Assessment of Collagen and Fibroblast Properties via Label-Free Higher Harmonic Generation Microscopy in Three-Dimensional Models of Osteogenesis Imperfecta and Ehlers-Danlos Syndrome
Abstract
1. Introduction
2. Results
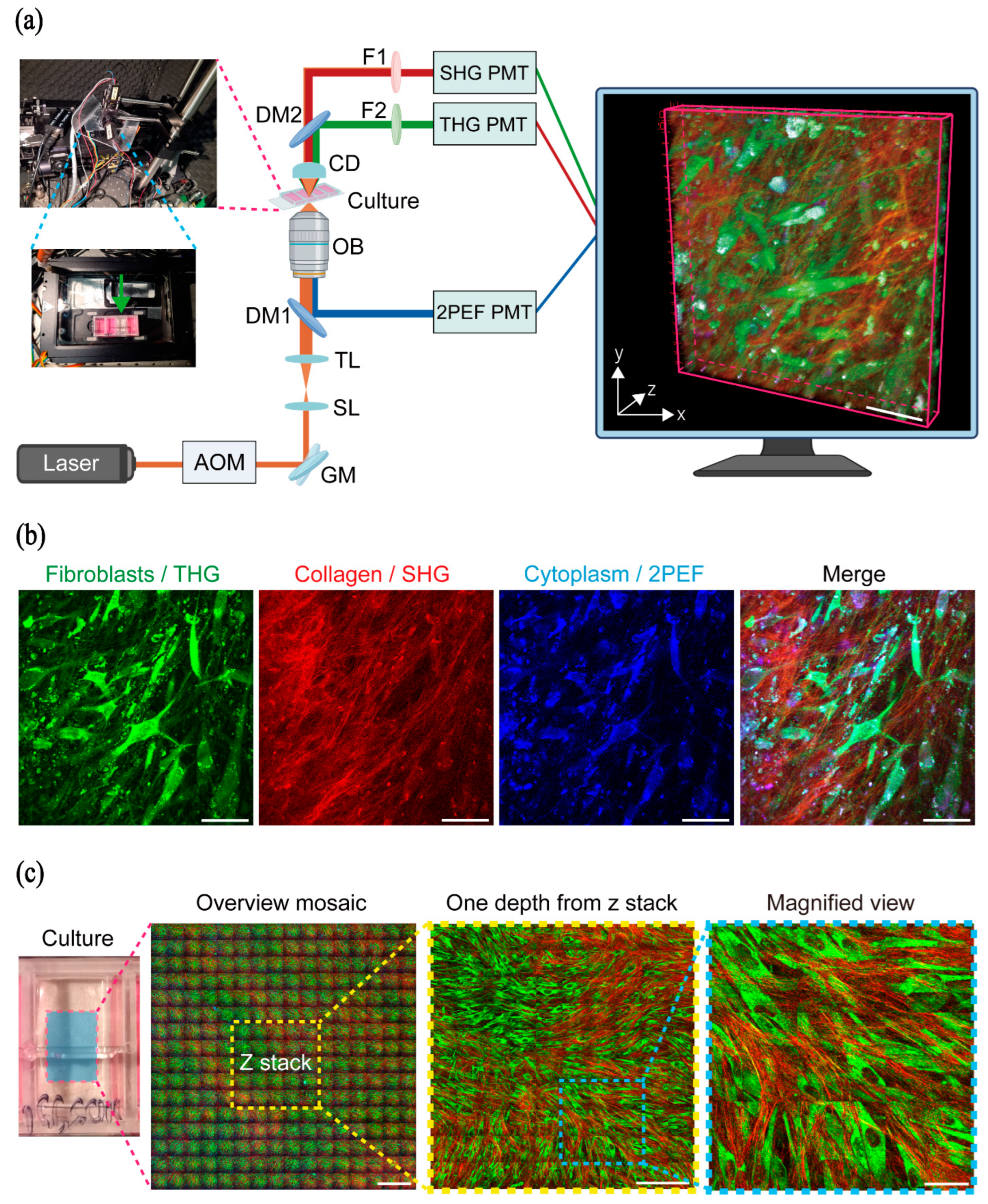
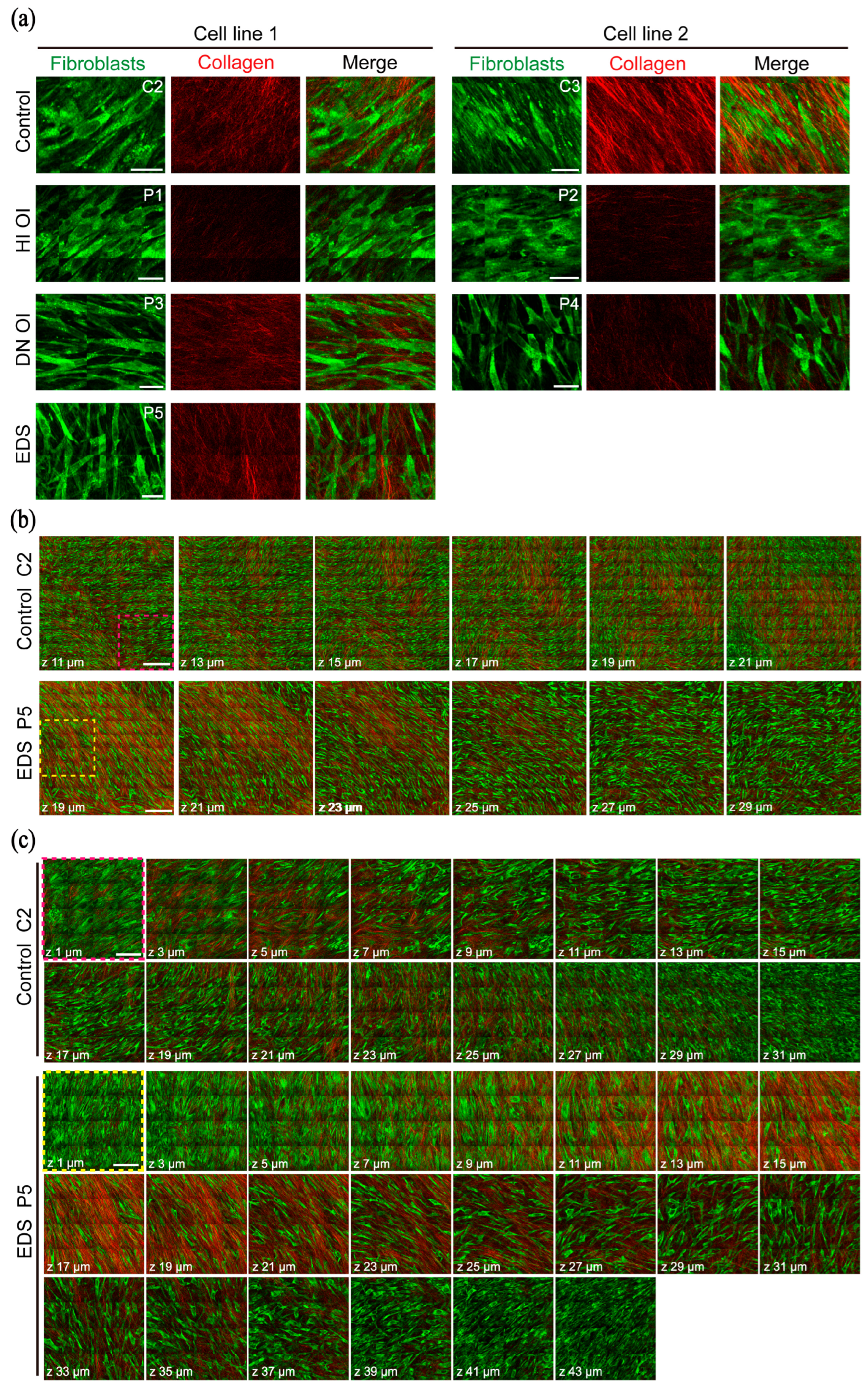
2.1. HHGM Images: Qualitative Assessment of 3D Cell Cultures
2.1.1. Visualization of Fibroblasts and Collagen in HHGM Images
2.1.2. Comparison of Fibroblast and Collagen Morphology
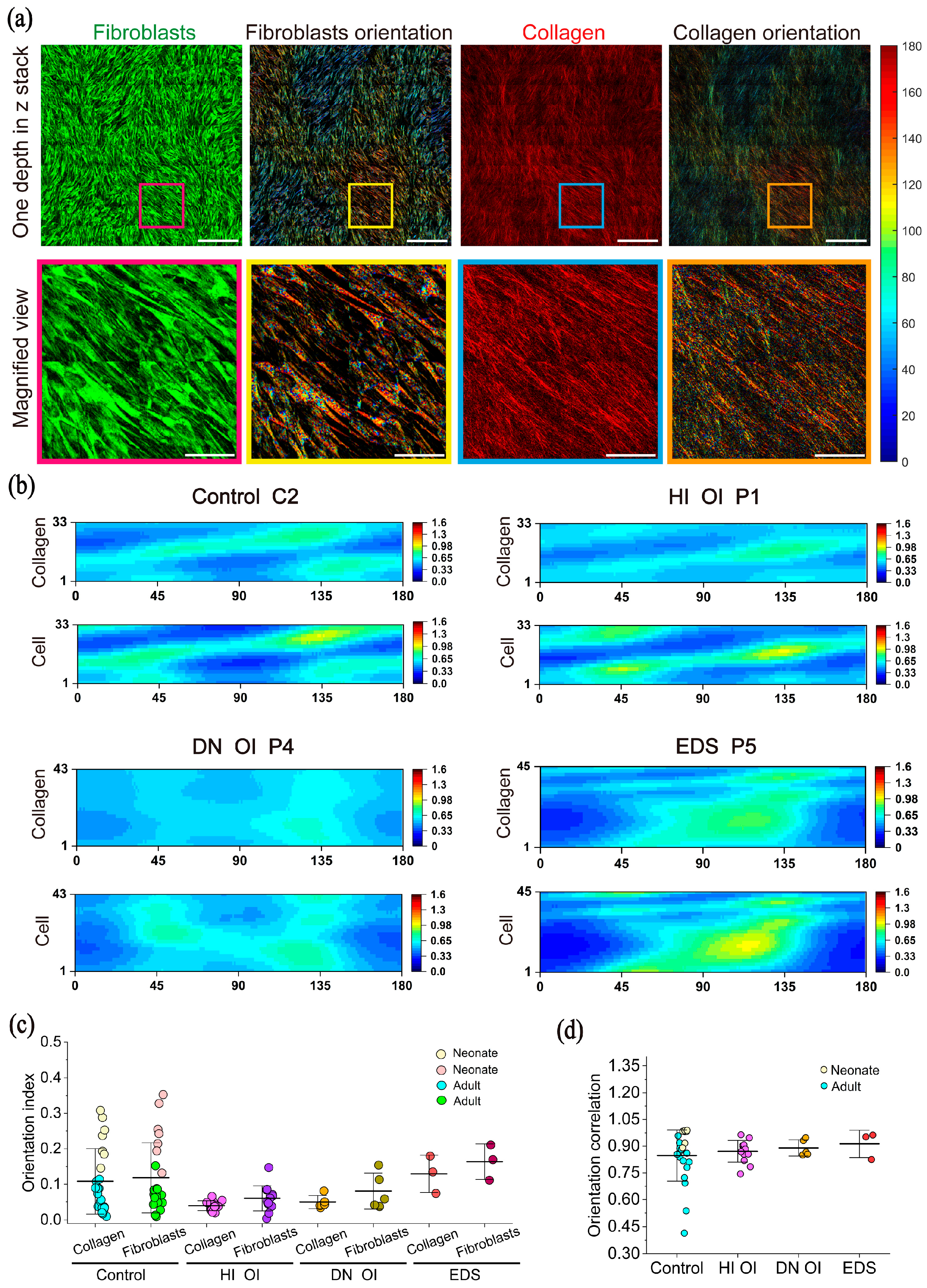
2.1.3. Counterclockwise Shift in Fibroblast Orientation
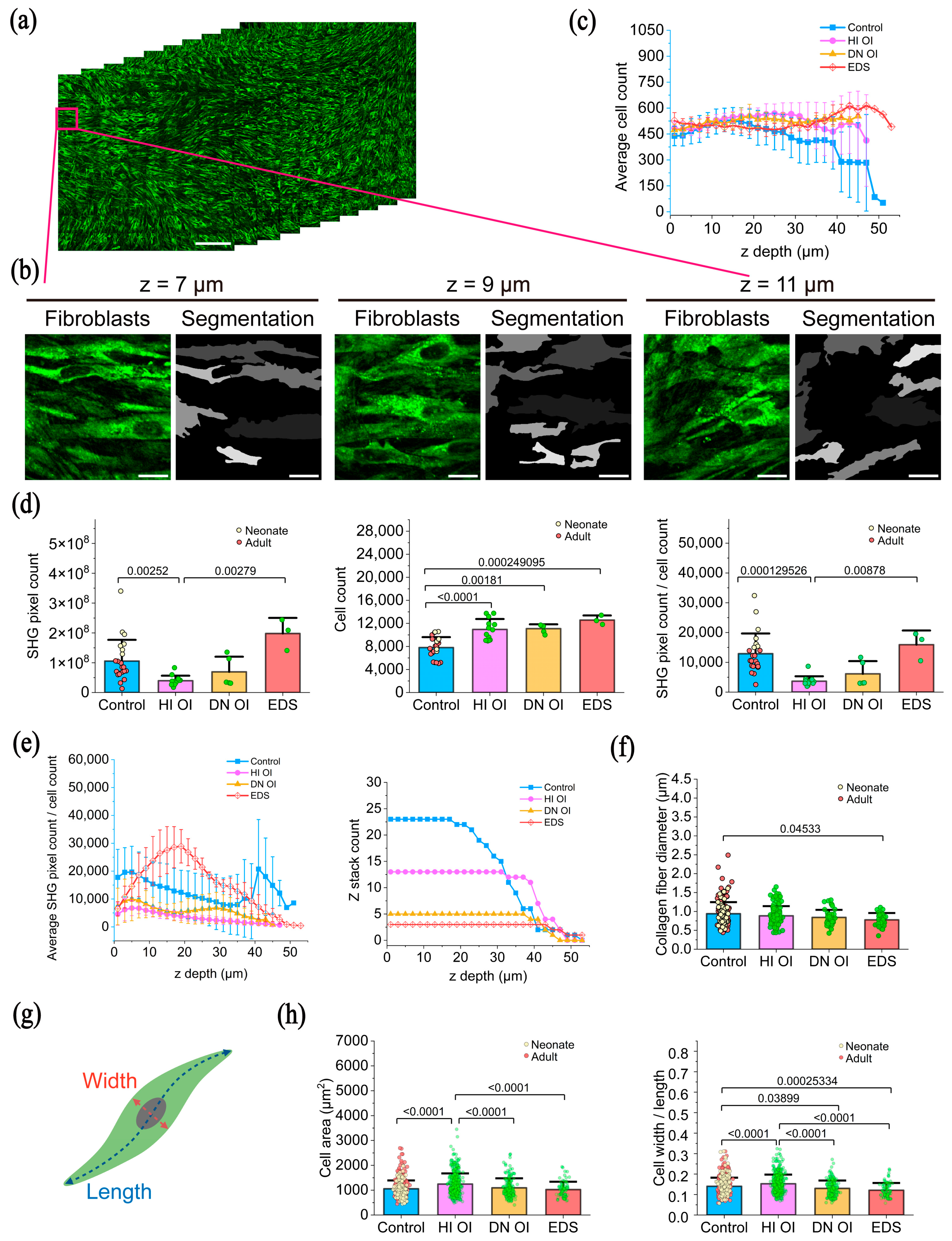
2.2. Quantification of HHGM Images
2.2.1. Cell Density
2.2.2. Collagen Content
2.2.3. Diameter of Collagen Fibers
2.2.4. Fibroblast Morphology
2.2.5. Three-Dimensional Orientation of Fibroblasts and Collagen
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Three-Dimensional Fibroblast Culture
4.3. HHGM Imaging
4.4. Fibroblast Segmentation
4.5. Collagen Amount and Collagen Fiber Diameter
4.6. Fibroblast Area and Shape
4.7. Orientation Calculation
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jovanovic, M.; Guterman-Ram, G.; Marini, J.C. Osteogenesis Imperfecta: Mechanisms and Signaling Pathways Connecting Classical and Rare OI Types. Endocr. Rev. 2022, 43, 61–90. [Google Scholar] [CrossRef] [PubMed]
- Jovanovic, M.; Marini, J.C. Update on the Genetics of Osteogenesis Imperfecta. Calcif. Tissue Int. 2024, 115, 891–914. [Google Scholar] [CrossRef]
- Micha, D.; Pals, G.; Smit, T.H.; Ghazanfari, S. An in Vitro Model to Evaluate the Properties of Matrices Produced by Fibroblasts from Osteogenesis Imperfecta and Ehlers-Danlos Syndrome Patients. Biochem. Biophys. Res. Commun. 2020, 521, 310–317. [Google Scholar] [CrossRef]
