Application of Artificial Intelligence Technology in Plant MicroRNA Research: Progress, Challenges, and Prospects
Abstract
1. Introduction
1.1. Biological Significance of Plant miRNAs
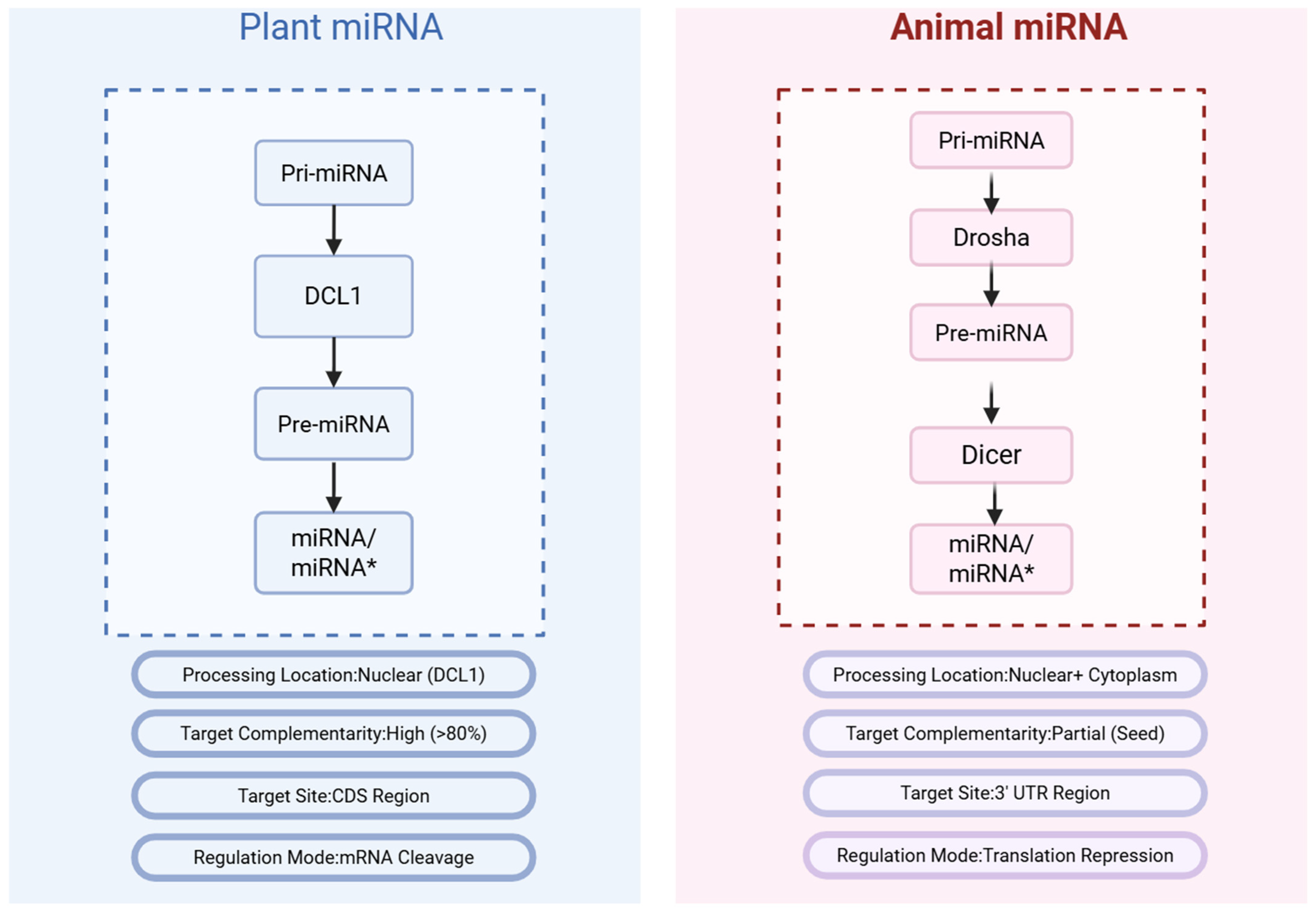
1.2. Essential Differences Between Plant and Animal miRNA Systems
1.3. Limitations of Traditional Research Methods
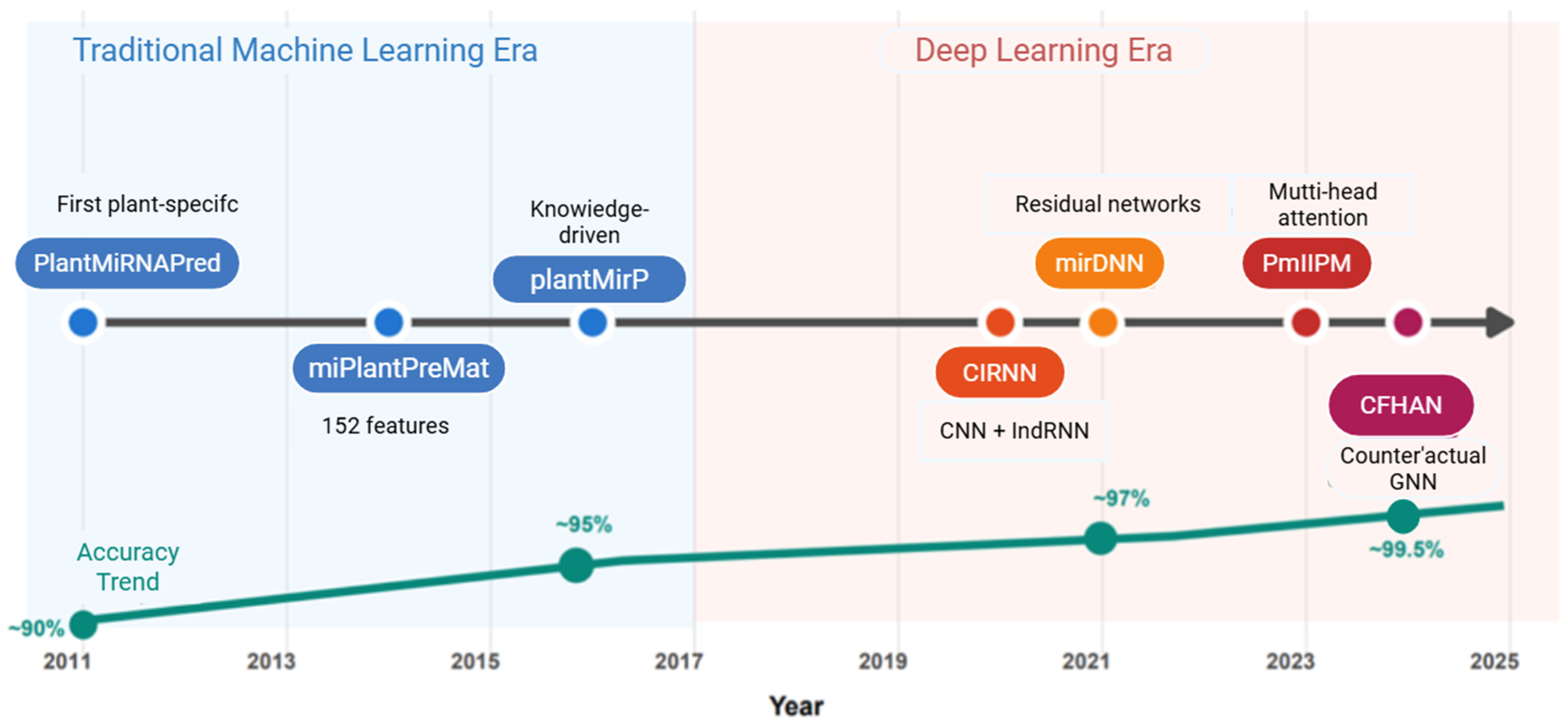
1.4. Necessity and Development History of Artificial Intelligence Technology
2. Traditional Machine Learning Methods
2.1. Support Vector Machines: The Foundation Tool for Plant miRNA Prediction
2.2. Random Forests and Ensemble Learning: Leveraging Multiple Perspectives
2.3. Data Imbalance: A Pervasive Challenge and Its Solutions
2.4. Systematic Comparison of Prediction Criteria and Weighting Schemes Across Tools
2.5. Practical Accessibility and Long-Term Sustainability of Traditional Machine Learning Tools
3. Deep Learning Methods
3.1. Convolutional Neural Networks: Extracting Local Patterns with Global Understanding
3.2. Recurrent Neural Networks: Modeling Sequential Dependencies
3.3. Transformer Architecture: Leveraging Attention Mechanisms for Long-Range Interactions
3.4. Graph Neural Networks: Analyzing Network Topology
3.5. Systematic Comparison of Deep Learning Architectures
3.6. Systematic Advantages and Persistent Challenges of Deep Learning
3.7. Critical Assessment of Deep Learning Tool Ecosystem and Reproducibility
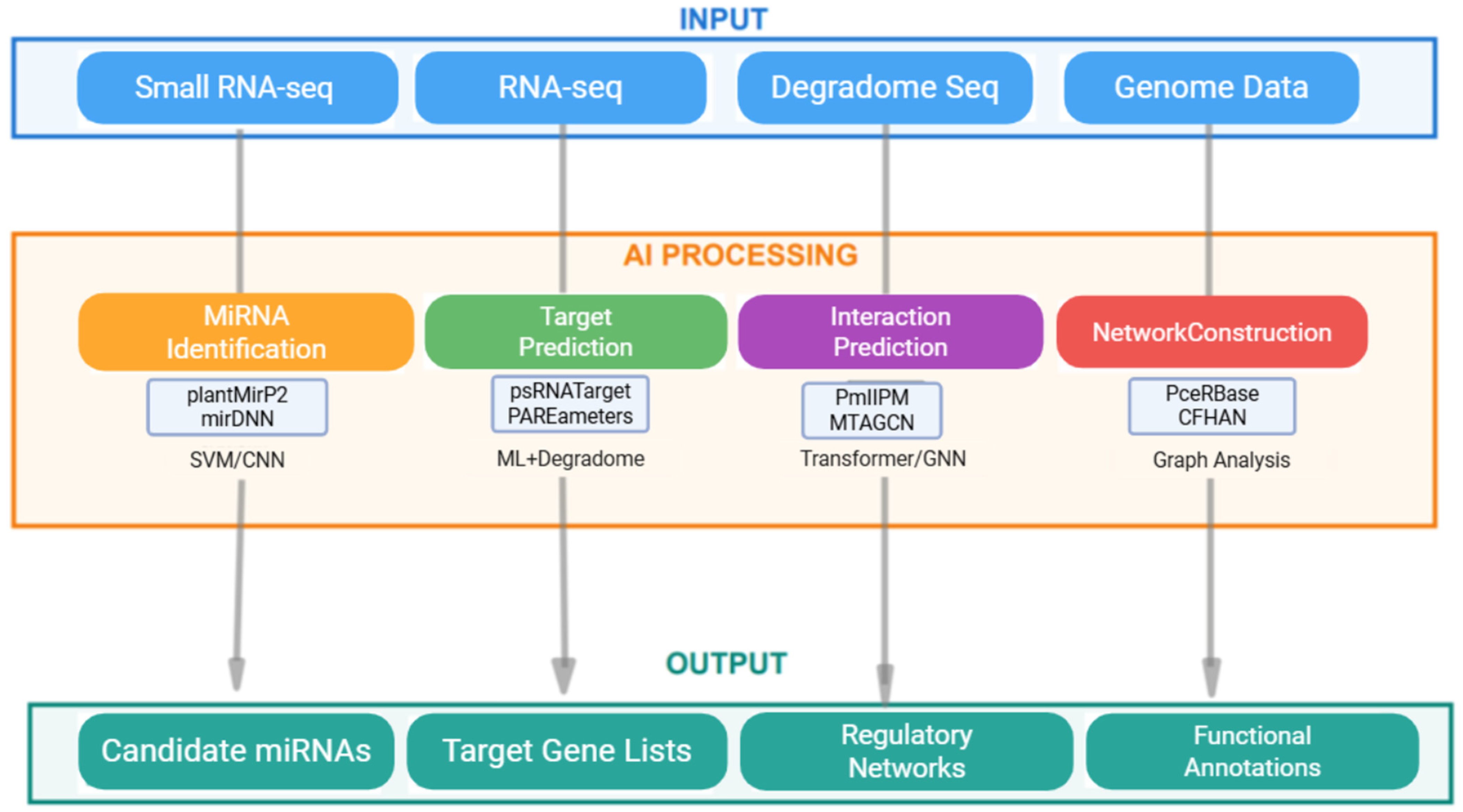
4. Application Scenarios: From Identification to Networks
4.1. Evolution of Target Gene Prediction Methods
4.2. Exploration of Multi-Omics Data Integration
4.3. Competing Endogenous RNA Networks: Another Dimension of Regulation
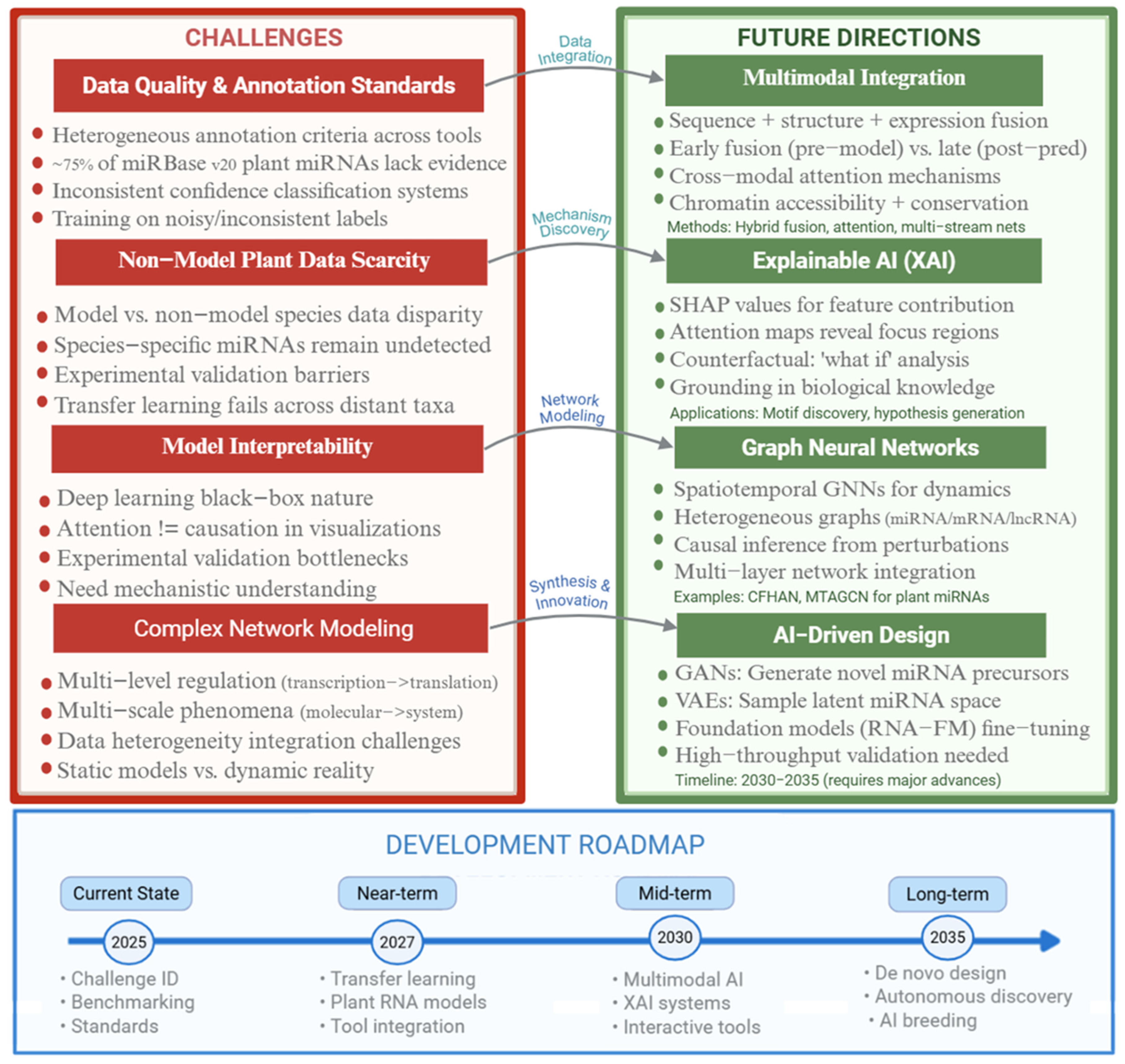
5. Challenges and Prospects
5.1. Data Quality and Annotation Standards
5.2. Non-Model Plant Data Scarcity: A Critical Bottleneck
5.3. Model Interpretability Challenges
5.4. Complex Regulatory Network Modeling
5.5. Future Development Directions
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bartel, D.P. MicroRNAs: Target Recognition and Regulatory Functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Jones-Rhoades, M.W.; Bartel, D.P.; Bartel, B. MicroRNAS and Their Regulatory Roles in Plants. Annu. Rev. Plant Biol. 2006, 57, 19–53. [Google Scholar] [CrossRef]
- Voinnet, O. Origin, Biogenesis, and Activity of Plant MicroRNAs. Cell 2009, 136, 669–687. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Poethig, R.S. Temporal Regulation of Shoot Development in Arabidopsis Thaliana by miR156 and Its Target SPL3. Development 2006, 133, 3539–3547. [Google Scholar] [CrossRef] [PubMed]
- Chen, X. Small RNAs and Their Roles in Plant Development. Annu. Rev. Cell Dev. Biol. 2009, 25, 21–44. [Google Scholar] [CrossRef] [PubMed]
- Sunkar, R.; Li, Y.-F.; Jagadeeswaran, G. Functions of MicroRNAs in Plant Stress Responses. Trends Plant Sci. 2012, 17, 196–203. [Google Scholar] [CrossRef]
- Axtell, M.J.; Meyers, B.C. Revisiting Criteria for Plant MicroRNA Annotation in the Era of Big Data. Plant Cell 2018, 30, 272–284. [Google Scholar] [CrossRef]
- Li, Z.; Xu, R.; Li, N. MicroRNAs from plants to animals, do they define a new messenger for communication? Nutr. Metab. 2018, 15, 68. [Google Scholar] [CrossRef]
- Dai, X.; Zhuang, Z.; Zhao, P.X. psRNATarget: A Plant Small RNA Target Analysis Server (2017 Release). Nucleic Acids Res. 2018, 46, W49–W54. [Google Scholar] [CrossRef]
- Rogers, K.; Chen, X. Biogenesis, Turnover, and Mode of Action of Plant MicroRNAs. Plant Cell 2013, 25, 2383–2399. [Google Scholar] [CrossRef]
- Axtell, M.J.; Westholm, J.O.; Lai, E.C. Vive la Différence: Biogenesis and Evolution of MicroRNAs in Plants and Animals. Genome Biol. 2011, 12, 221. [Google Scholar] [CrossRef] [PubMed]
