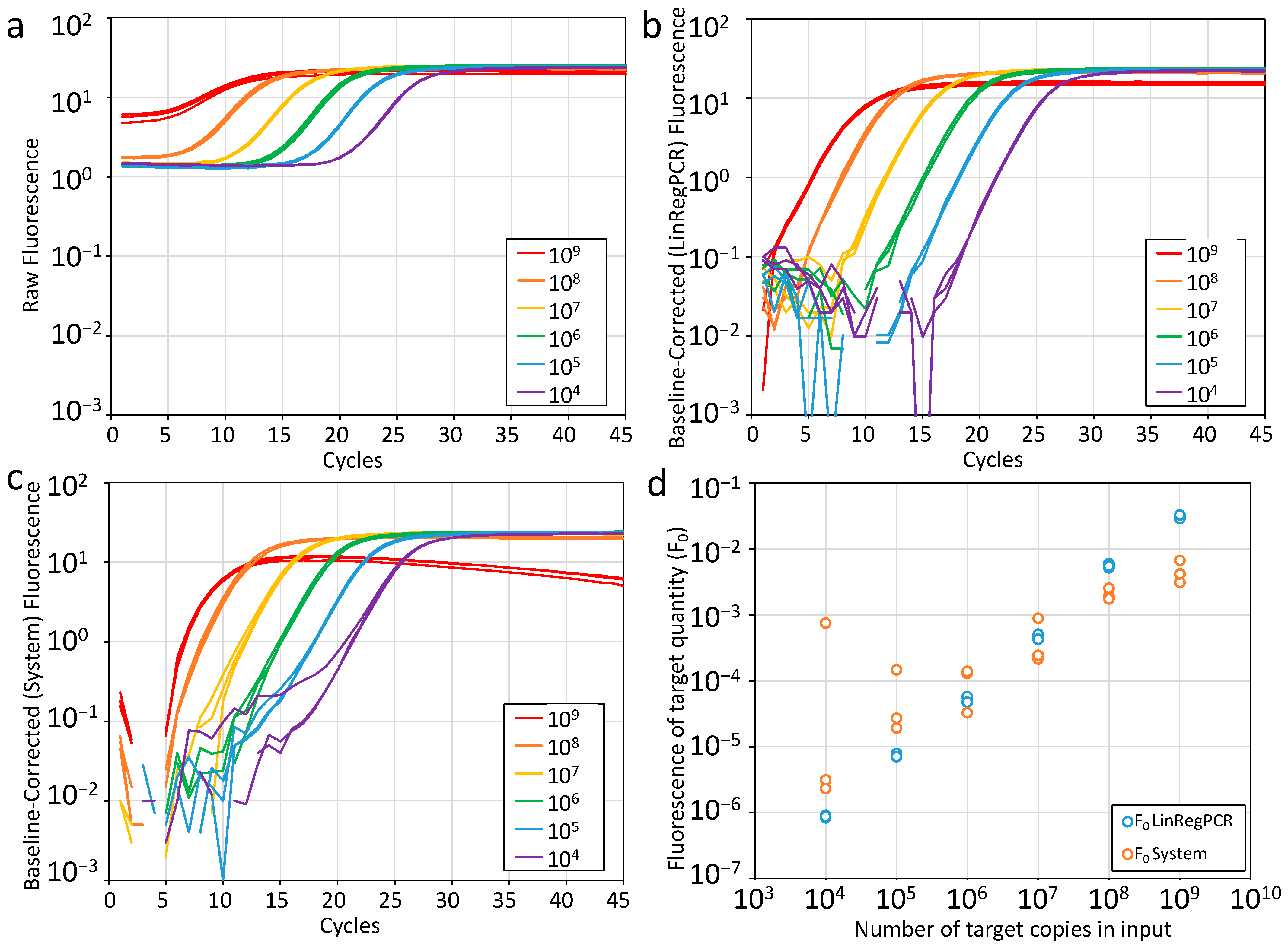
2.4. LinRegPCR Approach to qPCR Data Analysis
2.4.1. Baseline Subtraction Revisited
2.4.2. Quantification Threshold Setting
2.4.3. PCR Efficiency Determined from Amplification Curves
2.4.4. Reporting Results with F0 Values
3. From F0 to Ncopy
3.1. Ncopy Using a Calibration Curve
3.2. Ncopy Using Single Standard Calibration
3.3. Ncopy Using a Rule of Thumb
3.4. Ncopy Using the Limiting Component Approach
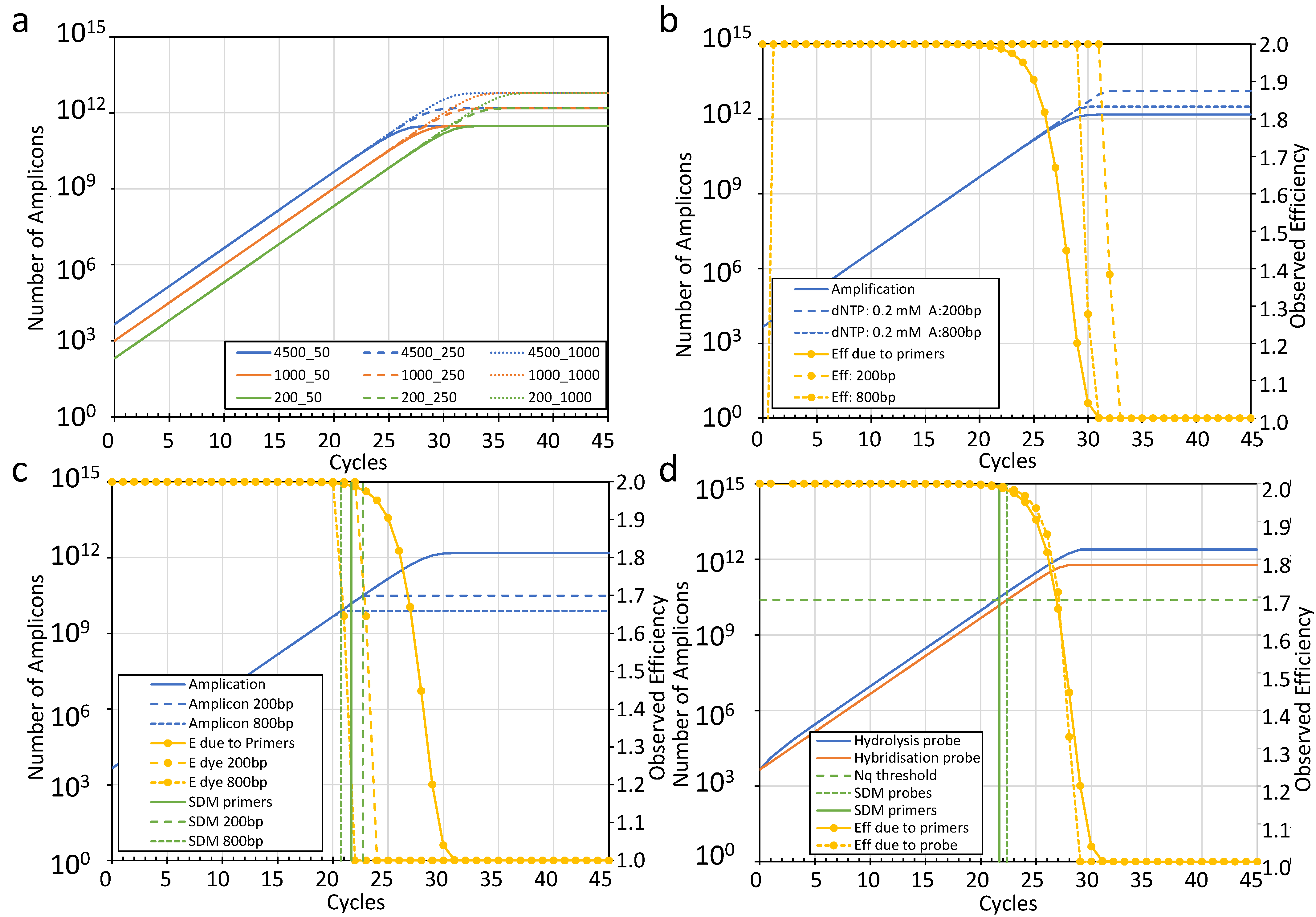
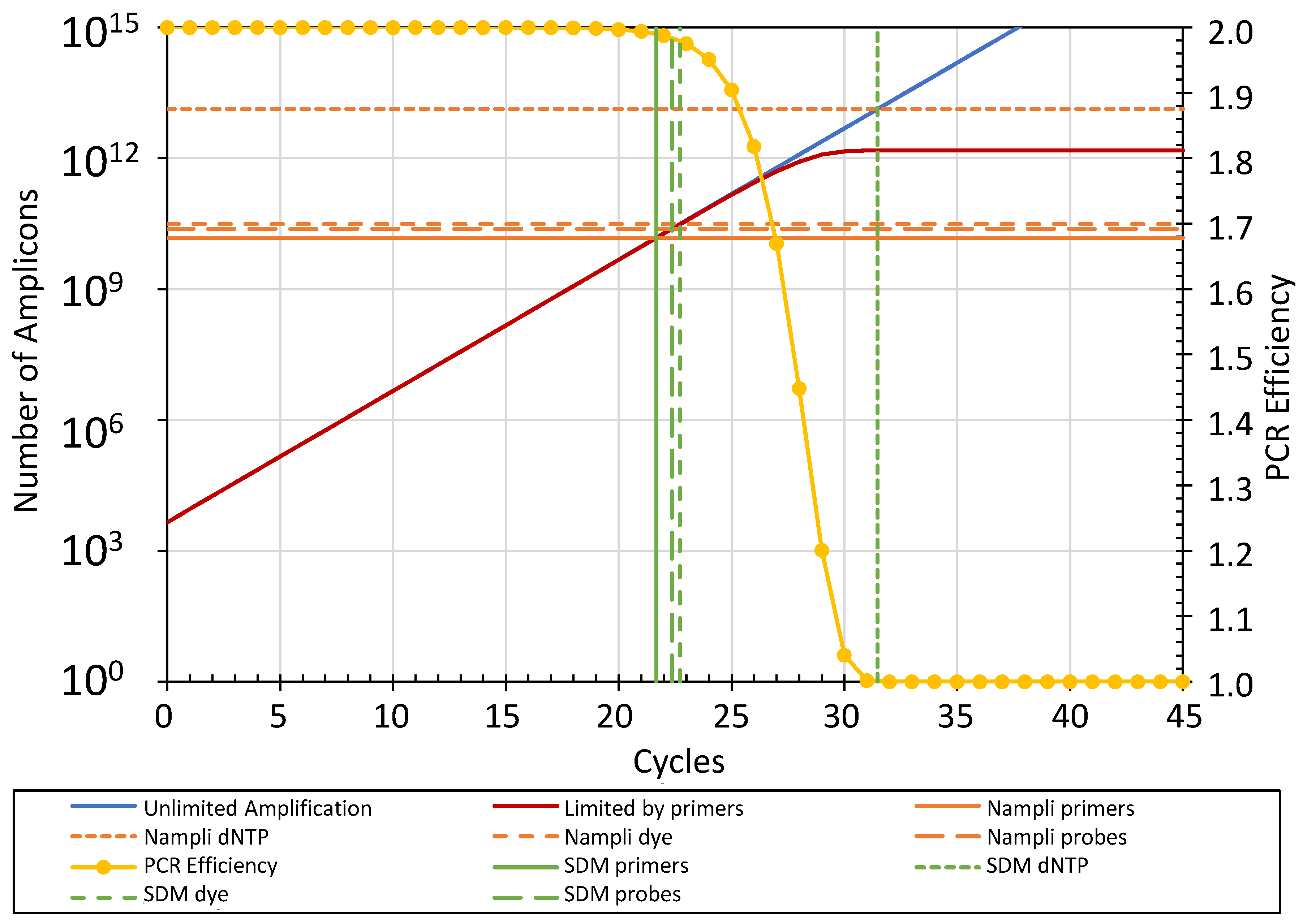
3.4.1. Principle of the Limiting Component Approach
3.4.2. Determining the SDM of an Amplification Curve
3.4.3. Components Limiting Amplicon Amplification
- Primers
- dNTPs
3.4.4. Components Limiting Amplicon Detection
- DNA-binding dyes
- Hybridization probes
- Hydrolysis probes
3.4.5. Calculating Ncopy Using the Limiting Components Approach
3.4.6. Relative Expression and Normalization Using Ncopy
4. Discussion and General Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| PCR | Polymerase Chain Reaction |
| qPCR | quantitative Polymerase Chain Reaction |
| toi | target of interest |
| ref | reference gene |
| unk | unknown sample |
| N0 | number of copies of the target at the start of the PCR |
| NC | number of copies of the target after C cycles |
| C | number of cycles |
| E | efficiency of the amplification reaction |
| F0 | fluorescence of the target molecules at the start of the PCR |
| FC | fluorescence of the copies of the target after C cycles |
| Fq | quantification threshold |
| Cq | quantification cycle |
| ΔCq | difference between the Cq values of toi and ref |
| ΔΔCq | difference between the ΔCq values of control and treated groups |
| Eamc | amplification-curve-derived PCR efficiency |
| Estc | standard-curve-derived PCR efficiency |
| SDM | maximum of the second derivative of the amplification curve |
| RelExp | relative expression |
| FoldDiff | fold difference |
| Ncopy | number of target copies at the start of the PCR |
| Nampli | maximum number of amplicons due to a limiting reaction component |
| KH | hybridization constant |
| [A] | concentration of the amplicon |
| [P] | concentration of the primers |