- Bowen, J.M.; Hernandez, M.; Johnson, D.S.; Green, C.; Kammin, T.; Baker, D.; Keigwin, S.; Makino, S.; Taylor, N.; Watson, O.; et al. Diagnosis and Management of Vascular Ehlers-Danlos Syndrome: Experience of the UK National Diagnostic Service, Sheffield. Eur. J. Hum. Genet. 2023, 31, 749–760. [Google Scholar] [CrossRef]
- Doolan, B.J.; Lavallee, M.E.; Hausser, I.; Schubart, J.R.; Michael Pope, F.; Seneviratne, S.L.; Winship, I.M.; Burrows, N.P. Extracutaneous Features and Complications of the Ehlers-Danlos Syndromes: A Systematic Review. Front. Med. 2023, 10, 1053466. [Google Scholar] [CrossRef]
- Tarnutzer, K.; Siva Sankar, D.; Dengjel, J.; Ewald, C.Y. Collagen Constitutes about 12% in Females and 17% in Males of the Total Protein in Mice. Sci. Rep. 2023, 13, 4490. [Google Scholar] [CrossRef]
- Van Dijk, F.S.; Byers, P.H.; Dalgleish, R.; Malfait, F.; Maugeri, A.; Rohrbach, M.; Symoens, S.; Sistermans, E.A.; Pals, G. EMQN Best Practice Guidelines for the Laboratory Diagnosis of Osteogenesis Imperfecta. Eur. J. Hum. Genet. 2012, 20, 11–19. [Google Scholar] [CrossRef]
- Griesbach, J.; de Leeuw, A.; Minacci, T.; Kodiyan, B.; Ndarugendamwo, T.; Lim, P.J.; Rohrbach, M.; Rubert, M.; Rüger, M.; Giunta, C.; et al. 3D-Bioprinted Patient-Specific Organotypic Bone Model Mimicking Mineralization Dysregulation in FKBP10 -Related Osteogenesis Imperfecta. bioRxiv 2024. [Google Scholar] [CrossRef]
- Borile, G.; Sandrin, D.; Filippi, A.; Anderson, K.I.; Romanato, F. Label-Free Multiphoton Microscopy: Much More than Fancy Images. Int. J. Mol. Sci. 2021, 22, 2657. [Google Scholar] [CrossRef] [PubMed]