- Jones-Rhoades, M.W.; Bartel, D.P. Computational Identification of Plant MicroRNAs and Their Targets, Including a Stress-Induced miRNA. Mol. Cell 2004, 14, 787–799. [Google Scholar] [CrossRef] [PubMed]
- German, M.; Pillay, M.; Jeong, D.; Hetawal, A.; Luo, S.; Janardhanan, P.; Kannan, V.; Rymarquis, L.A.; Nobuta, K.; German, R.; et al. Global identification of microRNA–target RNA pairs by parallel analysis of RNA ends. Nat. Biotechnol. 2008, 26, 941–946. [Google Scholar] [CrossRef] [PubMed]
- Addo-Quaye, C.; Eshoo, T.W.; Bartel, D.P.; Axtell, M.J. Endogenous siRNA and miRNA Targets Identified by Sequencing of the Arabidopsis Degradome. Curr. Biol. 2008, 18, 758–762. [Google Scholar] [CrossRef]
- Axtell, M.J. Classification and Comparison of Small RNAs from Plants. Annu. Rev. Plant Biol. 2013, 64, 137–159. [Google Scholar] [CrossRef]
- Axtell, M.J.; Bowman, J.L. Evolution of Plant MicroRNAs and Their Targets. Trends Plant Sci. 2008, 13, 343–349. [Google Scholar] [CrossRef]
- Kozomara, A.; Griffiths-Jones, S. miRBase: Annotating High Confidence MicroRNAs Using Deep Sequencing Data. Nucleic Acids Res. 2014, 42, D68–D73. [Google Scholar] [CrossRef]
- Dai, X.; Zhao, P.X. psRNATarget: A Plant Small RNA Target Analysis Server. Nucleic Acids Res. 2011, 39, W155–W159. [Google Scholar] [CrossRef]
- Li, Q.; Liu, G.; Bao, Y.; Wu, Y.; You, Q. Evaluation and Application of Tools for the Identification of Known MicroRNAs in Plants. Appl. Plant Sci. 2021, 9, e11414. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2018, 47, D155–D162. [Google Scholar] [CrossRef]
- Kuang, Z.; Wang, Y.; Li, L.; Yang, X. miRDeep-P2: Accurate and Fast Analysis of the MicroRNA Transcriptome in Plants. Bioinformatics 2019, 35, 2521–2522. [Google Scholar] [CrossRef] [PubMed]
- Xuan, P.; Guo, M.; Liu, X.; Huang, Y.; Li, W.; Huang, Y. PlantMiRNAPred: Efficient Classification of Real and Pseudo Plant Pre-miRNAs. Bioinformatics 2011, 27, 1368–1376. [Google Scholar] [CrossRef] [PubMed]
- Fan, D.; Yao, Y.; Yi, M. PlantMirP2: An Accurate, Fast and Easy-To-Use Program for Plant Pre-miRNA and miRNA Prediction. Genes 2021, 12, 1280. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.; Liu, D.; Sun, C.; Luan, Y. Prediction of Plant Pre-microRNAs and Their MicroRNAs in Genome-Scale Sequences Using Structure-Sequence Features and Support Vector Machine. BMC Bioinform. 2014, 15, 423. [Google Scholar] [CrossRef]
- Yao, Y.; Ma, C.; Deng, H.; Liu, Q.; Zhang, J.; Yi, M. plantMirP: An Efficient Computational Program for the Prediction of Plant Pre-miRNA by Incorporating Knowledge-Based Energy Features. Mol. Biosyst. 2016, 12, 3124–3131. [Google Scholar] [CrossRef]
- Tseng, K.-C.; Chiang-Hsieh, Y.-F.; Pai, H.; Chow, C.-N.; Lee, S.-C.; Zheng, H.-Q.; Kuo, P.-L.; Li, G.-Z.; Hung, Y.-C.; Lin, N.-S.; et al. microRPM: A MicroRNA Prediction Model Based Only on Plant Small RNA Sequencing Data. Bioinformatics 2018, 34, 1108–1115. [Google Scholar] [CrossRef]
- Rawal, H.C.; Ali, S.; Mondal, T.K. miRPreM and tiRPreM: Improved Methodologies for the Prediction of miRNAs and tRNA-Induced Small Non-Coding RNAs for Model and Non-Model Organisms. Brief. Bioinform. 2022, 23, bbab448. [Google Scholar] [CrossRef]
- Rousseau, A.-J.; Geubbelmans, M.; Burzykowski, T.; Valkenborg, D. Deep Learning. Am. J. Orthod. Dentofac. Orthop. 2024, 165, 369–371. [Google Scholar] [CrossRef]
- Ching, T.; Himmelstein, D.S.; Beaulieu-Jones, B.K.; Kalinin, A.A.; Do, B.T.; Way, G.P.; Ferrero, E.; Agapow, P.-M.; Zietz, M.; Hoffman, M.M.; et al. Opportunities and Obstacles for Deep Learning in Biology and Medicine. J. R. Soc. Interface 2018, 15, 20170387. [Google Scholar] [CrossRef]
- Eraslan, G.; Avsec, Ž.; Gagneur, J.; Theis, F.J. Deep Learning: New Computational Modelling Techniques for Genomics. Nat. Rev. Genet. 2019, 20, 389–403. [Google Scholar] [CrossRef]