| [H] | concentration of the hybrids |
| Nprimer | number of primer molecules |
| NdNTP | number of dNTP molecules |
| AdNTP | number of dNTP molecules needed to synthesize one amplicon |
| Ndye | number of DNA-binding dye molecules |
| Adye | number of dye molecules needed to stain one amplicon |
| SGI | SYBR Green I |
| Nprobe | number of probe molecules |
References
- Higuchi, R.; Fockler, C.; Dollinger, G.; Watson, R. Kinetic PCR analysis: Real-time monitoring of DNA amplification reactions. Biotechnology 1993, 11, 1026–1030. [Google Scholar] [CrossRef]
- Wittwer, C.T.; Herrmann, M.G.; Moss, A.A.; Rasmussen, R.P. Continuous fluorescence monitoring of rapid cycle DNA amplification. Biotechniques 1997, 22, 130–138. [Google Scholar] [CrossRef]
- Derveaux, S.; Vandesompele, J.; Hellemans, J. How to do successful gene expression analysis using real-time PCR. Methods 2010, 50, 227–230. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Ruijter, J.M.; van den Hoff, M.J.B.; Kubista, M.; Pfaffl, M.W.; Shipley, G.L.; Tran, N.; Rodiger, S.; Untergasser, A.; Mueller, R.; et al. MIQE 2.0: Revision of the Minimum Information for Publication of Quantitative Real-Time PCR Experiments Guidelines. Clin. Chem. 2025, 71, 634–651. [Google Scholar] [CrossRef]
- Larkin, S.E.; Holmes, S.; Cree, I.A.; Walker, T.; Basketter, V.; Bickers, B.; Harris, S.; Garbis, S.D.; Townsend, P.A.; Aukim-Hastie, C. Identification of markers of prostate cancer progression using candidate gene expression. Br. J. Cancer 2012, 106, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A. Why the need for qPCR publication guidelines?—The case for MIQE, Methods. Methods 2010, 50, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): Trends and problems. J. Mol. Endocrinol. 2002, 29, 23–39. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Tichopad, A.; Kitchen, R.; Riedmaier, I.; Becker, C.; Stahlberg, A.; Kubista, M. Design and optimization of reverse-transcription quantitative PCR experiments. Clin. Chem. 2009, 55, 1816–1823. [Google Scholar] [CrossRef]