- Kok, S.D.; Schaap, P.M.R.; van Dommelen, L.; van Huizen, L.M.G.; Dickhoff, C.; van Dijkum, E.M.N.; Engelsman, A.F.; van der Valk, P.; Groot, M.L. Compact Portable Higher Harmonic Generation Microscopy for the Real Time Assessment of Unprocessed Thyroid Tissue. J. Biophotonics 2024, 17, e202300079. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.H.; Huang, J.R.; Wang, Y.K.; Lin, K.H. Three-Dimensional Fibroblast Morphology on Compliant Substrates of Controlled Negative Curvature. Integr. Biol. 2013, 5, 1447–1455. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Pouli, D.; Sood, D.; Sundarakrishnan, A.; Hui Mingalone, C.K.; Arendt, L.M.; Alonzo, C.; Quinn, K.P.; Kuperwasser, C.; Zeng, L.; et al. Automated Quantification of Three-Dimensional Organization of Fiber-like Structures in Biological Tissues. Biomaterials 2017, 116, 34–47. [Google Scholar] [CrossRef] [PubMed]
- Plikus, M.V.; Wang, X.; Sinha, S.; Forte, E.; Thompson, S.M.; Herzog, E.L.; Driskell, R.R.; Rosenthal, N.; Biernaskie, J.; Horsley, V. Fibroblasts: Origins, Definitions, and Functions in Health and Disease. Cell 2021, 184, 3852–3872. [Google Scholar] [CrossRef] [PubMed]
- Fan, Q.; Zheng, Y.; Wang, X.; Xie, R.; Ding, Y.; Wang, B.; Yu, X.; Lu, Y.; Liu, L.; Li, Y.; et al. Dynamically Re-Organized Collagen Fiber Bundles Transmit Mechanical Signals and Induce Strongly Correlated Cell Migration and Self-Organization. Angew. Chem. Int. Ed. 2021, 60, 11858–11867. [Google Scholar] [CrossRef]