- Zou, J.; Huss, M.; Abid, A.; Mohammadi, P.; Torkamani, A.; Telenti, A. A Primer on Deep Learning in Genomics. Nat. Genet. 2019, 51, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Camacho, D.M.; Collins, K.M.; Powers, R.K.; Costello, J.C.; Collins, J.J. Next-Generation Machine Learning for Biological Networks. Cell 2018, 173, 1581–1592. [Google Scholar] [CrossRef]
- Min, S.; Lee, B.; Yoon, S. Deep Learning in Bioinformatics. Brief. Bioinform. 2017, 18, 851–869. [Google Scholar] [CrossRef] [PubMed]
- Angermueller, C.; Pärnamaa, T.; Parts, L.; Stegle, O. Deep Learning for Computational Biology. Mol. Syst. Biol. 2016, 12, 878. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Rives, A.; Meier, J.; Sercu, T.; Goyal, S.; Lin, Z.; Liu, J.; Guo, D.; Ott, M.; Zitnick, C.L.; Ma, J.; et al. Biological Structure and Function Emerge from Scaling Unsupervised Learning to 250 Million Protein Sequences. Proc. Natl. Acad. Sci. USA 2021, 118, e2016239118. [Google Scholar] [CrossRef]
- Tripathi, N.; Hérisson, J.; Faulon, J.-L. Machine Learning in Predictive Biocatalysis: A Comparative Review of Methods and Applications. Biotechnol. Adv. 2025, 84, 108698. [Google Scholar] [CrossRef]
- Hao, P.-Y.; Chiang, J.-H.; Chen, Y.-D. Possibilistic Classification by Support Vector Networks. Neural Netw. 2022, 149, 40–56. [Google Scholar] [CrossRef]
- Mantero, A.; Ishwaran, H. Unsupervised Random Forests. Stat. Anal. Data Min. 2021, 14, 144–167. [Google Scholar] [CrossRef]
- Cui, H.; Zhai, J.; Ma, C. miRLocator: Machine Learning-Based Prediction of Mature MicroRNAs within Plant Pre-miRNA Sequences. PLoS ONE 2015, 10, e0142753. [Google Scholar] [CrossRef]
- Zhang, Z.; Jiang, L.; Wang, J.; Gu, P.; Chen, M. MTide: An Integrated Tool for the Identification of miRNA-Target Interaction in Plants. Bioinformatics 2015, 31, 290–291. [Google Scholar] [CrossRef]
- Naseem, A.; Khan, Y.D. An Intelligent Model for Prediction of Abiotic Stress-Responsive MicroRNAs in Plants Using Statistical Moments Based Features and Ensemble Approaches. Methods 2024, 228, 65–79. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Wu, H.; Wang, W.; Ma, W.; Sun, X.; Lu, Z. MiPred: Classification of Real and Pseudo MicroRNA Precursors Using Random Forest Prediction Model with Combined Features. Nucleic Acids Res. 2007, 35, W339–W344. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Liao, M.; Gao, Y.; Ji, R.; He, Z.; Zou, Q. Improved and Promising Identification of Human MicroRNAs by Incorporating a High-Quality Negative Set. IEEE/ACM Trans. Comput. Biol. Bioinform. 2014, 11, 192–201. [Google Scholar] [CrossRef]
- Dablain, D.; Krawczyk, B.; Chawla, N.V. DeepSMOTE: Fusing Deep Learning and SMOTE for Imbalanced Data. IEEE Trans. Neural Netw. Learn. Syst. 2023, 34, 6390–6404. [Google Scholar] [CrossRef]
- Lertampaiporn, S.; Thammarongtham, C.; Nukoolkit, C.; Kaewkamnerdpong, B.; Ruengjitchatchawalya, M. Heterogeneous Ensemble Approach with Discriminative Features and Modified-SMOTEbagging for Pre-miRNA Classification. Nucleic Acids Res. 2013, 41, e21. [Google Scholar] [CrossRef]
- Alptekin, B.; Akpinar, B.A.; Budak, H. A Comprehensive Prescription for Plant miRNA Identification. Front. Plant Sci. 2017, 7, 2058. [Google Scholar] [CrossRef]
- Hammond, R.K.; Gupta, P.; Patel, P.; Meyers, B.C. miRador: A fast and precise tool for the prediction of plant miRNAs. Plant Physiol. 2023, 191, 894–903. [Google Scholar] [CrossRef]
- Vanek, A.; Griffiths-Jones, S.; Meyers, B.C.; Shahid, S.; Axtell, M.J. miRScore: A rapid and precise microRNA validation tool. PLOS Comput. Biol. 2025, 21, e1013663. [Google Scholar] [CrossRef]
- Srivastava, P.K.; Moturu, T.R.; Pandey, P.; Baldwin, I.T.; Pandey, S.P. A comparison of performance of plant miRNA target prediction tools and the characterization of features for genome-wide target prediction. BMC Genom. 2014, 15, 348. [Google Scholar] [CrossRef]