- Bivins, A.; Kaya, D.; Bibby, K.; Simpson, S.L.; Bustin, S.A.; Shanks, O.C.; Ahmed, W. Variability in RT-qPCR assay parameters indicates unreliable SARS-CoV-2 RNA quantification for wastewater surveillance. Water Res. 2021, 203, 117516. [Google Scholar] [CrossRef]
- Ruiz-Villalba, A.; Ruijter, J.M.; van den Hoff, M.J.B. Use and Misuse of Cq in qPCR Data Analysis and Reporting. Life 2021, 11, 496. [Google Scholar] [CrossRef]
- Bustin, S.; Nolan, T. Talking the talk, but not walking the walk: RT-qPCR as a paradigm for the lack of reproducibility in molecular research. Eur. J. Clin. Investig. 2017, 47, 756–774. [Google Scholar] [CrossRef] [PubMed]
- Ramakers, C.; Ruijter, J.M.; Lekanne Deprez, R.H.; Moorman, A.F.M. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 2003, 339, 62–66. [Google Scholar] [CrossRef] [PubMed]
- Ruijter, J.M.; Ramakers, C.; Hoogaars, W.M.; Karlen, Y.; Bakker, O.; van den Hoff, M.J.; Moorman, A.F. Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009, 37, e45. [Google Scholar] [CrossRef]
- Ruijter, J.M.; Barnewall, R.J.; Marsh, I.B.; Szentirmay, A.N.; Quinn, J.C.; van Houdt, R.; Gunst, Q.D.; van den Hoff, M.J.B. Efficiency-correction is required for accurate qPCR analysis and reporting. Clin. Chem. 2021, 67, 829–842. [Google Scholar] [CrossRef]
- Karsai, A.; Muller, S.; Platz, S.; Hauser, M.T. Evaluation of a homemade SYBR green I reaction mixture for real-time PCR quantification of gene expression. Biotechniques 2002, 32, 790–792, 794–796. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Paepe, A.; Speleman, F. Elimination of primer-dimer artifacts and genomic coamplification using a two-step SYBR green I real-time RT-PCR. Anal. Biochem. 2002, 303, 95–98. [Google Scholar] [CrossRef]
- Ruijter, J.M.; Lorenz, P.; Tuomi, J.M.; Hecker, M.; van den Hoff, M.J. Fluorescent-increase kinetics of different fluorescent reporters used for qPCR depend on monitoring chemistry, targeted sequence, type of DNA input and PCR efficiency. Mikrochim. Acta 2014, 181, 1689–1696. [Google Scholar] [CrossRef]
- Rasmussen, R. Quantification on the LightCycler instrument. In Rapid Cycle Real-Time PCR: Methods and Applications; Meuer, S., Wittwer, C., Nakagawara, K., Eds.; Springer: Berlin/Heidelberg, Germany, 2001; pp. 21–34. [Google Scholar]