- Yeung, T.; Georges, P.C.; Flanagan, L.A.; Marg, B.; Ortiz, M.; Funaki, M.; Zahir, N.; Ming, W.; Weaver, V.; Janmey, P.A. Effects of Substrate Stiffness on Cell Morphology, Cytoskeletal Structure, and Adhesion. Cell Motil. Cytoskelet. 2005, 60, 24–34. [Google Scholar] [CrossRef]
- Ishihara, S.; Kurosawa, H.; Haga, H. Stiffness-Modulation of Collagen Gels by Genipin-Crosslinking for Cell Culture. Gels 2023, 9, 148. [Google Scholar] [CrossRef]
- Nijhuis, W.H.; Eastwood, D.M.; Allgrove, J.; Hvid, I.; Weinans, H.H.; Bank, R.A.; Sakkers, R.J. Current Concepts Review Current Concepts in Osteogenesis Imperfecta: Bone Structure, Biomechanics and Medical Management. J. Child. Orthop. 2019, 13, 1–11. [Google Scholar] [CrossRef]
- Celli, M.; Iacovino, C.; Febbo, A.; Lotti, L.V.; Miraglia, E.; Celli, L.; Roberti, V.; Sernicola, A.; Zambrano, A.; Turchetti, A.; et al. Ultrastructure Study of Skin Fibroblasts in Patients with Ehlers-Danlos Syndrome (EDS): Preliminary Results. Clin. Ter. 2020, 171, e431–e436. [Google Scholar] [CrossRef]
- Hadar, N.; Porgador, O.; Cohen, I.; Levi, H.; Dolgin, V.; Yogev, Y.; Sued-Hendrickson, S.; Shelef, I.; Didkovsky, E.; Eskin-Schwartz, M.; et al. Heterozygous THBS2 Pathogenic Variant Causes Ehlers–Danlos Syndrome with Prominent Vascular Features in Humans and Mice. Eur. J. Hum. Genet. 2024, 32, 550–557. [Google Scholar] [CrossRef]
- Malfait, F.; Francomano, C.; Byers, P.; Belmont, J.; Berglund, B.; Black, J.; Bloom, L.; Bowen, J.M.; Brady, A.F.; Burrows, N.P.; et al. The 2017 International Classification of the Ehlers–Danlos Syndromes. Am. J. Med. Genet. C Semin. Med. Genet. 2017, 175, 8–26. [Google Scholar] [CrossRef]