- Thody, J.; Moulton, V.; Mohorianu, I. PAREameters: A Tool for Computational Inference of Plant miRNA-mRNA Targeting Rules Using Small RNA and Degradome Sequencing Data. Nucleic Acids Res. 2020, 48, 2258–2270. [Google Scholar] [CrossRef] [PubMed]
- Persson Hodén, K.; Hu, X.; Martínez, G.; Dixelius, C. smartPARE: An R Package for Efficient Identification of True mRNA Cleavage Sites. Int. J. Mol. Sci. 2021, 22, 4267. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Shen, H.-B. Predicting RNA–protein binding sites and motifs through combining local and global deep convolutional neural networks. Bioinformatics 2018, 34, 3427–3436. [Google Scholar] [CrossRef] [PubMed]
- Yones, C.; Raad, J.; Bugnon, L.A.; Milone, D.H.; Stegmayer, G. High Precision in MicroRNA Prediction: A Novel Genome-Wide Approach with Convolutional Deep Residual Networks. Comput. Biol. Med. 2021, 134, 104448. [Google Scholar] [CrossRef]
- Zhang, P.; Meng, J.; Luan, Y.; Liu, C. Plant miRNA-lncRNA Interaction Prediction with the Ensemble of CNN and IndRNN. Interdiscip. Sci. 2020, 12, 82–89. [Google Scholar] [CrossRef]
- Hochreiter, S.; Schmidhuber, J. Long Short-Term Memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef]
- Bu, Y.; Zheng, J.; Jia, C. An Efficient Deep Learning Based Predictor for Identifying miRNA-Triggered phasiRNA Loci in Plant. Math. Biosci. Eng. 2023, 20, 6853–6865. [Google Scholar] [CrossRef]
- Kang, Q.; Meng, J.; Shi, W.; Luan, Y. Ensemble Deep Learning Based on Multi-Level Information Enhancement and Greedy Fuzzy Decision for Plant miRNA-lncRNA Interaction Prediction. Interdiscip. Sci. 2021, 13, 603–614. [Google Scholar] [CrossRef]
- Akerman, O.; Isakov, H.; Levi, R.; Psevkin, V.; Louzoun, Y. Counting Is Almost All You Need. Front. Immunol. 2022, 13, 1031011. [Google Scholar] [CrossRef]
- Tang, X.; Ji, L. Predicting Plant miRNA-lncRNA Interactions via a Deep Learning Method. IEEE Trans. Nanobiosci. 2023, 22, 728–733. [Google Scholar] [CrossRef]
- Li, H.; Meng, J.; Wang, Z.; Luan, Y. PmiProPred: A Novel Method Towards Plant miRNA Promoter Prediction Based on CNN-Transformer Network and Convolutional Block Attention Mechanism. Int. J. Biol. Macromol. 2025, 302, 140630. [Google Scholar] [CrossRef] [PubMed]
- Scarselli, F.; Gori, M.; Tsoi, A.C.; Hagenbuchner, M.; Monfardini, G. The Graph Neural Network Model. IEEE Trans. Neural Netw. 2009, 20, 61–80. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Ning, Z.; Zhu, X.; Zhang, Y.; Liu, C.; Jiang, S.; Yuan, Z.; Zhang, H. Plant lncRNA-miRNA Interaction Prediction Based on Counterfactual Heterogeneous Graph Attention Network. Interdiscip. Sci. 2025, 17, 244–256. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Xiang, Y.; Wang, X.; Xue, W.; Yue, Z. MTAGCN: Predicting miRNA-Target Associations in Camellia sinensis var. assamica Through Graph Convolution Neural Network. BMC Bioinform. 2022, 23, 271. [Google Scholar] [CrossRef]
- Bugnon, L.A.; Yones, C.; Milone, D.H.; Stegmayer, G. Deep Neural Architectures for Highly Imbalanced Data in Bioinformatics. IEEE Trans. Neural Netw. Learn. Syst. 2020, 31, 2857–2867. [Google Scholar] [CrossRef]
- Zhang, S.; Zhou, J.; Hu, H.; Gong, H.; Chen, L.; Cheng, C.; Zeng, J. A deep learning framework for modeling structural features of RNA-binding protein targets. Nucleic Acids Res. 2015, 44, e32. [Google Scholar] [CrossRef]
- Zhang, W.; Zhang, P.; Sun, W.; Xu, J.; Liao, L.; Cao, Y.; Han, Y. Improving plant miRNA-target prediction with self-supervised k-mer embedding and spectral graph convolutional neural network. PeerJ 2024, 12, e17396. [Google Scholar] [CrossRef]
- Wen, M.; Cong, P.; Zhang, Z.; Lu, H.; Li, T. DeepMirTar: A deep-learning approach for predicting human miRNA targets. Bioinformatics 2018, 34, 3781–3787. [Google Scholar] [CrossRef]