- Rebrikov, D.V.; Trofimov, D.I. Real-time PCR: A review of approaches to data analysis. Appl. Biochem. Microbiol. 2006, 42, 455–463. [Google Scholar] [CrossRef]
- Ruijter, J.M.; Pfaffl, M.W.; Zhao, S.; Spiess, A.N.; Boggy, G.; Blom, J.; Rutledge, R.G.; Sisti, D.; Lievens, A.; De Preter, K.; et al. Evaluation of qPCR curve analysis methods for reliable biomarker discovery: Bias, resolution, precision, and implications. Methods 2013, 59, 32–46. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Nolan, T.; Hands, R.E.; Bustin, S.A. Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 2006, 1, 1559–1582. [Google Scholar] [CrossRef]
- Ruijter, J.M.; Ruiz-Villalba, A.; van den Hoff, M.J.B. Cq Values Do Not Reflect Nucleic Acid Quantity in Biological Samples. Clin. Chem. 2021, 68, 7–9. [Google Scholar] [CrossRef]
- Whale, A.S.; von der Heide, E.K.; Kohlenberg, M.; Brinckmann, A.; Baedker, S.; Karalay, O.; Fernandez-Gonzalez, A.; Busby, E.J.; Bustin, S.A.; Hauser, H.; et al. Digital PCR can augment the interpretation of RT-qPCR Cq values for SARS-CoV-2 diagnostics. Methods 2022, 201, 5–14. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Walker, N.J. Tech.Sight. A technique whose time has come. Science 2002, 296, 557–559. [Google Scholar] [CrossRef]
- Ginzinger, D.G. Gene quantification using real-time quantitative PCR: An emerging technology hits the mainstream. Exp. Hematol. 2002, 30, 503–512. [Google Scholar] [CrossRef] [PubMed]
- Peirson, S.N.; Butler, J.N.; Foster, R.G. Experimental validation of novel and conventional approaches to quantitative real-time PCR data analysis. Nucleic Acids Res. 2003, 31, e73. [Google Scholar] [CrossRef] [PubMed]
- Lekanne Deprez, R.H.; Fijnvandraat, A.C.; Ruijter, J.M.; Moorman, A.F.M. Sensitivity and accuracy of quantitative real-time polymerase chain reaction using SYBR green I depends on cDNA synthesis conditions. Anal. Biochem. 2002, 307, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Grgicak, C.M.; Urban, Z.M.; Cotton, R.W. Investigation of reproducibility and error associated with qPCR methods using Quantifiler(R) Duo DNA quantification kit. J. Forensic Sci. 2010, 55, 1331–1339. [Google Scholar] [CrossRef]