- Koenig, S.N.; Cavus, O.; Williams, J.; Bernier, M.; Tonniges, J.; Sucharski, H.; Dew, T.; Akel, M.; Baker, P.; Madiai, F.; et al. New Mechanistic Insights to PLOD1-Mediated Human Vascular Disease. Transl. Res. 2022, 239, 1–17. [Google Scholar] [CrossRef]
- Baumann, M.; Giunta, C.; Krabichler, B.; Rüschendorf, F.; Zoppi, N.; Colombi, M.; Bittner, R.E.; Quijano-Roy, S.; Muntoni, F.; Cirak, S.; et al. Mutations in FKBP14 Cause a Variant of Ehlers-Danlos Syndrome with Progressive Kyphoscoliosis, Myopathy, and Hearing Loss. Am. J. Hum. Genet. 2012, 90, 201–216. [Google Scholar] [CrossRef] [PubMed]
- Sarathchandra, P.; Pope, F.M.; Ali, S.Y. Morphometric Analysis of Type I Collagen Fibrils in the Osteoid of Osteogenesis Imperfecta. Calcif. Tissue Int. 1999, 65, 390–395. [Google Scholar] [CrossRef]
- Jones, C.J.P.; Cummings, C.; Ball, J.; Beighton, P. Collagen Defect of Bone in Osteogenesis Imperfecta (Type I). An Electron Microscopic Study. Clin. Orthop. Relat. Res. 1984, 183, 208–214. [Google Scholar] [CrossRef]
- Charvolin, J.; Sadoc, J.-F. Conjecture on the Lateral Growth of Type I Collagen Fibrils. Biophys. Rev. Lett. 2014, 9, 225–238. [Google Scholar] [CrossRef]
- Mateu, R.; Ẑivicová, V.; Krejí, E.D.; Grim, M.; Strnad, H.; Vlek, A.; Kolá, M.; Lacina, L.; Gál, P.; Borský, J.; et al. Functional Differences between Neonatal and Adult Fibroblasts and Keratinocytes: Donor Age Affects Epithelial-Mesenchymal Crosstalk in Vitro. Int. J. Mol. Med. 2016, 38, 1063–1074. [Google Scholar] [CrossRef]
- Fisher, G.J.; Wang, B.; Cui, Y.; Shi, M.; Zhao, Y.; Quan, T.; Voorhees, J.J. Skin Aging from the Perspective of Dermal Fibroblasts: The Interplay between the Adaptation to the Extracellular Matrix Microenvironment and Cell Autonomous Processes. J. Cell Commun. Signal 2023, 17, 523–529. [Google Scholar] [CrossRef] [PubMed]