- Maleki, F.; Ovens, K.; Gupta, R.; Reinhold, C.; Spatz, A.; Forghani, R. Generalizability of Machine Learning Models: Quantitative Evaluation of Three Methodological Pitfalls. Radiol. Artif. Intell. 2022, 5, e220028. [Google Scholar] [CrossRef]
- Hossen, M.I.; Awrangjeb, M.; Pan, S.; Mamun, A.A. Transfer learning in agriculture: A review. Artif. Intell. Rev. 2025, 58, 97. [Google Scholar] [CrossRef]
- Karim, M.R.; Islam, T.; Shajalal, M.; Beyan, O.; Lange, C.; Cochez, M.; Rebholz-Schuhmann, D.; Decker, S. Explainable AI for Bioinformatics: Methods, Tools and Applications. Brief. Bioinform. 2023, 24, bbad236. [Google Scholar] [CrossRef] [PubMed]
- Murdoch, W.J.; Singh, C.; Kumbier, K.; Abbasi-Asl, R.; Yu, B. Definitions, Methods, and Applications in Interpretable Machine Learning. Proc. Natl. Acad. Sci. USA 2019, 116, 22071–22080. [Google Scholar] [CrossRef] [PubMed]
- Rudin, C. Stop Explaining Black Box Machine Learning Models for High Stakes Decisions and Use Interpretable Models Instead. Nat. Mach. Intell. 2019, 1, 206–215. [Google Scholar] [CrossRef] [PubMed]
- Gunning, D.; Stefik, M.; Choi, J.; Miller, T.; Stumpf, S.; Yang, G.-Z. XAI-Explainable Artificial Intelligence. Sci. Robot. 2019, 4, eaay7120. [Google Scholar] [CrossRef]
- Yang, L.; Lu, L.; Liu, C.; Zhang, J.; Guo, K.; Zhang, N.; Zhou, F.; Zhao, Y. Interactive Exploration of CNN Interpretability via Coalitional Game Theory. Sci. Rep. 2025, 15, 9261. [Google Scholar] [CrossRef]
- Sheng, N.; Huang, L.; Gao, L.; Cao, Y.; Xie, X.; Wang, Y. A survey of computational methods and databases for lncRNA-miRNA interaction prediction. IEEE/ACM Trans. Comput. Biol. Bioinform. 2023, 20, 2810–2826. [Google Scholar] [CrossRef]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA Hypothesis: The Rosetta Stone of a Hidden RNA Language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef]
- Enright, A.J.; John, B.; Gaul, U.; Tuschl, T.; Sander, C.; Marks, D.S. MicroRNA Targets in Drosophila. Genome Biol. 2003, 5, R1. [Google Scholar] [CrossRef]
- Allen, E.; Xie, Z.; Gustafson, A.M.; Carrington, J.C. microRNA-Directed Phasing during Trans-Acting siRNA Biogenesis in Plants. Cell 2005, 121, 207–221. [Google Scholar] [CrossRef]
- Axtell, M.J. ShortStack: Comprehensive Annotation and Quantification of Small RNA Genes. RNA 2013, 19, 740–751. [Google Scholar] [CrossRef]
- Meng, J.; Shi, L.; Luan, Y. Plant MicroRNA-Target Interaction Identification Model Based on the Integration of Prediction Tools and Support Vector Machine. PLoS ONE 2014, 9, e103181. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hou, C.; Wu, M.; Lu, S.; Hsing, Y.I.; Chen, H.M. Beyond cleaved small RNA targets: Unraveling the complexity of plant RNA degradome data. BMC Genom. 2014, 15, 15. [Google Scholar] [CrossRef] [PubMed]
- Brousse, C.; Liu, Q.; Beauclair, L.; Delarue, M.; Crespi, M.; Szécsi, J. A Non-Canonical Plant microRNA Target Site. Nucleic Acids Res. 2014, 42, 5270–5279. [Google Scholar] [CrossRef] [PubMed]
- Libbrecht, M.W.; Noble, W.S. Machine Learning Applications in Genetics and Genomics. Nat. Rev. Genet. 2015, 16, 321–332. [Google Scholar] [CrossRef]
- Peng, Y.; Croce, C.M. The Role of MicroRNAs in Human Cancer. Signal Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef]
- Hasin, Y.; Seldin, M.; Lusis, A. Multi-Omics Approaches to Disease. Genome Biol. 2017, 18, 83. [Google Scholar] [CrossRef]
- Pinu, F.R.; Beale, D.J.; Paten, A.M.; Kouremenos, K.; Swarup, S.; Schirra, H.J.; Wishart, D. Systems Biology and Multi-Omics Integration: Viewpoints from the Metabolomics Research Community. Metabolites 2019, 9, 76. [Google Scholar] [CrossRef]
- Barabási, A.-L.; Gulbahce, N.; Loscalzo, J. Network Medicine: A Network-Based Approach to Human Disease. Nat. Rev. Genet. 2011, 12, 56–68. [Google Scholar] [CrossRef]