- Suslov, O.; Steindler, D.A. PCR inhibition by reverse transcriptase leads to an overestimation of amplification efficiency. Nucleic Acids Res. 2005, 33, e181. [Google Scholar] [CrossRef]
- Casado-Martin, L.; Hernandez, M.; Yeramian, N.; Gonzalez-Pena, M.J.; Eiros, J.M.; Rodriguez-Lazaro, D. Wastewater-based epidemiology for monitoring enteric viruses: A case study in Valladolid, Spain (2020–2021). Front. Microbiol. 2025, 16, 1586478. [Google Scholar] [CrossRef]
- Svec, D.; Tichopad, A.; Novosadova, V.; Pfaffl, M.W.; Kubista, M. How good is a PCR efficiency estimate: Recommendations for precise and robust qPCR efficiency assessments. Biomol. Detect. Quantif. 2015, 3, 9–16. [Google Scholar] [CrossRef]
- Huggett, J.F.; Novak, T.; Garson, J.A.; Green, C.; Morris-Jones, S.D.; Miller, R.F.; Zumla, A. Differential susceptibility of PCR reactions to inhibitors: An important and unrecognised phenomenon. BMC Res. Notes 2008, 1, 70. [Google Scholar] [CrossRef]
- Karlen, Y.; McNair, A.; Perseguers, S.; Mazza, C.; Mermod, N. Statistical significance of quantitative PCR. BMC Bioinform. 2007, 8, 131. [Google Scholar] [CrossRef]
- Rutledge, R.G.; Stewart, D. Assessing the performance capabilities of LRE-based assays for absolute quantitative real-time PCR. PLoS ONE 2010, 5, e9731. [Google Scholar] [CrossRef]
- Guescini, M.; Sisti, D.; Rocchi, M.B.; Stocchi, L.; Stocchi, V. A new real-time PCR method to overcome significant quantitative inaccuracy due to slight amplification inhibition. BMC Bioinform. 2008, 9, 326. [Google Scholar] [CrossRef]
- Spiess, A.N.; Deutschmann, C.; Burdukiewicz, M.; Himmelreich, R.; Klat, K.; Schierack, P.; Rodiger, S. Impact of smoothing on parameter estimation in quantitative DNA amplification experiments. Clin. Chem. 2015, 61, 379–388. [Google Scholar] [CrossRef]
- Zhao, S.; Fernald, R.D. Comprehensive algorithm for quantitative real-time polymerase chain reaction. J. Comput. Biol. 2005, 12, 1047–1064. [Google Scholar] [CrossRef]
- Boggy, G.J.; Woolf, P.J. A mechanistic model of PCR for accurate quantification of quantitative PCR data. PLoS ONE 2010, 5, e12355. [Google Scholar] [CrossRef]