- Nair, A.K.; Gautieri, A.; Chang, S.W.; Buehler, M.J. Molecular Mechanics of Mineralized Collagen Fibrils in Bone. Nat. Commun. 2013, 4, 1724. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Lubbersen, N.B.; Spies, S.; Dijkhuis, A.; van Dorp, M.; Dickhoff, C.; Annema, J.; Duitman, J.W.; Groot, M.L. Label-Free 4D Live Imaging of Immune–Stromal Interactions in Human Alveolar Tissue Using Higher Harmonic Generation Microscopy. Eur. Respir. J. 2025, 66, 2501530, ahead of print. [Google Scholar] [CrossRef]
- Meijns, N.R.C.; Blokker, M.; Idema, S.; t Hart, B.A.; Veta, M.; Ettema, L.; van Iersel, J.; Zhang, Z.; Schenk, G.J.; Groot, M.L.; et al. Dynamic Imaging of Myelin Pathology in Physiologically Preserved Human Brain Tissue Using Third Harmonic Generation Microscopy. PLoS ONE 2025, 20, e0310663, Erratum in PLoS ONE 2025, 20, e0329374. [Google Scholar] [CrossRef]
- Uchugonova, A.; König, K.; Bueckle, R.; Isemann, A.; Tempea, G. Targeted Transfection of Stem Cells with Sub-20 Femtosecond Laser Pulses. Opt. Express 2008, 16, 9357–9364. [Google Scholar] [CrossRef]
- Yu, Q.; Heikal, A.A. Two-Photon Autofluorescence Dynamics Imaging Reveals Sensitivity of Intracellular NADH Concentration and Conformation to Cell Physiology at the Single-Cell Level. J. Photochem. Photobiol. B 2009, 95, 46–57. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; González, P.J.; Van Haasterecht, L.; Soylu, A.; Mihailovski, M.; Van Zuijlen, P.; Groot, M.L. Uniaxial Mechanical Stretch Properties Correlated with Three-Dimensional Microstructure of Human Dermal Skin. Biomech. Model Mechanobiol. 2024, 23, 911–925. [Google Scholar] [CrossRef] [PubMed]