- Ritchie, M.D.; Holzinger, E.R.; Li, R.; Pendergrass, S.A.; Kim, D. Methods of Integrating Data to Uncover Genotype-Phenotype Interactions. Nat. Rev. Genet. 2015, 16, 85–97. [Google Scholar] [CrossRef]
- Bersanelli, M.; Mosca, E.; Remondini, D.; Giampieri, E.; Sala, C.; Castellani, G.; Milanesi, L. Methods for the Integration of Multi-Omics Data: Mathematical Aspects. BMC Bioinform. 2016, 17 (Suppl. S2), 15. [Google Scholar] [CrossRef]
- Mattick, J.S.; Rinn, J.L. Discovery and Annotation of Long Noncoding RNAs. Nat. Struct. Mol. Biol. 2015, 22, 5–7. [Google Scholar] [CrossRef] [PubMed]
- Quinn, J.J.; Chang, H.Y. Unique Features of Long Non-coding RNA Biogenesis and Function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Chew, G.-L.; Pauli, A.; Schier, A.F. Conservation of uORF Repressiveness and Sequence Features in Mouse, Human and Zebrafish. Nat. Commun. 2016, 7, 11663. [Google Scholar] [CrossRef] [PubMed]
- Lucero, L.; Ferrero, L.; Fonouni-Farde, C.; Ariel, F. Functional classification of plant long noncoding RNAs: A transcript is known by the company it keeps. New Phytol. 2021, 229, 1251–1260. [Google Scholar] [CrossRef]
- Franco-Zorrilla, J.M.; Valli, A.; Todesco, M.; Mateos, I.; Puga, M.I.; Rubio-Somoza, I.; Leyva, A.; Weigel, D.; García, J.A.; Paz-Ares, J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 2007, 39, 1033–1037. [Google Scholar] [CrossRef]
- Todesco, M.; Rubio-Somoza, I.; Paz-Ares, J.; Weigel, D. A Collection of Target Mimics for Comprehensive Analysis of MicroRNA Function in Arabidopsis thaliana. PLoS Genet. 2010, 6, e1001031. [Google Scholar] [CrossRef]
- Tay, Y.; Rinn, J.; Pandolfi, P.P. The Multilayered Complexity of ceRNA Crosstalk and Competition. Nature 2014, 505, 344–352. [Google Scholar] [CrossRef]
- Thomson, D.W.; Dinger, M.E. Endogenous MicroRNA Sponges: Evidence and Controversy. Nat. Rev. Genet. 2016, 17, 272–283. [Google Scholar] [CrossRef]
- Cesana, M.; Cacchiarelli, D.; Legnini, I.; Santini, T.; Sthandier, O.; Chinappi, M.; Tramontano, A.; Bozzoni, I. A Long Noncoding RNA Controls Muscle Differentiation by Functioning as a Competing Endogenous RNA. Cell 2011, 147, 358–369. [Google Scholar] [CrossRef]
- Karreth, F.A.; Tay, Y.; Perna, D.; Ala, U.; Tan, S.M.; Rust, A.G.; DeNicola, G.; Webster, K.A.; Weiss, D.; Perez-Mancera, P.A.; et al. In Vivo Identification of Tumor-Suppressive PTEN ceRNAs in an Oncogenic BRAF-Induced Mouse Model of Melanoma. Cell 2011, 147, 382–395. [Google Scholar] [CrossRef]
- Kang, Q.; Meng, J.; Su, C.; Luan, Y. Mining Plant Endogenous Target Mimics from miRNA-lncRNA Interactions Based on Dual-Path Parallel Ensemble Pruning Method. Brief. Bioinform. 2022, 23, bbab440. [Google Scholar] [CrossRef]
- Yuan, C.; Meng, X.; Li, X.; Illing, N.; Ingle, R.A.; Wang, J.; Chen, M. PceRBase: A Database of Plant Competing Endogenous RNA. Nucleic Acids Res. 2017, 45, D1009–D1014. [Google Scholar] [CrossRef] [PubMed]
- Ala, U.; Karreth, F.A.; Bosia, C.; Pagnani, A.; Taulli, R.; Léopold, V.; Tay, Y.; Provero, P.; Zecchina, R.; Pandolfi, P.P. Integrated Transcriptional and Competitive Endogenous RNA Networks Are Cross-Regulated in Permissive Molecular Environments. Proc. Natl. Acad. Sci. USA 2013, 110, 7154–7159. [Google Scholar] [CrossRef] [PubMed]
- Bosson, A.D.; Zamudio, J.R.; Sharp, P.A. Endogenous miRNA and Target Concentrations Determine Susceptibility to Potential ceRNA Competition. Mol. Cell 2014, 56, 347–359. [Google Scholar] [CrossRef] [PubMed]
- Denzler, R.; Agarwal, V.; Stefano, J.; Bartel, D.P.; Stoffel, M. Assessing the ceRNA Hypothesis with Quantitative Measurements of miRNA and Target Abundance. Mol. Cell 2014, 54, 766–776. [Google Scholar] [CrossRef]