- Lievens, A.; Van Aelst, S.; Van den Bulcke, M.; Goetghebeur, E. Enhanced analysis of real-time PCR data by using a variable efficiency model: FPK-PCR. Nucleic Acids Res. 2012, 40, e10. [Google Scholar] [CrossRef]
- Tichopad, A.; Dilger, M.; Schwarz, G.; Pfaffl, M.W. Standardized determination of real-time PCR efficiency from a single reaction set-up. Nucleic Acids Res. 2003, 31, e122. [Google Scholar] [CrossRef]
- Untergasser, A.; Ruijter, J.M.; Benes, V.; van den Hoff, M.J.B. Web-based LinRegPCR: Application for the visualization and analysis of (RT)-qPCR amplification and melting data. BMC Bioinform. 2021, 22, 398. [Google Scholar] [CrossRef]
- Ruiz-Villalba, A.; Mattiotti, A.; Gunst, Q.D.; Cano-Ballesteros, S.; van den Hoff, M.J.; Ruijter, J.M. Reference genes for gene expression studies in the mouse heart. Sci. Rep. 2017, 7, 24. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034-1. [Google Scholar] [CrossRef]
- Hellemans, J.; Mortier, G.; De, P.A.; Speleman, F.; Vandesompele, J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007, 8, R19. [Google Scholar] [CrossRef]
- Ruiz-Villalba, A.; van Pelt-Verkuil, E.; Gunst, Q.D.; Ruijter, J.M.; van den Hoff, M.J. Amplification of nonspecific products in quantitative polymerase chain reactions (qPCR). Biomol. Detect. Quantif. 2017, 14, 7–18. [Google Scholar] [CrossRef]
- Ririe, K.M.; Rasmussen, R.P.; Wittwer, C.T. Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal. Biochem. 1997, 245, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Cleveland, M.H.; He, H.J.; Milavec, M.; Bae, Y.K.; Vallone, P.M.; Huggett, J.F. Digital PCR for the characterization of reference materials. Mol. Asp. Med. 2024, 96, 101256. [Google Scholar] [CrossRef]
- Huggett, J.F.; Foy, C.A.; Benes, V.; Emslie, K.; Garson, J.A.; Haynes, R.; Hellemans, J.; Kubista, M.; Mueller, R.D.; Nolan, T.; et al. The digital MIQE guidelines: Minimum Information for Publication of Quantitative Digital PCR Experiments. Clin. Chem. 2013, 59, 892–902. [Google Scholar] [CrossRef]
- Wang, Y.; Keith, M.; Leyme, A.; Bergelson, S.; Feschenko, M. Monitoring long-term DNA storage via absolute copy number quantification by ddPCR. Anal. Biochem. 2019, 583, 113363. [Google Scholar] [CrossRef]