- Spies, S.; Nazarian, E.; Annavarapu, S.; Collini, P.; Coulomb L’Hermine, A.; D’Hooghe, E.; Kobos, J.; Morcrette, G.; Morini, M.A.; Popov, S.D.; et al. Multi-Observer Study on Diagnostic Accuracy of Pediatric Renal Tumors Imaged with Higher-Harmonic-Generation Microscopy. Cancers 2025, 17, 1693. [Google Scholar] [CrossRef]
- Zhang, Z.; de Munck, J.C.; Verburg, N.; Rozemuller, A.J.; Vreuls, W.; Cakmak, P.; van Huizen, L.M.G.; Idema, S.; Aronica, E.; de Witt Hamer, P.C.; et al. Quantitative Third Harmonic Generation Microscopy for Assessment of Glioma in Human Brain Tissue. Adv. Sci. 2019, 6, 1900163. [Google Scholar] [CrossRef] [PubMed]
| Group | Samples ID | Gender | Age | Diagnosis | Pathogenic Variant |
|---|---|---|---|---|---|
| Control | C1 | Male | 44 | Healthy control | NA |
| Control | C2 | Male | 34 | Healthy control | NA |
| Control | C3 | Male | 0 | Healthy control | NA |
| Control | C4 | Male | 0 | Healthy control | NA |
| HI OI | P1 | Female | 29 | Osteogenesis Imperfecta type 1 | NM_000088.4(COL1A1):c.495T>A, p.(Tyr165*) |
| HI OI | P2 | Male | 38 | Osteogenesis Imperfecta type 1 | NM_000088.4(COL1A1):c.2784del, p.(Gly929Alafs*179) |
| DN OI | P3 | Female | 0 | Osteogenesis Imperfecta type 4 | NM_000088.4(COL1A1):c.1678G>A, p.(Gly560Ser) |
| DN OI | P4 | Female | 0 | Osteogenesis Imperfecta type 3 | NM_000089.4(COL1A2):c.2113_2121del, p.(Ala705_Pro707del) |