- Wang, J.; Liu, X.; Wu, H.; Ni, P.; Gu, Z.; Qiao, Y.; Chen, N.; Sun, F.; Fan, Q. CREB Up-Regulates Long Non-Coding RNA, HULC Expression Through Interaction with MicroRNA-372 in Liver Cancer. Nucleic Acids Res. 2010, 38, 5366–5383. [Google Scholar] [CrossRef]
- Guo, Z.; Kuang, Z.; Zhao, Y.; Deng, Y.; He, H.; Wan, M.; Tao, Y.; Wang, D.; Wei, J.; Li, L.; et al. PmiREN2.0: From data annotation to functional exploration of plant microRNAs. Nucleic Acids Res. 2021, 50, D1475–D1482. [Google Scholar] [CrossRef]
- Palatnik, J.F.; Allen, E.; Wu, X.; Schommer, C.; Schwab, R.; Carrington, J.C.; Weigel, D. Control of Leaf Morphogenesis by MicroRNAs. Nature 2003, 425, 257–263. [Google Scholar] [CrossRef]
- Barabási, A.-L.; Oltvai, Z.N. Network Biology: Understanding the Cell’s Functional Organization. Nat. Rev. Genet. 2004, 5, 101–113. [Google Scholar] [CrossRef]
- Karlebach, G.; Shamir, R. Modelling and Analysis of Gene Regulatory Networks. Nat. Rev. Mol. Cell Biol. 2008, 9, 770–780. [Google Scholar] [CrossRef]
- Zitnik, M.; Nguyen, F.; Wang, B.; Leskovec, J.; Goldenberg, A.; Hoffman, M.M. Machine Learning for Integrating Data in Biology and Medicine: Principles, Practice, and Opportunities. Inf. Fusion 2019, 50, 71–91. [Google Scholar] [CrossRef] [PubMed]
- Alipanahi, B.; Delong, A.; Weirauch, M.T.; Frey, B.J. Predicting the Sequence Specificities of DNA- and RNA-Binding Proteins by Deep Learning. Nat. Biotechnol. 2015, 33, 831–838. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Chaudhary, K.; Garmire, L.X. More Is Better: Recent Progress in Multi-Omics Data Integration Methods. Front. Genet. 2017, 8, 84. [Google Scholar] [CrossRef]
- Montesinos-López, O.A.; Chavira-Flores, M.; Kiasmiantini; Crespo-Herrera, L.; Saint Pierre, C.; Li, H.; Fritsche-Neto, R.; Al-Nowibet, K.; Montesinos-López, A.; Crossa, J. A Review of Multimodal Deep Learning Methods for Genomic-Enabled Prediction in Plant Breeding. Genetics 2024, 228, iyae161. [Google Scholar] [CrossRef]
- Eraslan, G.; Simon, L.M.; Mircea, M.; Mueller, N.S.; Theis, F.J. Single-Cell RNA-Seq Denoising Using a Deep Count Autoencoder. Nat. Commun. 2019, 10, 390. [Google Scholar] [CrossRef]
- Lopez, R.; Regier, J.; Cole, M.B.; Jordan, M.I.; Yosef, N. Deep Generative Modeling for Single-Cell Transcriptomics. Nat. Methods 2018, 15, 1053–1058. [Google Scholar] [CrossRef]
- Bhati, D.; Neha, F.; Amiruzzaman, M. A Survey on Explainable Artificial Intelligence (XAI) Techniques for Visualizing Deep Learning Models in Medical Imaging. J. Imaging 2024, 10, 239. [Google Scholar] [CrossRef]
- Quinn, K.N.; Wilber, H.; Townsend, A.; Sethna, J.P. Chebyshev Approximation and the Global Geometry of Model Predictions. Phys. Rev. Lett. 2019, 122, 158302. [Google Scholar] [CrossRef]
- Singh, R.; Lanchantin, J.; Sekhon, A.; Qi, Y. Attend and Predict: Understanding Gene Regulation by Selective Attention on Chromatin. Adv. Neural Inf. Process. Syst. 2017, 30, 6785–6795. [Google Scholar]
- Watson, D.S. Interpretable machine learning for genomics. Hum Genet. 2022, 141, 1499–1513. [Google Scholar] [CrossRef]
- Agharbaoui, Z.; Leclercq, M.; Remita, M.A.; Badawi, M.A.; Lord, E.; Houde, M.; Danyluk, J.; Diallo, A.B.; Sarhan, F. An Integrative Approach to Identify Hexaploid Wheat miRNAome Associated with Development and Tolerance to Abiotic Stress. BMC Genom. 2015, 16, 339. [Google Scholar] [CrossRef] [PubMed]
- Murmu, S.; Sinha, D.; Chaurasia, H.; Sharma, S.; Das, R.; Jha, G.K.; Archak, S. A Review of Artificial Intelligence-Assisted Omics Techniques in Plant Defense: Current Trends and Future Directions. Front. Plant Sci. 2024, 15, 1292054. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Pan, S.; Chen, F.; Long, G.; Zhang, C.; Yu, P.S. A Comprehensive Survey on Graph Neural Networks. IEEE Trans. Neural Netw. Learn. Syst. 2021, 32, 4–24. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.M.; Shah, N.; Iqbal, J.; El-Ghandour, N.M.F.; Vukic, M.; Lawton, M.; Morcos, J.J.; Matos, B.; El-Abbadi, N.; Samii, A.; et al. Evaluating Artificial Intelligence Models for Rupture Risk Prediction in Unruptured Intracranial Aneurysms: A Focus on Vessel Geometry and Hemodynamic Insights. Neurosurg. Rev. 2025, 48, 539. [Google Scholar] [CrossRef]