- Hays, A.; Wissel, M.; Colletti, K.; Soon, R.; Azadeh, M.; Smith, J.; Doddareddy, R.; Chalfant, M.; Adamowicz, W.; Ramaswamy, S.S.; et al. Recommendations for Method Development and Validation of qPCR and dPCR Assays in Support of Cell and Gene Therapy Drug Development. AAPS J. 2024, 26, 24. [Google Scholar] [CrossRef]
- Green, M.R.; Sambrook, J. Quantifying and Storing RNA. Cold Spring Harb. Protoc. 2020, 2020, 101709. [Google Scholar] [CrossRef]
- Vermeulen, J.; de Preter, K.; Naranjo, A.; Vercruysse, L.; Van Roy, N.; Hellemans, J.; Swerts, K.; Bravo, S.; Scaruffi, P.; Tonini, G.P.; et al. Predicting outcomes for children with neuroblastoma using a multigene-expression signature: A retrospective SIOPEN/COG/GPOH study. Lancet Oncol. 2009, 10, 663–671. [Google Scholar] [CrossRef]
- Brankatschk, R.; Bodenhausen, N.; Zeyer, J.; Burgmann, H. Simple absolute quantification method correcting for quantitative PCR efficiency variations for microbial community samples. Appl. Environ. Microbiol. 2012, 78, 4481–4489. [Google Scholar] [CrossRef] [PubMed]
- Shipley, G. Assay Design for Real-Time qPCR. In PCR Technology: Current Innovations, 3rd ed.; Nolan, T., Bustin, S.A., Eds.; CRC Press: London, UK; New York, NY, USA, 2013; pp. 177–199. [Google Scholar]
- de Ronde, M.W.; Ruijter, J.M.; Lanfear, D.; Bayes-Genis, A.; Kok, M.; Creemers, E.; Pinto, Y.M.; Pinto-Sietsma, S.J. Practical data handling pipeline improves performance of qPCR-based circulating miRNA measurements. RNA 2017, 23, 811–821. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Liao, P.; Chen, Z.; Chen, H.; Wu, Y.; Li, S.; Deng, Y.; He, N. Improvement and Application of qPCR (Real-Time Quantitative Polymerase Chain Reaction) Data Processing Method for Home-Made Integrated Nucleic Acid Detection System. J. Nanosci. Nanotechnol. 2020, 20, 7369–7375. [Google Scholar] [CrossRef] [PubMed]
- Spiess, A.N.; Feig, C.; Ritz, C. Highly accurate sigmoidal fitting of real-time PCR data by introducing a parameter for asymmetry. BMC Bioinform. 2008, 9, 221. [Google Scholar] [CrossRef] [PubMed]