| EDS | P5 | Male | 11 | Ehlers–Danlos Syndrome subtype 8 | NM_000302.4(PLOD1):c.1651-2A>G, p.? a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, Y.; Bo, Q.; Zhang, Z.; van Haasterecht, L.; Kloen, P.; Rustemeyer, T.; Ventura, L.; Zhytnik, L.; Eekhoff, E.M.W.; Micha, D.; et al. Assessment of Collagen and Fibroblast Properties via Label-Free Higher Harmonic Generation Microscopy in Three-Dimensional Models of Osteogenesis Imperfecta and Ehlers-Danlos Syndrome. Int. J. Mol. Sci. 2025, 26, 11848. https://doi.org/10.3390/ijms262411848
Ma Y, Bo Q, Zhang Z, van Haasterecht L, Kloen P, Rustemeyer T, Ventura L, Zhytnik L, Eekhoff EMW, Micha D, et al. Assessment of Collagen and Fibroblast Properties via Label-Free Higher Harmonic Generation Microscopy in Three-Dimensional Models of Osteogenesis Imperfecta and Ehlers-Danlos Syndrome. International Journal of Molecular Sciences. 2025; 26(24):11848. https://doi.org/10.3390/ijms262411848
Chicago/Turabian StyleMa, Yuanyuan, Qiyu Bo, Zhiqing Zhang, Ludo van Haasterecht, Peter Kloen, Thomas Rustemeyer, Laura Ventura, Lidiia Zhytnik, Elisabeth M. W. Eekhoff, Dimitra Micha, and et al. 2025. "Assessment of Collagen and Fibroblast Properties via Label-Free Higher Harmonic Generation Microscopy in Three-Dimensional Models of Osteogenesis Imperfecta and Ehlers-Danlos Syndrome" International Journal of Molecular Sciences 26, no. 24: 11848. https://doi.org/10.3390/ijms262411848
APA StyleMa, Y., Bo, Q., Zhang, Z., van Haasterecht, L., Kloen, P., Rustemeyer, T., Ventura, L., Zhytnik, L., Eekhoff, E. M. W., Micha, D., & Groot, M. L. (2025). Assessment of Collagen and Fibroblast Properties via Label-Free Higher Harmonic Generation Microscopy in Three-Dimensional Models of Osteogenesis Imperfecta and Ehlers-Danlos Syndrome. International Journal of Molecular Sciences, 26(24), 11848. https://doi.org/10.3390/ijms262411848