- Shi, H.; Zhang, X.; Tang, L.; Liu, L. Heterogeneous Graph Neural Network for lncRNA-Disease Association Prediction. Sci. Rep. 2022, 12, 17519. [Google Scholar] [CrossRef]
- Wein, S.; Malloni, W.M.; Tomé, A.M.; Frank, S.M.; Henze, G.-I.; Wüst, S.; Greenlee, M.W.; Lang, E.W. A Graph Neural Network Framework for Causal Inference in Brain Networks. Sci. Rep. 2021, 11, 8061. [Google Scholar] [CrossRef]
- Liu, J.; Yang, M.; Yu, Y.; Xu, H.; Wang, T.; Li, K.; Zhou, X. Advancing Bioinformatics with Large Language Models: Components, Applications and Perspectives. arXiv 2025, arXiv:2401.04155. [Google Scholar]
- Yu, H.; Yang, H.; Sun, W.; Yan, Z.; Yang, X.; Zhang, H.; Ding, Y.; Li, K. An Interpretable RNA Foundation Model for Exploring Functional RNA Motifs in Plants. Nat. Mach. Intell. 2024, 6, 1616–1625. [Google Scholar] [CrossRef]
- Pla, A.; Zhong, X.; Rayner, S. miRAW: A deep learning-based approach to predict microRNA targets by analyzing whole microRNA transcripts. PLoS Comput. Biol. 2018, 14, e1006185. [Google Scholar] [CrossRef]
- AlSaad, R.; Abd-Alrazaq, A.; Boughorbel, S.; Ahmed, A.; Renault, M.-A.; Damseh, R.; Sheikh, J. Multimodal Large Language Models in Health Care: Applications, Challenges, and Future Outlook. J. Med. Internet Res. 2024, 26, e59505. [Google Scholar] [CrossRef]
| Criterion | Stringent (High-Confidence) | Moderate | Relaxed (Discovery) |
|---|---|---|---|
| MFE threshold | ≤−25 kcal/mol | ≤−20 kcal/mol | ≤−18 kcal/mol |
| Stem paired bases | ≥18 bp | ≥16 bp | ≥14 bp |
| Expression (RPM) | ≥10 | ≥5 | ≥3 |
| Read alignment precision | ≥90% | ≥80% | ≥75% |
| miRNA* detection | Required | Preferred | Optional |
| 2-nt 3′ overhang | Strict enforcement | Allowed deviation | Not enforced |
| Independent datasets | ≥2 | 1 | Computational only |
| Example tools | miRador [48], miRScore [49] | plantMirP2 [23] | miRDeep-P2 [21] |
| Architecture | Tool | Key Parameters | Input | Training * | GPU Memory † | RAM | Accuracy |
|---|---|---|---|---|---|---|---|
| CNN | mirDNN [55] | 3 conv blocks (64 → 128 → 256) | Seq + Struct | 2–6 h | 8–12 GB | 16 GB | 87–99% |
| CNN-RNN | CIRNN [56] | CNN 32 filters; IndRNN 64 units | Sequence | 4–8 h | 8–12 GB | 8 GB | 85–95% |
| LSTM | DIGITAL [58] | 2 layers × 128 units | Sequence | 3–5 h | 4–8 GB | 16 GB | 94–98% |
| Transformer | PmlIPM [61] | 8 heads; 128-dim; 6 layers | Seq pairs | 12–24 h | 16 GB ‡ | 32 GB | 90–97% |
| GNN | CFHAN [64] | 3 GCN + 3 attention | Graph | 8–16 h | 8–16 GB § | 32–64 GB | 97–99% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, R.; Zhang, H. Application of Artificial Intelligence Technology in Plant MicroRNA Research: Progress, Challenges, and Prospects. Int. J. Mol. Sci. 2025, 26, 11854. https://doi.org/10.3390/ijms262411854
Yang R, Zhang H. Application of Artificial Intelligence Technology in Plant MicroRNA Research: Progress, Challenges, and Prospects. International Journal of Molecular Sciences. 2025; 26(24):11854. https://doi.org/10.3390/ijms262411854
Chicago/Turabian StyleYang, Ruilin, and Hanma Zhang. 2025. "Application of Artificial Intelligence Technology in Plant MicroRNA Research: Progress, Challenges, and Prospects" International Journal of Molecular Sciences 26, no. 24: 11854. https://doi.org/10.3390/ijms262411854
APA StyleYang, R., & Zhang, H. (2025). Application of Artificial Intelligence Technology in Plant MicroRNA Research: Progress, Challenges, and Prospects. International Journal of Molecular Sciences, 26(24), 11854. https://doi.org/10.3390/ijms262411854