- Lawyer, F.C.; Stoffel, S.; Saiki, R.K.; Chang, S.Y.; Landre, P.A.; Abramson, R.D.; Gelfand, D.H. High-level expression, purification, and enzymatic characterization of full-length Thermus aquaticus DNA polymerase and a truncated form deficient in 5′ to 3′ exonuclease activity. PCR Methods Appl. 1993, 2, 275–287. [Google Scholar] [CrossRef]
- Gevertz, J.L.; Dunn, S.M.; Roth, C.M. Mathematical model of real-time PCR kinetics. Biotechnol. Bioeng. 2005, 92, 346–355. [Google Scholar] [CrossRef]
- Watson, J.D.; Crick, F.H. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature 1953, 171, 737–738. [Google Scholar] [CrossRef] [PubMed]
- Dragan, A.I.; Pavlovic, R.; McGivney, J.B.; Casas-Finet, J.R.; Bishop, E.S.; Strouse, R.J.; Schenerman, M.A.; Geddes, C.D. SYBR Green I: Fluorescence properties and interaction with DNA. J. Fluoresc. 2012, 22, 1189–1199. [Google Scholar] [CrossRef] [PubMed]
- Zipper, H.; Brunner, H.; Bernhagen, J.; Vitzthum, F. Investigations on DNA intercalation and surface binding by SYBR Green I, its structure determination and methodological implications. Nucleic Acids Res. 2004, 32, e103. [Google Scholar] [CrossRef] [PubMed]
- Mao, F.; Leung, W.Y.; Xin, X. Characterization of EvaGreen and the implication of its physicochemical properties for qPCR applications. BMC Biotechnol. 2007, 7, 76. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruijter, J.M.; van den Hoff, M.J.B. Analysis of qPCR Data: From PCR Efficiency to Absolute Target Quantity. Int. J. Mol. Sci. 2025, 26, 11885. https://doi.org/10.3390/ijms262411885
Ruijter JM, van den Hoff MJB. Analysis of qPCR Data: From PCR Efficiency to Absolute Target Quantity. International Journal of Molecular Sciences. 2025; 26(24):11885. https://doi.org/10.3390/ijms262411885
Chicago/Turabian StyleRuijter, Jan M., and Maurice J. B. van den Hoff. 2025. "Analysis of qPCR Data: From PCR Efficiency to Absolute Target Quantity" International Journal of Molecular Sciences 26, no. 24: 11885. https://doi.org/10.3390/ijms262411885
APA StyleRuijter, J. M., & van den Hoff, M. J. B. (2025). Analysis of qPCR Data: From PCR Efficiency to Absolute Target Quantity. International Journal of Molecular Sciences, 26(24), 11885. https://doi.org/10.3390/ijms262411885






